Summary
The protein kinase KIS is made by the juxtaposition of a unique kinase domain and a C-terminal domain with a U2AF Homology Motif (UHM), a sequence motif for protein interaction initially identified in the heterodimeric pre-mRNA splicing factor U2AF. This domain of KIS is closely related to the C-terminal UHM domain of the U2AF large subunit, U2AF65. KIS phosphorylates the splicing factor SF1, which in turn enhances SF1 binding to U2AF65 and the 3′ splice site, an event known to take place at an early step of spliceosome assembly. Here, the analysis of the subcellular localization of mutated forms of KIS indicates that the kinase domain of KIS is the necessary domain for its nuclear localization. As in the case of U2AF65, the UHM containing C-terminal domain of KIS is required for binding to the splicing factors SF1 and SF3b155. The efficiency of KIS binding to SF1 and SF3b155 is similar to that of U2AF65 in pull-down assays. These results further support the functional link of KIS with splicing factors. Interestingly, when compared to other UHM containing proteins, KIS presents a different specificity for the UHM docking sites that are present in the N-terminal region of SF3b155, thus providing a new insight into the variety of interactions mediated by UHM domains.
Keywords: Kinase KIS, Protein phosphorylation, U2AF-homology-motif, SF1, SF3b155
Introduction
Protein kinases are essential regulators of cell functions and constitute one of the largest and most functionally diverse gene families1. By adding phosphate groups to substrate proteins, they regulate their activity, localization and overall function. In addition to variations among the catalytic domains, the ability of protein kinases to serve diverse functions relies on regulatory domains, expression levels, and subcellular localization. We initially identified the vertebrate protein kinase KIS as a potential regulator of RNA metabolism, as it presents the unique property among protein kinases of possessing an RRM (RNA Recognition Motif), the most common type of single stranded RNA binding motif2 (See Fig. 1 for the domain organization of the proteins in this study). Although KIS is ubiquitously expressed, higher levels are detected in the nervous system3–6. A dramatic increase in KIS expression levels during development suggests that this kinase may serve regulatory functions in both the developing and adult brain5. Interestingly, a fine mapping analysis of the chromosome 1 region 1q23.3 has identified an association between polymorphisms located in the UHMK1 gene, encoding the KIS protein, and schizophrenia7,8. In support of this potential implication of the UHMK1 gene in the etiology of this disease, KIS expression is higher in discrete regions of the brain more particularly related to schizophrenia, including the ventral tegmental area, substantia nigra compacta, hippocampus and cortex5,6. Altogether, expression data suggests a ubiquitous function for KIS that is particularly significant in the nervous system, where KIS might participate in the control of protein expression. Additional functions of KIS in regulating the peptidylglycine alpha-amidating monooxygenase6 and in cell cycle control through the regulation of the stability of the cdk regulator p27kip19–11 have also been proposed.
Figure 1. Domains organization of the proteins used in this study.
(a) KIS is formed by the juxtaposition of a serine/threonine kinase domain and a C-terminal noncanonical RNA Recognition Motif of the UHM type (U2AF Homology Motif). This C-terminal domain is highly homologous to the C-terminal UHM domain of U2AF65.
(b) In addition to its C-terminal UHM domain necessary for interaction with SF117,22,23 and SF3b155 29,30,33, U2AF65 possesses two classical RRM domains involved in RNA binding16, while its N-terminal RS domain also contacts the branch site during spliceosome assembly47. Heterodimerisation with U2AF35 involves an ULM (UHM-Ligand Motif) indicated by a W for the crucial tryptophan residue that is required for UHM binding.
(c) SF1 is expressed as multiple spliced forms differing by their C-terminal proline rich domains48. The most abundant SF1HL1 isoform of HeLa cells is presented. The C-terminal proline rich region is proposed to mediate protein interactions in particular with WW motifs containing proteins as FBP1149 and CA15050. The N-terminal ULM domain (W) is required for binding to U2AF65. An extended KH-QUA2 domain mediates binding of SF1 to RNA19–21,23. In between the RNA and U2AF65 binding regions two serine residues within a SPSP conserved motif are mostly in a phosphorylated state in cells and substrate for KIS in vitro and in vivo (ref 36 and our unpublished results).
(d) SF3b155 is part of the proteins that form the complex SF3b an integral part of U2snRNP and of the U11/U12 di-snRNP51. The C-terminus of SF3b155 contains 22 tandem helical repeats (HEAT repeats)26 and was shown by electron cryomicroscopy to form a structural element of the outer shell of the complex SF3b enclosing p14 another component of SF3b31. Nevertheless, a minimal peptide comprising amino-acids [396–424] of SF3b155 binds p14 30,32,51,52. The N-terminal domain of SF3b155 contains seven potential ULMs in the [190–344] region necessary for U2AF65 binding. In addition numerous TP dipeptides in this region constitute potential phosphorylation sites in agreement with SF3b155 being phosphorylated in vivo26 and in vitro by CyclinE-cdk253. Interaction of the nuclear phosphatase 1 inhibitor NIPP1 is dependent on phosphorylation of the N-terminus of SF3b15554.
Since our first identification of KIS, recent progress in the characterization of "kinomes" of various species including mouse12 and sea urchin13 have further established that the KIS kinase domain sequence shares at most 30% identical residues with other kinase domains. However a few sequence features within the kinase domain are predictive for proline-directed kinases (i.e. kinases that preferentially phosphorylate residues followed by proline)14. In agreement with this observation KIS phosphorylates serine containing peptides within MBP and synapsin I in a proline-directed manner in vitro14. However the efficiency of phosphorylation of these proteins is relatively low. More recently we identified the splicing factor SF1 as a strongly preferred substrate for KIS in vitro and in vivo (ref 36 and our unpublished data). The identification of the two phosphorylated serine residues within an SPSP motif of SF1 further demonstrated the proline-directed specificity of KIS. To summarize, the kinase domain of KIS is not closely related to other members of the "kinome", but KIS specificity for proline-directed residues most probably relies on shared structural features among proline-directed kinases.
Beside its peculiar kinase domain, a second characteristic feature of KIS is the presence in its COOH-terminus of a 101 amino acid domain with strong sequence homology with the C-terminal noncanonical RRM of the essential splicing factor U2AF65, which interacts with U2AF35 to form the U2AF heterodimer15,16. U2AF and SF1 cooperatively associate with consensus sequences near the 3′ end of introns at an early step of splicing17. This recognition involves the binding of U2AF65 to the polypyrimidine tract15,18, while SF1 binds the upstream branchpoint sequence19–21. The U2AF65-SF1 interaction mainly relies on interactions between the C-terminal RRM-like domain of U2AF65 and an N-terminal linear peptide of SF122,23. The solution structure of this complex revealed a similar overall fold for the C-terminal domain of U2AF65 and typical RRM domains: two α-helices (A and B) packed against a four-stranded, antiparallel β-sheet22. However when compared to classical RRM domains, an additional C-terminal α-helix (helix C) occludes the canonical RNA binding site on the β-sheet, in agreement with the absence of detectable RNA affinity for this domain22. The interaction with the SF1 N-terminal peptide relies mainly on the insertion of a key tryptophan residue of SF1 (tryptophan 22) in a hydrophobic pocket of the U2AF65-RRM. Additionally, important electrostatic interactions occur between positively charged residues N-terminal to this tryptophan of SF1 and the negatively charged helix A of the RRM22. A crystallographic study had previously shown that a similar interaction occurs between the noncanonical RRM domain of U2AF35 and a tryptophan containing peptide of U2AF6524. Thus, based on these observations it was hypothesized that the atypical RRMs with sequence features similar to those of U2AF35 and U2AF65 would comprise a novel family of protein-interaction domains, called "U2AF Homology Motifs" (UHMs), and further that these UHMs would interact with tryptophan-containing peptides called "UHM-Ligand Motifs" (ULMs)25.
