Abstract
A common pathobiological feature of malignant gliomas is the insidious infiltration of single tumor cells into the brain parenchyma, rendering these deadly tumors virtually incurable with available therapies. In this study, we report that ADP-ribosylation factor 6 (ARF6), a Ras superfamily small GTPase, is abundantly expressed in invasive human glioma cells. Cellular depletion of ARF6 by siRNA decreased Rac1 activation, impaired HGF- and serum-stimulated glioma cell migration in vitro, and markedly decreased the invasive capacity of invasive glioma in the brain. Furthermore, ectopic expression of ARF6 in glioma cells promoted cell migration via the activation of Rac1. Upon stimulation of glioma cells with HGF, We show that IQGAP1 is recruited to and overlaps with ARF6 at the leading edge of migrating cells. However, cellular depletion of ARF6 abrogated this recruitment of IQGAP1 and attenuated the formation of surface protrusions. ARF6 forms complexes with Rac1 and IQGAP1 in glioma cells upon HGF stimulation, and knockdown of IQGAP1 significantly inhibits ARF6-induced Rac1 activation and cell migration. Taken together, these data suggest that ARF6-mediated Rac1 activation is essential for glioma cell invasion, via a signaling pathway that requires IQGAP1.
Keywords: ARF6, IQGAP1, Rac1, glioma, invasion
Introduction
Malignant human gliomas display rapid proliferation, extensive neovascularization, apoptosis resistance and diffuse invasion into the surrounding brain parenchyma, and eventually gives rise to recurrent tumors despite radical surgery, irrespective of their histological type and grade of malignancy (1, 2). Glioma cell invasion is a highly complex process, and mechanisms regulating cell motility may represent key elements of the invasive cascade (3). ADP-ribosylation factor 6 (ARF6), which belongs to the ARF family of small GTP-binding proteins with multiple roles in fundamental biological processes such as actin remodeling (4-6), has been shown to play an important role in tumor cell invasion (4-6). Studies showed that ARF6 is expressed at high levels in highly invasive breast cancer cells and ARF6 activation increases the invasive potential of melanoma cells and breast cancer cells (7-9). In gliomas, exogenous expression of EFA6A, a guanine nucleotide exchange factor (GEF) that promotes ARF6 activation, enhanced cell motility and invasiveness in in vitro assays for invasion and migration (10).
The requirement of ARF6 for cell spreading, Rac1-induced ruffling, cell migration and wound healing has been recently described (5, 6). Although ARF6-dependent modulation of Rac1 activation appears to be important, molecular mechanisms by which ARF6 regulates cell motility remain unclear. Several studies showed that ARF6 stimulates actin reorganization and membrane ruffling, promoting the acquisition of a migratory phenotype through activation of Rac1 (11-13). Rac1 is critical for changes that facilitate cell motility. Rac1 directs actin assembly that results in the formation of lamellipodia at the leading edge of migrating cells, and thus is a key player in cell movement (14). Depletion of Rac1 by siRNA decreases cell migration and invasion in tumor cells including glioma cells (2, 15, 16). Although Rac1 has been described as a downstream target of ARF6, the ARF6-dependent Rac1 regulation appears to be complex and varies with cell types. For example, in epithelial cells an ARF6-GEF, ARNO promotes Rac1 activation by recruiting a Rac1-GEF complex, Dock180/ELMO1 (17). A Rac1-GEF, β-PIX is relocated to the plasma membrane and Rac1 activation is enhanced in both ARF6-depleted HEK 293 and HepG2 cells (18). In addition, in NIH 3T3 cells, another key regulator of cell motility, IQ-domain GTPase-activating protein 1 (IQGAP1) found in the macropinosomes induced by ARF6 near sites of membrane ruffling (19). IQGAP1 plays important roles in various aspects of cell physiology, particularly in cell migration through its interaction with various proteins (20-22). Although IQGAP1 has been reported to function as either a target or a regulator of Rac1, IQGAP1 regulates cell adhesion and migration by binding to Rac1 and stabilizing Rac1-GTP (22). Similar to ARF6 activation modulated by upstream growth factors (7, 8, 11, 23), IQGAP1 participates in signaling cascades that are also stimulated by growth factors, such as EGF and HGF (20, 22). IQGAP1 localizes at the leading edge of migrating cells and knockdown of IQGAP1 in breast cancer cells decreases cell motility and invasion (24). In human cancers, IQGAP1 is overexpressed in colorectal carcinomas and associated with invasion fronts (25).