The interaction between SF1 and U2AF during the initial step of spliceosome assembly is transient. In the following step, when U2snRNP comes in contact with the branch site, the U2snRNP component SF3b15526,27, also known as SAP155, interacts with the UHM domain of U2AF65 in place of SF128,29. This interaction involves the N-terminal domain of SF3b155 which is thought to serve as a scaffold for proteins30, while its larger C-terminal region has a structural role for U2snRNP function31. In the N-terminal domain of SF3b155, several ULM domains have been shown to exhibit significant affinities for the UHM domain of U2AF65 suggesting different possible modes of SF3b155 binding to U2AF65 30,32,33. Therefore, remarkably, the first steps of spliceosome assembly appear to involve at least three different "UHM-ULM" interactions.
Based on the conservation of sequence features of U2AF65 and U2AF35 UHMs involved in binding to their ULMs, a small family of UHM-containing proteins was suggested to include URP, SPF45, CAPER, KIS and PUF6025. In agreement with this, the UHM domain of SPF45, a splicing factor known to affect alternative splicing of pre-mRNAs, has recently been shown to lack RNA affinity34 and to bind the ULM domains of SF1, U2AF65 and SF3b15535. The structure of SPF45-UHM in complex with an ULM containing peptide of SF3b155 confirms the proposed UHM-ULM interactions, and documents shared structural features of these interactions35. Notably, the UHM domains of KIS and U2AF65 clearly exhibit the closest relationship, with about 40% sequence identity4,25. This suggests that a common ancestor unique for these domains existed and that common characteristics of these UHMs for KIS and U2AF65 function have been conserved.
We previously reported the ability of KIS to interact with the splicing factor SF1 and to phosphorylate SF1 on two highly conserved serine residues36. Our present data indicate that the UHM domain of KIS is dispensable for its nuclear localization but is necessary for KIS to interact with SF1 and SF3b155. Interestingly the efficiencies of KIS binding to SF1 and SF3b155 in pull-down experiments are similar to that of U2AF65 binding. Analysis of mutants of KIS, SF1 and SF3b155 further indicates that these interactions involve UHM-ULM contacts. Finally we observe an intriguing different specificity of KIS and U2AF65 for their binding to the SF3b155 ULMs, which highlights the diversity of interactions mediated by UHM domains.
Results
The kinase domain of KIS is required for its nuclear accumulation
We previously observed that KIS was enriched in the nuclei of transfected cells4. This observation is consistent with the proposed RNA-related action of KIS, suggested by the presence of the RRM-like domain located C-terminal to the kinase core. According to its molecular mass of 47 kDa, KIS theoretically should not passively diffuse through nuclear pores37. Thus an active mechanism is required for the nuclear accumulation of KIS. Nevertheless, a purely cytoplasmic protein is predicted by analysis of the KIS sequence: the pSORTII38 and Nucpred39 algorithms yield very low nuclear localization prediction scores (− 0.47 and 0.07 respectively); similarly, the predictNLS algorithm40 failed to identify a nuclear localization signal (NLS) within the KIS sequence. In spite of these theoretical predictions, significant amounts of KIS are observed in the nucleus, with some cells showing only nuclear localization. We thus hypothesize that KIS, lacking its own NLS, enters the nucleus via co-transport with an NLS-containing protein.
To test the possibility that the UHM-dependent binding of KIS to splicing factors mediates its nuclear accumulation, we transfected CHO cells with a series of constructs for expression of various KIS mutants. The KIS variants were expressed as fusion proteins with the hemagglutinin (HA) epitope at the N-terminus to provide handles for detection by immunofluorescence. Immunoblot analyses of transfected cell extracts using anti-HA antibodies confirmed that tagged proteins with the expected molecular sizes were expressed, and the amounts of transfected DNA were adjusted to achieve similar expression levels for each construct. We next compared the subcellular distribution of the mutant forms of KIS by immunofluorescence staining with anti HA antibodies (Fig. 2). Immunofluorescence using antibodies against a C-terminal epitope of KIS independently confirmed the observed distribution for constructs containing the C-terminal KIS domain (data not shown).
Figure 2. Subcellular localization of KIS and mutated forms in CHO cells.
(a–h) Different HA-tagged forms of KIS were transiently expressed in Chinese Hamster Ovary (CHO) cells and their subcellular localization was analysed by indirect immunofluorescence microscopy using an anti HA antibody. Typical views with an X60 objective and corresponding to similar exposure times are presented (bar = 10 µm). The primary structure of each mutant is depicted at the bottom of each picture. In contrast to most constructs which presented a variety of situations with either only nuclear or nuclear and cytoplasmic localizations, the two mutants with major deletions of the kinase domain (KIS[212–419] and KIS[293–419]) never presented a nuclear enrichment (e,f).
(i) The mean values and standard deviation of the quantification of three experiments are presented. In transfected cells, nuclear enrichment was considered when a clear contrasted brighter staining of the nucleus compared to the cytoplasm was observed.
For each construct, we quantified the portion of transfected cells showing nuclear enrichment, i.e. contrasted higher staining of the nucleus compared with the cytoplasm (Fig. 2i). As previously observed in HEK293 cells, overexpressed KIS was enriched in the nucleus of CHO cells (Fig. 2a). KIS was excluded from nucleoli and generally diffusely distributed in the nucleoplasm. Deletion of either the entire UHM to leave residues [1–309], or a 41-amino acid region within the UHM (mutant Δ[369–409]) did not significantly alter the intracellular distribution of KIS (Fig. 2g–i). In contrast, all mutations within the kinase domain reduced the nuclear enrichment of KIS (Fig. 2b–f and 2i). The most significant effect was observed in the absence of residues [1–211] of the protein, as exemplified by the absence of detectable nuclear enrichment for KIS constructs [212–419] or [293–419] (Fig. 2e and 2f). Altogether our data show that the KIS kinase domain but not its UHM is necessary for nuclear import and/or retention of KIS.
Analysis of the binding of KIS to splicing factors SF1 and SF3b155
We previously reported the property of KIS to interact with and phosphorylate the splicing factor SF136. This interaction was dependent on the presence of the UHM domain of KIS. By analogy to the binding of U2AF65 with SF1, this interaction is likely to involve the "UHM-Ligand Motif" (ULM) domain of SF1. We therefore tested whether, like U2AF65, KIS has the ability to interact with the ULM-containing N-terminus of SF3b155. Using a two-hybrid system in yeast we could detect the interaction between KIS fragment [120–419] and a fragment of SF3b155 (residues [171–775]29) encompassing the U2AF65 interacting domain (Fig. 3a). This fragment of SF3b155 also interacted with U2AF65 and U2AF35 as described29. As negative controls, no interaction signal was detected between SF3b155 and lamin C, stathmin or ras, or between KIS, U2AF35 or U2AF65 and tsg101, a known partner of stathmin in the two-hybrid system3. Thus the interaction between SF3b155 and KIS appeared specific and independent of the N-terminal part of KIS encompassing the small lobe of the kinase domain4.
Figure 3. Interaction of KIS with SF3b155.