In the current study, we examined the involvement of ARF6 in glioma cell invasion using in vitro, ex vivo and in vivo models. Our data demonstrate that ARF6 is abundantly expressed in various highly invasive glioma cell lines, and is required for enhanced glioma cell migration. Knockdown of ARF6 expression by siRNAs suppressed glioma cell invasion ex vivo and in the brain of mice. Furthermore, ARF6 mediates Rac1 activation in glioma cells upon external stimulation with serum and HGF. We show that IQGAP1 is required for the ARF6-mediated activation of Rac1, a signaling pathway that appears to be essential for glioma cell invasion.
Materials and Methods
Glioma cell lines, antibodies and reagents
Human LN18, LN229, U118MG, U87MG, T98G glioma cells were obtained from American Type Culture Collection (Rockville, MD). U251MG, U373MG, LN215, LNZ308 glioma cells were from our collection (26). SNB19 cells were from Dr. Y-H Zhou (University of California, Irvine). D54MG and A172 cells were from Dr. D. Bigner (Duke University). Normal human astrocytes (NHA) were from Lonza, Allendale, NJ. The following reagents were used in our studies: mouse anti-ARF6 (3A-1), rabbit anti-IQGAP1 (H-109), mouse anti-Dock180 (H-4), goat anti-β-PIX (L-17) and goat anti-β-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-ERK1/2 and anti-p-ERK1/2 antibodies (Cell Signaling, Danvers, MA), a mouse anti-Rac1 antibody (BD Bioscience PharMingen, San Diego, CA), rabbit anti-ELMO1 antibody (27), an Alexa Fluor 488-conjugated rabbit anti-GFP antibody (Molecular Probes, Eugene, OR), and Rac1/Cdc42 Activation Assay kit (Upstate Technology, Lake Placid, NY). Cell culture media and other reagents were from Hyclone (Salt Lake City, UT), Invitrogen/BRL, (Grand Island, NY), Sigma Chemicals (St. Louis, MO) and Fisher Scientific (Hanover Park, IL).
Vectors, siRNAs and transfections
Human wild type ARF6 (WT), constitutively active ARF6 (Q67L) in pcDNA3.1 or pcDNA3.1/neo (7) were separately transfected into U87MG cells using an Effectene Reagent (Qiagen, Valencia, CA). G418 (900 μg/ml)-resistant cells were selected and expression of ARF6 in various cells was determined by immunoblotting (IB) with a mouse anti-ARF6 antibody. Small interfering RNAs (siRNAs) were synthesized by Dharmacon (Lafayette, CO). The target sequences were ARF6: 5′- GCACCGCAUUAUCAAUGACCG-3′ (18), Rac1: 5′-AAGGAGAUUGGUGCUGUAAAA-3′ (15), and IQGAP1: 5′-AAAGUUCUACGGGAAGUAA -3′ (28).
Transfection of glioma cells with the aforementioned siRNA was performed using Lipofectamine 2000 as previously described (16). Mock transfection was done in parallel using the Stealth RNAi Negative Control Med GC (Invitrogen). After 24 hr, the siRNA/lipid complexes were removed, and the cells were maintained in complete medium for an additional 48 hr. The inhibition of protein expression was determined by IB analyses.
An ARF6 siRNA was constructed into a pG-SUPER/GFP vector (from Dr. G. Borisy at Northwestern University, Chicago, IL) (29) to synthesize the 19-base pair double-stranded ARF6 target sequence as an ARF6 shRNA. Either pG-SUPER/ARF6-GFP or pG-SUPER/GFP was co-transfected with pcDNA3.1/neo vector at a ratio of 10 to 1 into SNB19 cells using an Effectene Reagent. G418 (700mg/ml)-resistant cell clones were reselected by GFP-expression via fluorescence activated cell sorting (FCAS) and knockdown of ARF6 in these cells was validated by IB analyses.
Immunoblotting (IB), co-immunoprecipitation (co-IP) and Rac1 activation assays
IB and Rac1 activation analyses were performed as previously described (16). Co-IP followed by IB analyses for association of ARF6 and IQGAP1 were performed as previously described (30) using an anti-ARF6 antibody or an isotype-matched IgG control for IP and indicated antibodies for IB analyses.