(a) KIS interacts with SF3b155 in the two-hybrid assay. Different interactions were tested in the two-hybrid system in yeast. Cells expressing the bait plasmids were mated to cells expressing the prey plasmids and expression of the reporter genes HIS3 that allows growth on medium lacking histidine and of the reporter gene lacZ revealed by the activity of beta-galactosidase are presented. KIS (fragment [120–419]), U2AF65 and U2AF35 interacted with SF3b155 (fragment [171–775]29) but not with a fragment of tsg101 used as a negative control3.
(b) Co-precipitation of KIS with SF3b155. GST-pull-down assays were performed to test the binding of KIS to SF3b155. Extracts of HEK293 cells that had been transfected with a KIS expressing vector were mixed with GST or with GST fused to a N-terminal part of SF3b155 (SF3b155n: amino-acids [1–493]) and pull-down were performed with glutathione beads. 20% of the extract used as “input” and the proteins retained on the beads were analysed by SDS PAGE and immunoblotting with an anti-KIS polyclonal antibody (left panel). A similar experiment was conducted using recombinant KIS partially purified from bacteria (right panel). A truncated form of KIS most probably originating from internal cleavage by thrombin is also detected (asterix). The picture shows typical results obtained in duplicate in two experiments.
(c) Preferential binding of KIS to SF1 and SF3b155. KIS was in vitro translated using [35S]-methionine in a reticulocyte lysate, then diluted in interaction buffer containing 1 µg/µL BSA and mixed with about 20 pmoles of each GST-fusion protein to test their interaction by co-precipitation on glutathione beads. The proteins bound to the beads (lanes 2–7) and 2% of the amount of the diluted solution of KIS used for each interaction (input 2%, lane 8) were separated by SDS PAGE. The gel was stained with Coomassie blue (top panel) revealing the GST-fusion proteins (lanes 2–7) and BSA as the major protein in the KIS dilution (input 2%, lane 8). Once dried, the gel was analysed using a Phosphorimager to quantify the binding of KIS to each of the GST-fusion protein as described in the Material and Methods section (bottom panel). SF1f: fragment [1–255] of human SF1; CTD: C-terminus of polymerase II; p27: p27kip1. The mean values of three experiments with standard deviation are presented showing a clear preferential co-precipitation of KIS with SF1f and SF3b155n over the other GST-fusion proteins.
(d) Competition of SF1 and SF3b155 for binding to KIS. SF1f (produced by cleavage of GST-SF1f by precision protease (Amersham) and ion exchange purification) was used as a competitor in pull-down assays of KIS with GST-SF1f or GST-SF3b155n performed as in (c). A ten fold excess of SF1f greatly reduced the co-precipitation of in vitro translated KIS with both GST-fusion proteins. The mean values and standard deviation of duplicates are presented.
We next further analysed the SF3b155-KIS interaction using a pull-down assay with a GST-fused N-terminal domain of SF3b155 (residues [1–493], thereafter SF3b155n), and a protein extract of HEK293 cells overexpressing KIS as input. A significant amount of KIS was recovered on GST-SF3b155n beads but not on control GST beads (Fig. 3b left panel). To exclude the possibility that a factor in the cell lysate was necessary for bridging the SF3b155-KIS interaction, we also tested recombinant KIS produced in bacteria and observed a similar retention of this recombinant KIS on GST-SF1f (SF1 fragment [1–255]36) and GST-SF3b155n beads (Fig. 3b right panel). Therefore KIS expressed in human cells, as well as in yeast or bacterial systems, is capable of specifically interacting with the N-terminal domain of SF3b155.
To further assess the specificity of KIS binding to SF3b155, we expressed [35S]-methionine labelled KIS using a rabbit reticulocyte lysate and compared its binding to various bacterially produced GST-fusion proteins (Fig. 3c). The use of [35S]-labelled proteins allows bound fractions in pull-down assays to be precisely quantified, and circumvents the difficulty of producing high levels of soluble recombinant KIS or KIS fragments that would be necessary for other approaches. All the GST-fusion proteins were prepared in identical conditions. Purified proteins as well as in vitro translated products were treated with RNAse I to exclude any bridging of the proteins by RNA molecules in the assays (see Material and Methods). Using GST-fusion proteins at a concentration of 80 nM, we observed a high recovery of KIS on GST-SF3b155n ([1–493]) and GST-SF1f ([1–255]) (Fig. 3c, lanes 6 and 7). Interestingly, addition of a ten fold excess of the SF1f fragment (800 nM) in the binding reactions led to a dramatic reduction of KIS binding to GST-SF1f but also to GST-SF3b155n (Fig. 3d compare lanes 3 and 4 with lanes 1 and 2). Thus, SF1 and SF3b155 are likely to compete for binding to an overlapping site on KIS.
The apparently significant retention of KIS by SF1f and SF3b155n was compared with KIS retention by several control proteins. A relatively small amount of KIS was reproducibly retained on GST-p27kip1, which is a described interaction partner for KIS10 (Fig. 3c, lane 4). Background levels comparable to nonspecific KIS binding to GST alone were detected on stathmin, which interacts with KIS in the two-hybrid system3, and on a C-terminal, heptad repeat containing domain of polymerase II, which has been proposed to serve an analogous scaffolding function during transcription as SF3b155 in splicing30 (Fig. 3c, lanes 2, 3 and 5). However, we cannot exclude the possibility that under our conditions the GST tag sterically impedes significant levels of KIS retention by these proteins.
We previously reported that, in addition to binding KIS, SF1f is a very efficient substrate for KIS kinase activity. Given the similar interaction of SF3b155n and SF1f with KIS, we compared the phosphorylation of GST-SF3b155n with that of SF1f and GST-p27kip1, another known substrate for KIS10. Endogenous kinase activities present in a soluble extract of proliferating embryonic fibroblasts phosphorylated each of the substrates to a similar extent (Fig. 4, lanes 4–6). In contrast, with bacterially produced recombinant GST-KIS, much higher levels of phosphate incorporation were obtained using SF1f as a substrate (16 fold higher than that obtained with SF3b155n and two orders of magnitude over that with GST-p27kip1) (Fig. 4, lanes 1–3). Phosphopeptide and phosphoaminoacid mapping showed that multiple serine and threonine residues of SF3b155n were phosphorylated by KIS (data not shown). Thus, under these conditions SF1f serves as a much better substrate for KIS than SF3b155n.
Figure 4. KIS phosphorylates SF1f much more efficiently than SF3b155 in vitro.
In vitro kinase assays were performed using either recombinant GST-KIS or a soluble cell extract of mouse embryonic fibroblasts as a source of kinases as indicated. About 20 pmoles of each purified protein SF1f, GST-SF3b155n and GST-p27kip1 were used as substrates. Phosphorylation reactions were performed with 100 µM [γ-32P]ATP for 30 minutes and products were separated on SDS PAGE that was Coomassie stained (top) and analysed with a phosphorimager for quantification of the radioactivity (bottom). The amount of phosphate (pmoles) that was incorporated in each substrate is indicated beneath the lanes.
KIS domains involved in splicing factors interactions
To further compare the binding of KIS to SF1 and SF3b155 we used different mutated forms of KIS expressed in vitro in the reticulocyte lysate (Fig. 5). The retention of KIS on GST-SF1f and GST-SF3b155n beads was mainly affected by the complete deletion of the UHM (Fig. 5a, lanes 11 and 17 and Fig. 5b) or by a large deletion within the UHM (KISΔ[369–409], Fig. 5a, lanes 10 and 16). We also tested the effect of mutating acidic residues in the putative helix A of the KIS UHM. Interestingly, while mutation of glutamic acid 341 and aspartic acid 342 to lysine strongly reduced binding to SF1f as previously observed36 (Fig. 5a, lane 12), it had only a limited effect on the binding of KIS to SF3b155n (Fig. 5a, lane 18 and 5b). Therefore, despite similar efficiencies and UHM requirement for the KIS-SF1f and KIS-SF3b155n interactions, the use of mutant forms suggests differences in the detailed binding mechanisms.
Figure 5. The KIS UHM domain is necessary for interaction with SF1 and SF3b155.
(a) Full length KIS and U2AF65 and mutant forms of KIS as schematically presented were in vitro translated in the presence of [35S]-methionine and tested for their interaction with GST-SF1f (SF1 fragment [1–255]) and GST-SF3b155n (fragment [1–493]) in GST pull-down assays. The autoradiogram of the SDS PAGE gel of a typical experiment is presented. 2% of the inputs were loaded on the same gel (lanes 1 to 6) to allow the quantification of the retention of the labelled proteins on the beads as described for Fig. 3c. (b) Representation of the means of retention rate and standard deviation for four experiments.
We also tested the binding of a "kinase defective" mutant of KIS with a lysine to arginine mutation of residue 54 which, by analogy to other kinases, is predicted to be involved in phosphoryl transfer and stabilization of the active state of the kinase domain4,41. Surprisingly this mutant was less efficient in binding to SF1f and SF3b155n, suggesting that the kinase domain of KIS is also participating in these interactions (Fig. 5a, compare lanes 7, 9, 13 and 15). Finally, in these experiments we observed a similar retention of KIS and U2AF65 on GST-SF1f and GST-SF3b155n beads (Fig. 5a, lane 8 and 14, and Fig. 5b). The observed retention rates were consistent with equilibrium dissociation constants of the order of 100 nM, in agreement with the previously reported 50–100 nM range for the binding of the U2AF65 UHM to the SF1 ULM in similar pull-down assays22.
Importance of ULMs for KIS binding to SF1 and SF3b155
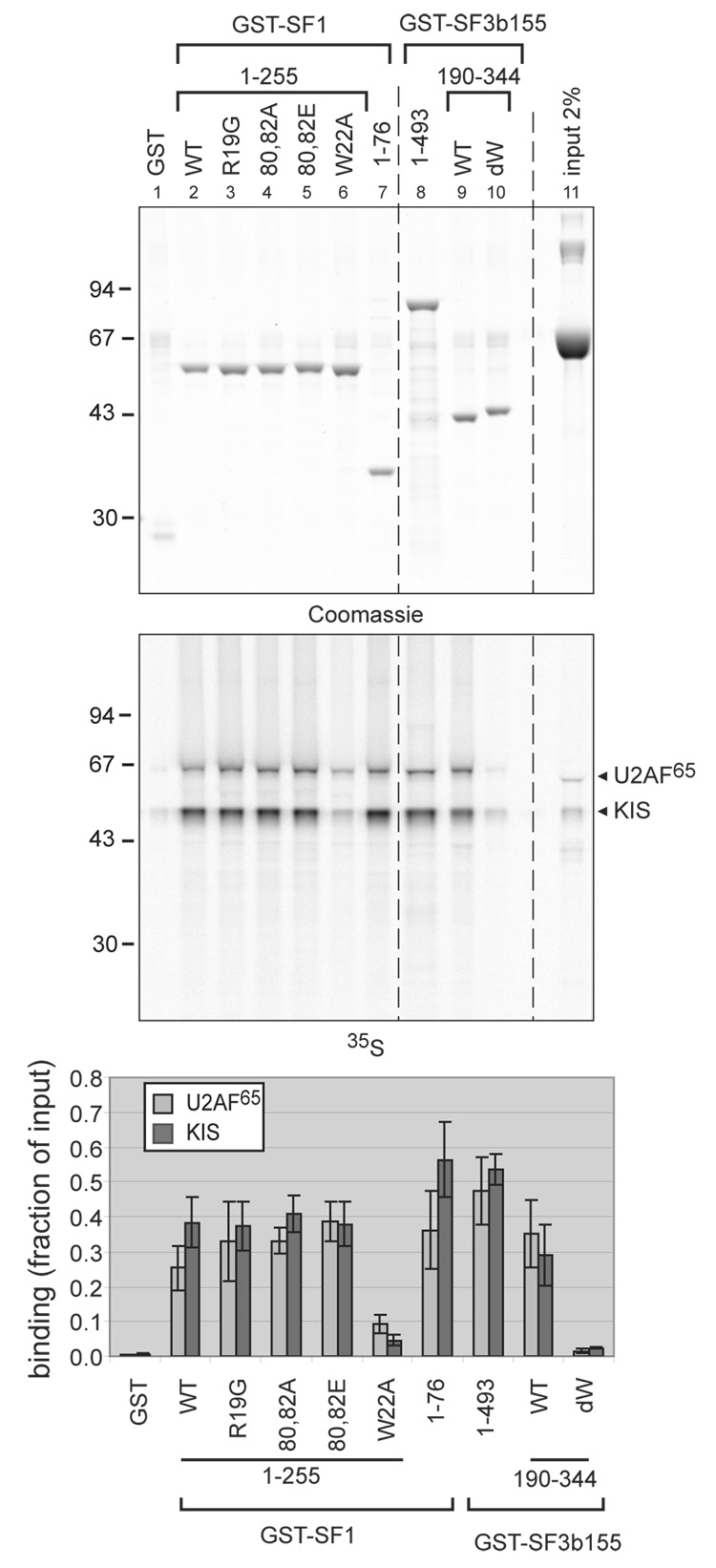
Although our results with the kinase defective form of KIS indicated a partial involvement of the kinase domain in the interaction of KIS with SF1 and SF3b155, mutations of the UHM domain had the most dramatic effect on these interactions, suggesting that, as previously demonstrated in the case of U2AF65, the ULM peptides within the SF1 and SF3b155 proteins are most important for their interaction with KIS. To further test this hypothesis we compared the effect of different SF1 and SF3b155 mutations within or outside their ULMs (Fig. 6). Reducing the SF1 fragment to amino-acids [1–76] did not modify the retention of KIS and U2AF65 (Fig. 6, lane 7). Similarly, mutation of arginine 19 to glycine, and of the KIS phophorylation site serines 80 and 82 to either alanine or glutamic acid did not significantly alter the interactions. In contrast, mutating tryptophan 22 of SF1f to alanine greatly reduced the binding of KIS and U2AF65 (Fig. 6, lane 6) as previously described for U2AF65 22. A similar effect was observed when using a mutant form of SF3b155r (fragment [190–344]) in which the tryptophan residues of the seven potential ULMs where mutated to alanine (SF3b155-dW33) (Fig. 6, lanes 9 and 10) in agreement with previous results in the case of U2AF65 30,33. Importantly, previous analyses indicated that these mutations of SF3b155r did not alter the stability of the overall protein fold33. Furthermore, the N-terminus of SF3b155 is predicted to be intrinsically unstructured30,32,33, in agreement with the lack of structural element determined by electron cryomicroscopy31, and ultracentrifugation and circular dichroism data30. Therefore tryptophan residues most probably do not play a crucial role in folding of this domain and the observed dramatic effects of their mutation are rather suggesting their direct involvement in KIS and U2AF65 binding.