Immunofluorescent staining
After 48 hr of transfection with siRNA for ARF6, IQGAP1 or a control, the cells were grown in chamber slides in the serum-free medium for 48 hr, then treated with or without 20 ng/ml HGF for 30 min. The cells on slides were fixed with 4% PFA and permeablized with 0.2% Triton X-100. The cells were then dual-stained with mouse anti-ARF6 and rabbit anti-IQGAP1 antibodies followed by incubation with both Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa Fluor 596-conjugated goat anti-rabbit IgG antibodies (Molecular Probes). Cell nuclei were stained with Hoechst.
Wound healing, migration, and ex vivo brain slice invasion assays and quantification
As previously described (16), various cells were seeded on 6-well plates with DMEM containing 10% FBS and grown to confluence. The cells were scratched with a sterile 200 μl pipette tip to create artificial wounds. At 0 and 24 hr after wounding, phase-contrast images of the wound healing process were photographed digitally using an inverted Olympus IX50 microscope with a 10X objective. Eight images per treatment were analyzed to determine averaging position of the migrating cells at the wound edges by digitally drawing lines using the Image-Pro Plus software (Media Cybernetics, Bethesda, MD). The cell migration distance was determined by measuring the width of the wound divided by two and by subtracting this value from the initial half-width of the wound (31). In vitro cell migration and ex vivo brain slice invasion assays were performed as previously described (16). Lateral cell migration on brain slices was assessed by epifluorescent examination of GFP-expressing cells using a stereomicroscope (SZX12, Olympus) at 10X magnification. Haptotatic cell migration assay was performed in the presence or absence of serum or HGF in the bottom wells by using Boyden Chambers through an 8-μm pore size membrane. Statistical analyses using one-way ANOVA followed by Newman-Keuls posttest were performed as previously described (16). P<0.05 was considered statistically significant.
Intracranial brain tumor xenografts
SNB19/ARF6 siRNA #10 and #12 cells, and SNB19/control siRNA cells (1 × 106 cells in 5 μl PBS) were stereotactically implanted into the brain of an individual nude mouse with five mice per group. The glioma-bearing mice were sacrificed two weeks post-implantation. Their brains were removed, processed and analyzed as previously described (30). The GFP signal of various SBN19 tumors was enhanced by immunostaining with an Alexa Fluor 488-conjugated rabbit anti-GFP antibody.
Results
Cellular depletion of ARF6 expression by siRNA suppresses glioma cell motility
To determine whether ARF6 plays a role in glioma cell motility, we examined the expression of ARF6 in various human glioma cell lines. As shown in Supplementary Fig. 1 (see supplementary data), all glioma cell lines as well as normal human astrocytes (NHA) examined expressed endogenous ARF6 proteins, albeit to varying degrees. LN229, SNB19 and U373MG cells, which are invasive glioma cell lines and express relatively higher levels of ARF6, were chosen to evaluate the impact of silencing ARF6 on FBS- or HGF-stimulated Rac1 activity and cell motility using siRNA knockdown (9). After 48 hr, treatment of various cells with ARF6 siRNA effectively suppressed ARF6 expression to levels <10% of those cells treated with control siRNA (Fig. 1A) and without affecting cell viability (data not shown). Knockdown of ARF6 by siRNA significantly impaired Rac1 activity stimulated by 10% FBS or 20 ng/ml HGF (Fig. 1A, lower panels). To test whether suppression of ARF6 affects glioma cell motility, we performed in vitro migration assays and found that knocking down ARF6 markedly inhibited the ability of LN229, SNB19 and U373MG cells to migrate toward chemoattractants, 10% FBS or 20 ng/ml HGF (Fig. 1B).
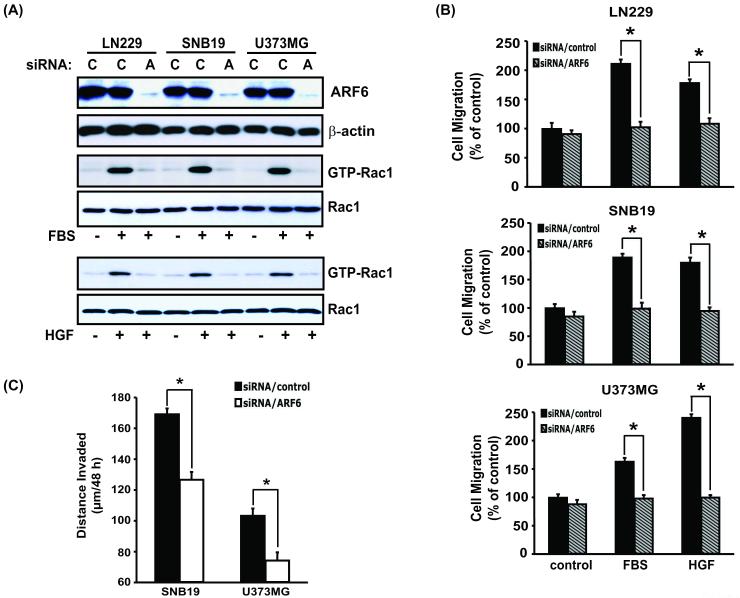
Figure 1. Suppression of ARF6 by siRNA inhibits glioma cell migration in vitro and cell invasion ex vivo.