Figure 6. Effect of mutations of SF1 and SF3b155 on binding to KIS and U2AF65.
Different mutant forms of GST-SF1 and GST-SF3b155 as indicated above the lanes, were compared in pull-down assays with a mixture of [35S]-labelled in vitro translated U2AF65 and KIS diluted in interaction buffer containing 1 µg/µL BSA as described for Fig. 3c. After SDS PAGE of the binding reactions, the gel was stained with Coomassie blue and digitalized with an infrared scanner (top panel). Retention of KIS and U2AF65 was quantified with a phosphorimager (middle panel) as explained in the Material and Methods section. The mean values of three experiments with standard deviation are presented (bottom panel), showing that mutations of tryptophan residues within the ULM peptides of the SF1f and SF3b155r proteins reduced KIS and U2AF65 co-precipitation.
We also observed that the use of the shorter fragment SF3b155r ([190–344]) already reduced the retention of KIS as compared to SF3b155n ([1–493]) (Fig. 6, lanes 8 and 9 and quantification below). The better interaction of KIS with SF3b155n compared to SF3b155r may be accounted for by the contribution made by SF3b155n elements outside the minimal SF3b155r domain including a potential additional ULM site (W388)30 or by a tertiary structural arrangement that presents the ULMs in an optimal manner for interactions with KIS.
To further analyse the relative contribution of each ULM within SF3b155r to its binding to KIS, we used the previously characterized constructs in which each tryptophan was independently reintroduced in the SF3b155-dW mutant33 (hereafter named SF3b155-W200 to SF3b155-W338 and collectively SF3b155-Wn). To accurately compare the binding of KIS and U2AF65 in the same conditions we used a mixture of both [35S]-labelled proteins as input (Fig. 7). In these conditions we observed a higher binding of U2AF65 to SF3b155-W338 (25% of the binding of wild type SF3b155r) followed by SF3b155-W200 (13% of wild type). KIS presented an opposite profile with a preferred binding to SF3b155-W200 (23% of wild type) followed by SF3b155-W338 (16% of wild type). In both cases the difference was statistically highly significant (one-way ANOVA Bonferroni post-hoc test p<0.001). Thus, interestingly closely related UHM domains like that of KIS and U2AF65 can have different specificities.
Figure 7. Different properties of SF3b155 ULMs in mediating interactions with U2AF65 and KIS.
(a) The binding of in vitro translated [35S]-labelled U2AF65 and KIS to the wild type SF3b155r (residues [190–344], lane 1), SF3b155r with the seven tryptophan mutated to alanine (lane 2) and the mutants in which each of the seven tryptophan residues were independently reintroduced (lanes 3–9) were tested. The experiments were performed as for Fig. 3c.
(b) The efficiency of binding was calculated as the ratio of binding to the wild type SF3b155r. The mean values of three experiments (each interaction reaction being performed in duplicate) are presented with standard deviation. One-way ANOVA statistical analysis was conducted with a Bonferroni post-hoc test yielding a p<0.001 for a significant difference of binding of U2AF65 on SF3b155-W338 compared to SF3b155-W200, and a significant difference of binding of KIS to SF3b155-W200 compared to SF3b155-W338.
(c) Alignment of the seven potential ULMs of SF3b155r with that of SF1. The key tryptophan residues are in boldface. Basic residues N-terminal to the key tryptophan are boxed in grey. Lysine 15 of SF1 that might contribute to the particular binding of SF1 to KIS is underlined (see Discussion). The best binding sites for KIS and U2AF65, ULM-W200 and ULM-W338 present a higher homology with that of SF1.
Discussion
The protein kinase KIS, composed of juxtaposed kinase and UHM-containing domains, is thought to be involved in splicing factor regulation as supported by its efficient phosphorylation of SF1 both in vitro and in vivo (ref 36 and our unpublished data). Our present results illustrate different roles for these two domains in promoting the nuclear localization of KIS and its interaction with splicing factors.
The UHM domain of KIS is not required for nuclear accumulation
The nuclear localization of KIS has been documented by immunofluorescence analyses4,10 as well as by cell fractionation experiments4. Here the observation of KIS localization by immunofluorescence in CHO cells revealed most cells presenting brighter staining in the nucleus. As KIS lacks detectable NLS sequences, it is likely that it enters the nucleus through interaction with an NLS-containing protein. In particular, SF3b155 nuclear localization depends on a monopartite NLS42 while SF1 possesses an N-terminal putative NLS43. However, whereas the UHM domain of KIS was required for its interaction with SF1 and SF3b155, it was dispensable for its nuclear accumulation. This suggests that neither SF1 nor SF3b155 are necessary for the nuclear accumulation of KIS. In contrast, deletions within the kinase domain of KIS led to characteristic exclusion from the nucleus. These data suggest that one or several interactions involving mainly residues [80–211] within its kinase domain allows KIS to be translocated or retained in the nucleus. Such partners of KIS await identification to shed light on the molecular mechanisms controlling KIS nuclear localization.
The UHM domain of KIS is necessary for interaction with SF1 and SF3b155
We previously determined the ability of KIS to interact in vitro with the splicing factor SF136. Using two-hybrid and pull-down experiments, we demonstrate here that KIS also binds to the splicing factor SF3b155. In addition to the high similarity of the KIS UHM with that of U2AF65, our data obtained with a variety of mutant forms of KIS, SF1 and SF3b155 strongly suggest that the interaction of KIS with SF1 and SF3b155 primarily relies on the interaction of the KIS UHM with ULMs of these splicing factors (See Fig. 8 for a schematic representation of major contacts among these proteins). Mutations to alanine of either tryptophan 22 of SF1f or the seven tryptophan residues within the putative ULMs of SF3b155r greatly reduced the interaction of these proteins with KIS. Reciprocally, the removal or internal deletion of 41 amino acids within the UHM domain of KIS dramatically inhibited its interaction with SF1f and SF3b155n. The KIS mutant with both negatively charged residues E341 and D342 replaced with lysine also lacked affinity for SF1f, but still interacted significantly with SF3b155n. Accordingly, acidic residues at the corresponding positions in helix A of U2AF65 UHM (E396 and E397) were shown to be in close proximity to lysine 15 of SF122 (underlined in Fig. 7c), while in contrast no basic residues are present at the corresponding position of the SF3b155 ULMs. Instead, the ULM sites of SF3b155 present diverse locations and numbers of basic residues, which may interact with remaining acidic residues in the KIS UHM (for example, E345 and D346). Thus, the different requirement for acidic residues at positions 341 and 342 of the putative helix A of the KIS UHM may reflect the greater extent of the stretch of basic residues in the N-terminus of the ULM of SF1 compared to that of SF3b155 (see alignment in Fig. 7c). However, we cannot exclude that additional contacts between KIS and SF3b155 may compensate for the reduction of electrostatic interactions with the KIS UHM caused by these mutations.
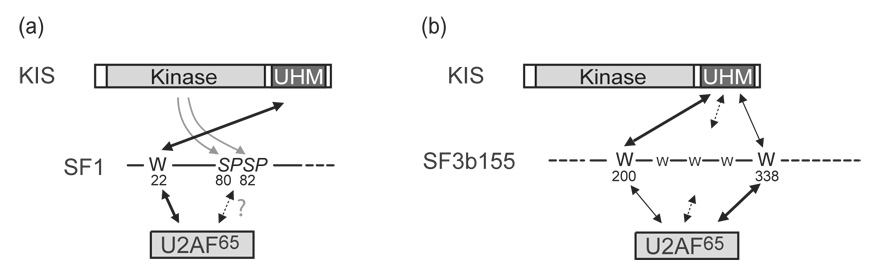
Figure 8.
Schematic representation for the comparison of interactions of KIS and U2AF65 with the N-terminal regions of SF1 and SF3b155. Contacts additional to those represented might contribute to these interactions.
(a) Thick arrows indicate the major contribution of W22 of SF1 for the binding of U2AF65 and KIS. Additionally KIS phosphorylates serines 80 and 82 which increases the binding of SF1 to U2AF65 potentially by direct contact (broken line arrow).
(b) Thick arrows indicate the predominant contribution of W200 and W338 for the binding of KIS and U2AF65 to the N-terminal region of SF3b155. Additional interactions depending on other tryptophan residues are indicated by broken lines arrows.
Interestingly, although there was some variation from one experiment to the other, we observed consistently comparable levels of retention of U2AF65 and KIS on GST-SF1f and GST-SF3b155n beads throughout our experiments. The similar binding of U2AF65 and KIS suggests that the high conservation during evolution of the UHM sequences in KIS and U2AF65 reflects the conserved property of these UHMs to interact with ULMs. It is thus likely that the UHM of KIS contributes to a network of UHM-ULM contacts as also recently proposed for the UHM-containing splicing factor SPF4535.
Potential involvement of interactions additional to the UHM-ULM contacts
We observed here that a "kinase defective" form of KIS with lysine 54 mutated to arginine (K54R) was less effective in binding to SF1 and SF3b155. This lysine residue in the small lobe of kinase domains is highly conserved and critical for the activity of protein kinases (see reference 41 and references therein). It is thought to have a conserved function in making contacts with the α- and β-phosphate groups of ATP and in stabilizing the active structure of the kinase. Although mutation of this lysine to histidine slightly decreases the thermal stability of the prototypical kinase family member cAMP-dependent kinase, the kinase remains predominantely folded under the conditions used for the assays described here41.Moreover, the mutation of lysine 54 to arginine is less expected to disrupt interdomain salt bridges, as was suggested for the histidine mutation of the cAMP-dependent kinase. Although we cannot exclude that the reduced interaction of the KIS[K54R] mutant is due to global unfolding, by analogy this possibility seems unlikely. Several alternative explanations for the reduced interactions with this kinase-domain mutant include direct contacts between the kinase domain of KIS and its partners SF1 and SF3b155, or indirect effects due to a modified interdomain arrangement of the kinase and UHM domains. Further experiments will address the interesting possibility that SF1 and SF3b155 preferentially bind KIS in its active conformation.
Several observations in the literature suggest that interactions outside the minimal UHM-ULM elements may frequently participate in the interactions between UHM and ULM containing proteins. For example, Cass and colleagues30 observed residual binding of SF3b155[1–430] to U2AF65 after removal of its UHM. Moreover, including sequences beyond the minimal SF1 ULM33 enhances binding to U2AF65, and this binding is enhanced by KIS-dependent phosphorylation outside the ULM36. The involvement of the kinase domain in KIS-SF1 and KIS-SF3b155 interactions observed here further supports a hypothesis that additional non-UHM-ULM regions commonly contribute to interactions between UHM and ULM containing proteins.
Comparison of KIS and U2AF65 specificities for binding ULM domains
Several ULM domains have previously been identified within the N-terminus of SF3b155 based on sequence similarity with the SF1 ULM30,32,33, designated here by the position of the corresponding tryptophan in the sequence. NMR binding studies have shown that addition of U2AF65-UHM to a solution of a SF3b155 fragment (residues [282–424]) mainly affected residues 331–346 of SF3b155 suggesting that ULM-W338 is the preferred binding site for U2AF65 in this region32. In addition, Cass and colleagues30 have shown that among different ULM peptides only those corresponding to ULM-W200 and ULM-W338 can displace the interaction of SF3b155[1–430] with U2AF65 in gel shift experiments. Furthermore, in pull-down assays, mutation of both residues severely reduced the binding of U2AF65 to this fragment of SF3b155, while single mutations led only to a slight decrease of U2AF65 binding30. Finally, data obtained by intrinsic tryptophan fluorescence shift using SF3b155[190–344] mutants in which all but one tryptophan residues have been mutated to alanine (SF3b155-Wn mutants), indicated a binding of the U2AF65-UHM to five ULM domains (corresponding to W200, W218, W232, W293 and W338) within SF3b155[190–344] with the highest affinity observed for SF3b155-W33833.
Consistent with these previous results, we observed that mutation of all tryptophans of the putative ULMs within SF3b155r ([190–344]) greatly reduced its binding to U2AF65. Our results, which were obtained with a different approach and with longer protein fragments are in agreement with previous reports but they emphasize the preference of U2AF65 for the ULM-W338 of SF3b155, followed by ULM-W200. Similarly, it has recently been suggested that the UHM-containing splicing factor SPF45 preferentially contacts ULM-W338 for binding to SF3b15535. Of note, the sequences of these two putative ULMs of SF3b155 are the most similar to those of SF1 (Fig. 7c).
When compared to U2AF65, KIS interacted significantly with the same two ULMs of SF3b155 in our experiments. However, KIS interestingly showed a clear preference for binding to SF3b155-W200 as compared to SF3b155-W338. Thus despite the particularly high sequence similarity of the UHMs of KIS and U2AF65, their specificities for binding to ULMs differ. However, as in our experiments we used full length KIS and U2AF65, it is possible that regions outside the UHM domains of these proteins also participate in their different preference for binding sites within SF3b155. It has been reported that serine 20 of SF1 (in position -2 relative to the key tryptophan) is a substrate for protein kinase G, and that this phosphorylation impedes the SF1-U2AF65 interaction43. Interestingly, a serine residue is also present in position -2 in the sequence of ULM-W338, but not in that of ULM-W200. Thus, the differing affinities of KIS and U2AF65 for these sites suggest a potential means for regulating interactions with KIS versus U2AF65 by phosphorylation of SF3b155.
Albeit different in terms of ULM preference, KIS and U2AF65 both require the integrity of several ULMs for binding with a maximal efficiency to SF3b155, in contrast to SF1, which possesses a single ULM. Therefore, despite in our conditions the similar retention of full length KIS or full length U2AF65 on GST-SF1f and GST-SF3b155n, their binding to SF1f is energetically favorable compared to their binding to each of the SF3b155-Wn. Accordingly, the apparent Kd measured for a single average binding site in the SF3b155r-(U2AF65-UHM) interaction is weaker than that of the SF1f-(U2AF65-UHM) interaction33. Our approach could not detect significant retention of U2AF65 and KIS on the other SF3b155-Wn domains in comparison with the SF3b155-dW mutant. As the sum of the retentions on the seven different SF3b155-Wn mutants was lower than that on the wild type SF3b155r, a synergistic effect of the tryptophan residues for the binding of U2AF65 and KIS to SF3b155 is a possibility that would need to be tested using complementary methods. Accordingly, an elevated local concentration of ULMs seated next to each other is available in the SF3b155 sequence.
In conclusion, our data further extend the UHM-ULM interaction network, and demonstrate for the first time that UHM-containing proteins exhibit distinct preferences for binding to various ULMs within the N-terminal domain of the scaffolding protein SF3b155. This might partly explain the conservation of multiple ULMs within this protein, and further suggests the possibility of simultaneous binding of different UHM-containing proteins to the SF3b155 scaffold with potentially regulatory action on splicing. Our analysis of KIS determinants for nuclear localization points to a different partner binding the kinase domain for KIS nuclear import or retention. Finally, combined with the evidence that KIS phosphorylates SF1 on two highly conserved residues in vitro and in vivo, the present data showing that KIS interacts with SF1 and SF3b155 in a comparable manner to U2AF65 further supports a role for KIS in the molecular functions of these splicing factors and in the proposed action of KIS in the control of protein expression in the nervous system, where perturbations are thought to contribute to the etiology of neurologic or psychiatric diseases.
Material and Methods
Two-hybrid
Two hybrid tests were performed with standard procedures by mating L40 cells expressing the prey VP16-fused proteins with AMR70 cells expressing fusion of the DNA binding domain of LexA with the bait proteins as described3. Diploid cells were then replicated on agar plates lacking tryptophan, leucine and histidine to test the expression of the His3 reporter, and to Whatman 40 filter papers covered agar plates lacking tryptophan and leucine to test the LacZ expression.
Constructs
For expression of KIS as a bait in the two-hybrid system, the whole coding sequence of rat KIS was subcloned in vector pVJL1044 to yield pLexA-KIS. As expression of full length KIS lead to autonomous expression of the reporter genes we prepared a construct for expression of a truncated form comprising amino acids [120–419]. For this a KIS cDNA Tsp509I-SacI fragment was ligated to pLexA-KIS previously digested by EcoRI and SacI to yield pLK-[120–419]. Other baits and prey plasmids were described previously3. Plasmid for the prey SF3b155-VP16 and the baits U2AF35 and U2AF65 were kindly provided by Dr. Robin Reed29 (Boston, USA).
Cell transfection and immunofluorescence analysis
Plasmids for expression in CHO cells were prepared by subcloning KIS rat cDNA downstream the SV40 promoter in the pECE vector. Constructions for expressing deleted forms of KIS were prepared by digestion with restriction enzymes, Klenow fragment treatment if required and ligation. CHO cells plated at a density of 200,000 cells in 35 mm diameter dishes were transfected the next day with 4µL lipofectamine reagent (Invitrogen) mixed with 1 µg of plasmid DNA in serum free medium. After 6 hours the medium was replaced by fresh medium containing 10% fetal calf serum. After 18 hours cells were fixed with 2% paraformaldehyde in Phosphate Buffer Saline for 20 minutes before permeabilization and incubation with primary antibody followed by Alexa 546 coupled anti-rabbit secondary antibody. Coverslips were mounted with Mowiol for observation with a Leica DM IRE2 epifluorescence microscope equipped with a micromax CCD camera (Roper Scientific).
Other plasmids
Plasmids for expression of human SF1 (residues [1–255]) (SF1f) in the pGEX6P-1 vector (Amersham Biosciences) was described36. Plasmid pSP64-U2AF65 16 for in vitro translation was kindly provided by Dr. Juan Valcarcel (Barcelona, Spain). Plasmid pSP64-KIS for expression of rat KIS from the same vector was previously described36. Plasmid for expression of GST-SF3b155n (amino acids [1–493] was kindly provided by Dr. Robin Reed (Boston, USA). Plasmids for GST-SF3b155r (amino acids [190–344]) of human SF3b155 and the tryptophan mutants were described33. Plasmid GST-p27kip1 was kindly provided by Dr. Elizabeth Nabel (Bethesda, USA). Plasmid for GST-CTD encoding the C-terminal domain of polymerase II was described45,46. Site-directed mutagenesis was performed using the Quickchange protocol from Stratagene (La Jolla, CA, USA). All constructs were checked for sequence accuracy by complete sequencing of the coding regions.
Protein expression and purification
GST-fusion proteins for use in pull-down assays were prepared from 100 mL cultures of BL21 cells transformed with the pGEX-6P or pGEX-KG derived constructions. After induction with 0.4 mM isopropyl thiobetagalactoside for 2 hours cells were harvested and frozen at −80°C until use. Frozen cell pellets were homogenized in GSB buffer (25 mM HEPES pH7.5, 100 mM KCl, 1 mM EDTA, 0.1% NP40 and 10% glycerol) with 1 mM DTT and antiprotease mix from Roche Diagnostics by sonication, and soluble material was purified on glutathione beads (Roche Diagnostics). After extensive washing of the beads, GST-fusion proteins were eluted with 40 mM reduced glutathione, and dialysed against 25 mM Hepes pH 7.5, 50 mM NaCl. Additionally for the experiment in Fig. 3b recombinant GST-KIS was cleaved with human thrombin (T4393, Sigma-Aldrich) at room temperature for 45 min, followed by inactivation of thrombin by addition of 1 mM PMSF (P7626, Sigma-Aldrich) and depletion of the GST fragment and uncleaved GST-KIS by incubation with glutathione beads. Protein concentrations were determined using the BCA method (Pierce) and solutions were snap frozen after adjusting to 25% glycerol and kept at −80°C until use.
GST pull-down
In vitro translations were performed using the TNT system (Amersham Biosciences), [35S]-methionine (NEN) and the appropriate plasmid DNAs. 25 µL of the in vitro translation reactions and of the GST-fusion proteins solutions were then treated with 250 ng of RNAse I (R4875, Sigma-Aldrich) for 5 minutes at room temperature, in conditions that were optimized to remove RNA as monitored by agarose gel electrophoresis and ethidium bromide staining. 0.3 µL of in vitro translation product or about 1 pmole of recombinant KIS (Fig. 3b) was mixed with 20 pmoles of GST-fusion proteins in 250 µL of interaction buffer containing 25 mM Hepes pH 7.5, 50 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% NP40, 1 mM DTT, antiprotease mix from Roche Diagnostics and 1 µg/µL BSA (or 0.1 µg/µL for experiment with recombinant KIS in Fig. 3b) as a non specific competitor. After a 90 min incubation at 4 °C with agitation, 10 µL of glutathione beads (Amersham Biosciences) were added for a further 30 min, beads were washed rapidly 5 times with interaction buffer, and proteins were analysed by SDS-PAGE, Coomassie blue staining and phosphorimaging or immunoblotting with a polyclonal anti-KIS antibody4. 10 pmoles of each GST-fusion proteins were loaded in parallel to allow the determination of the recovery of these proteins on the glutathione beads. Quantification of the Coomassie blue staining was achieved using a 700 nm infrared laser scanner (Odyssey, Li-Cor). The fraction of [35S]-labelled proteins bound to the GST-fusion proteins in the interaction mixture was then calculated as the fraction of radioactivity recovered on the beads divided by the fraction of GST-fusion proteins recovered on the beads.
Phosphorylation reactions
Phosphorylation reactions were performed essentially as described previously36. Briefly, 20 µL reactions contained approximately 20 pmoles of substrate in 50 mM MES pH 8.0, 10 mM MgCl2, 2 mM DTT, 2 mM EDTA, 25% glycerol, 10 µM [γ-P]ATP (5 nCi/pmole) (NEN). Phosphorylations by KIS were performed with about 10 ng of GST-KIS. The cell extract used as a source of kinase activities was prepared from proliferating mouse embryonic fibroblast by homogenisation in TrisHCl 20 mM pH7.5, 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 50 mM NaF, 10 mM b-glycerophosphate, 5 mM sodium pyrophosphate, 1 mM DTT, 0.5% Nonidet P40 and antiprotease mix from Roche Diagnostics. Reactions were allowed to proceed for 30 min at 30°C and analysed by SDS-PAGE, Coomassie blue staining and phosphorimaging.
Acknowledgements
We are grateful for the generous gifts of cDNA constructs from O. Bensaude, E. Nabel, R Reed and J. Valcárcel. We thank colleagues from INSERM UMR-S839 for stimulating discussions and support. We thank members of the Institut du Fer à Moulin Imaging Facility for help with microscopy imaging. This work was funded by the "Institut National de la Santé et de la Recherche Médicale", the "Université Pierre et Marie Curie", the "Association Française contre les Myopathies", the "Association pour la Recherche contre le Cancer" to A.S. and A.M, by the "NARSAD: The Mental Health Research Association" to A.M. (Independent Investigator Award) and by the National Institutes of Health grant R01-GM070503 to C.L.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunter T. Signaling--2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 3.Maucuer A, Camonis JH, Sobel A. Stathmin interaction with a putative kinase and coiled-coil-forming protein domains. Proc Natl Acad Sci U S A. 1995;92:3100–3104. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maucuer A, Ozon S, Manceau V, Gavet O, Lawler S, Curmi P, Sobel A. KIS is a protein kinase with an RNA recognition motif. J Biol Chem. 1997;272:23151–23156. doi: 10.1074/jbc.272.37.23151. [DOI] [PubMed] [Google Scholar]
- 5.Bieche I, Manceau V, Curmi PA, Laurendeau I, Lachkar S, Leroy K, Vidaud D, Sobel A, Maucuer A. Quantitative RT-PCR reveals a ubiquitous but preferentially neural expression of the KIS gene in rat and human. Brain Res Mol Brain Res. 2003;114:55–64. doi: 10.1016/s0169-328x(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell BD, Darlington DN, Penzes P, Johnson RC, Eipper BA, Mains RE. The novel kinase peptidylglycine alpha-amidating monooxygenase cytosolic interactor protein 2 interacts with the cytosolic routing determinants of the peptide processing enzyme peptidylglycine alpha-amidating monooxygenase. J Biol Chem. 1999;274:34646–34656. doi: 10.1074/jbc.274.49.34646. [DOI] [PubMed] [Google Scholar]
- 7.Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Moorey H, Morgan J, Punukollu B, Kandasami G, Curtis D, Gurling H. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry. 2007;61:873–879. doi: 10.1016/j.biopsych.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Puri V, McQuillin A, Datta S, Choudhury K, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Crombie C, Fraser G, Walker N, Moorey H, Ray MK, Sule A, Curtis D, Clair DS, Gurling H. Confirmation of the genetic association between the U2AF homology motif (UHM) kinase 1 UHMK1) gene and schizophrenia on chromosome 1q23.3. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.76. [DOI] [PubMed] [Google Scholar]
- 9.Crook MF, Olive M, Xue HH, Langenickel TH, Boehm M, Leonard WJ, Nabel EG. GA-binding protein regulates KIS gene expression, cell migration, and cell cycle progression. Faseb J. 2007 doi: 10.1096/fj.07-8573com. [DOI] [PubMed] [Google Scholar]
- 10.Boehm M, Yoshimoto T, Crook MF, Nallamshetty S, True A, Nabel GJ, Nabel EG. A growth factor-dependent nuclear kinase phosphorylates p27(Kip1) and regulates cell cycle progression. Embo J. 2002;21:3390–3401. doi: 10.1093/emboj/cdf343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrovic V, Costa RH, Lau LF, Raychaudhuri P, Tyner AL. FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem. 2008;283:453–460. doi: 10.1074/jbc.M705792200. [DOI] [PubMed] [Google Scholar]
- 12.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci U S A. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradham CA, Foltz KR, Beane WS, Arnone MI, Rizzo F, Coffman JA, Mushegian A, Goel M, Morales J, Geneviere AM, Lapraz F, Robertson AJ, Kelkar H, Loza-Coll M, Townley IK, Raisch M, Roux MM, Lepage T, Gache C, McClay DR, Manning G. The sea urchin kinome: a first look. Dev Biol. 2006;300:180–193. doi: 10.1016/j.ydbio.2006.08.074. [DOI] [PubMed] [Google Scholar]
- 14.Maucuer A, Le Caer JP, Manceau V, Sobel A. Specific Ser-Pro phosphorylation by the RNA-recognition motif containing kinase KIS. Eur J Biochem. 2000;267:4456–4464. doi: 10.1046/j.1432-1327.2000.01493.x. [DOI] [PubMed] [Google Scholar]
- 15.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 17.Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 19.Berglund JA, Fleming ML, Rosbash M. The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. Rna. 1998;4:998–1006. doi: 10.1017/s1355838298980499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrey SM, Voelker R, Berglund JA. An extended RNA binding site for the yeast branch point-binding protein and the role of its zinc knuckle domains in RNA binding. J Biol Chem. 2006;281:27443–27453. doi: 10.1074/jbc.M603137200. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 22.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 23.Rain JC, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. Rna. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 25.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt-Zachmann MS, Knecht S, Kramer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. Rna. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cass DM, Berglund JA. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- 31.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 32.Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Luhrmann R, Sattler M. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. Rna. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J Mol Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frenal K, Callebaut I, Wecker K, Prochnicka-Chalufour A, Dendouga N, Zinn-Justin S, Delepierre M, Tomavo S, Wolff N. Structural and functional characterization of the TgDRE multidomain protein, a DNA repair enzyme from Toxoplasma gondii. Biochemistry. 2006;45:4867–4874. doi: 10.1021/bi051948e. [DOI] [PubMed] [Google Scholar]
- 35.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat Struct Mol Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 36.Manceau V, Swenson M, Le Caer JP, Sobel A, Kielkopf CL, Maucuer A. Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. Febs J. 2006;273:577–587. doi: 10.1111/j.1742-4658.2005.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 38.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 39.Brameier M, Krings A, MacCallum RM. NucPred--predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–1160. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- 40.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer GH, Garrod S, Woods VL, Jr., Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Eilbracht J, Schmidt-Zachmann MS. Identification of a sequence element directing a protein to nuclear speckles. Proc Natl Acad Sci U S A. 2001;98:3849–3854. doi: 10.1073/pnas.071042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, Robinson PJ. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. Embo J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 45.Ahearn JM, Jr, Bartolomei MS, West ML, Cisek LJ, Corden JL. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987;262:10695–10705. [PubMed] [Google Scholar]
- 46.Peterson SR, Dvir A, Anderson CW, Dynan WS. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 47.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 48.Kramer A, Quentin M, Mulhauser F. Diverse modes of alternative splicing of human splicing factor SF1 deduced from the exon-intron structure of the gene. Gene. 1998;211:29–37. doi: 10.1016/s0378-1119(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 49.Lin KT, Lu RM, Tarn WY. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol Cell Biol. 2004;24:9176–9185. doi: 10.1128/MCB.24.20.9176-9185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstrohm AC, Albrecht TR, Sune C, Bedford MT, Garcia-Blanco MA. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol Cell Biol. 2001;21:7617–7628. doi: 10.1128/MCB.21.22.7617-7628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. Embo J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci U S A. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudrez A, Beullens M, Waelkens E, Stalmans W, Bollen M. Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J Biol Chem. 2002;277:31834–31841. doi: 10.1074/jbc.M204427200. [DOI] [PubMed] [Google Scholar]