(A), IB analyses of siRNA knockdown of ARF6 and the impact on Rac1 activation by FBS and HGF in indicated glioma cells. LN229, SNB19, and U373MG cells were transiently transfected with ARF6 siRNA or a control siRNA for 48 hr in media containing 10% FBS. Afterwards, the transfected cells were cultured in media containing no serum, 10% FBS or 20 ng/ml HGF for an additional 24 hr followed by IB with an anti-ARF6 antibody and analyses of GTP loading of Rac1 using a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. Data are representative of three independent experiments with similar results. (B), In vitro migration assays. LN229, SNB19, and U373MG cells were transiently transfected with ARF6 siRNA or control siRNA, followed by in vitro cell migration assays. Fifty μl of various siRNA-transfected cells in serum-free (SF) DMEM media containing 0.05% BSA was separately placed into the top compartment of a Boyden Chamber. Serum-free DMEM media containing 0.05% BSA with or without 20 ng/ml HGF or 10% FBS were placed in the bottom wells. The cells were allowed to migrate through an 8 μm-pore size membrane pre-coated with gelatin (10 μg/ml) for 16 hr at 37°C. Cell migration was determined as previously described (16). Control, media without serum or HGF. Columns, mean percentage of migrating control cells from three independent experiments in six replicates per pair per cell line (control siRNA-transfected and ARF6-siRNA-transfected cells); bars, SD. *, p < 0.05, one-way ANOVA followed by Newman-Keuls post hoc. (C), GFP-expressing SNB19 and U373MG cells were transiently transfected with ARF6 siRNA or control siRNA, followed by an ex vivo brain slice invasion assay. Depth of the GFP-expressing glioma cell invasion into a murine brain slice was determined by optical sectioning using a Zeiss LSM 510 confocal microscope. Columns, mean distance (μm) invaded in 48 hr from six independent experiments in six replicates per pair (control siRNA-transfected and ARF6-siRNA-transfected cells); bars, SD. *, p < 0.05, one-way ANOVA followed by Newman-Keuls post hoc.
To determine whether knockdown of ARF6 inhibits glioma cell invasion, we used a physiologically relevant murine brain ex vivo model (16, 31). GFP-expressing SNB19 and U373MG cells were transfected with either ARF6 siRNA or with control “scrambled” siRNA. After 48 hr, the cells were placed bilaterally onto the putamen of a 500-μm thick murine brain slice and allowed to invade into the brain tissue for an additional 48 hrs. As shown in Fig. 1C, analyses by confocal laser scanning microscopy revealed a significant inhibition of intrinsic invasiveness between control siRNA-treated and ARF6 siRNA-treated glioma cells. Taken together, these results indicate that ARF6 is required for glioma cell migration toward chemo-attractants in vitro and glioma cell invasion ex vivo.
Inhibition of ARF6 by siRNA suppresses glioma cell invasion in the brain
To directly assess the effect of knockdown of ARF6 on glioma cell invasion, we used a murine brain tumor in vivo model. We first generated a vector, pG-SUPER/ARF6/GFP that simultaneously expressed GFP and an ARF6 siRNA. This siRNA/GFP vector was co-transfected with a pcDNA3.0/(neo) vector into SNB19 cells. Previous studies demonstrated that orthotopic SNB19 glioma cells invade into normal brain parenchyma along the vasculature and white matter tracks in single cells and in cell clusters, reminiscent to the invasive phenotype of human primary glioma specimens (32). We selected several G418-resistant cell clones that stably express GFP and display effective suppression of ARF6 to similar extents as was seen with ARF6 siRNA (Fig. 1A). As shown in Fig. 2A, SNB19 ARF6 siRNA cell clone #4, #10, and #12 showed significant decrease in ARF6 expression and 10% FBS- or 20 ng/ml HGF-stimulated Rac1 activation than that with control siRNA (lower panels). The knockdown mediated by pG-SUPER/ARF6/GFP was maintained over more than three months without affecting cell viability under identical culture conditions (data not shown).
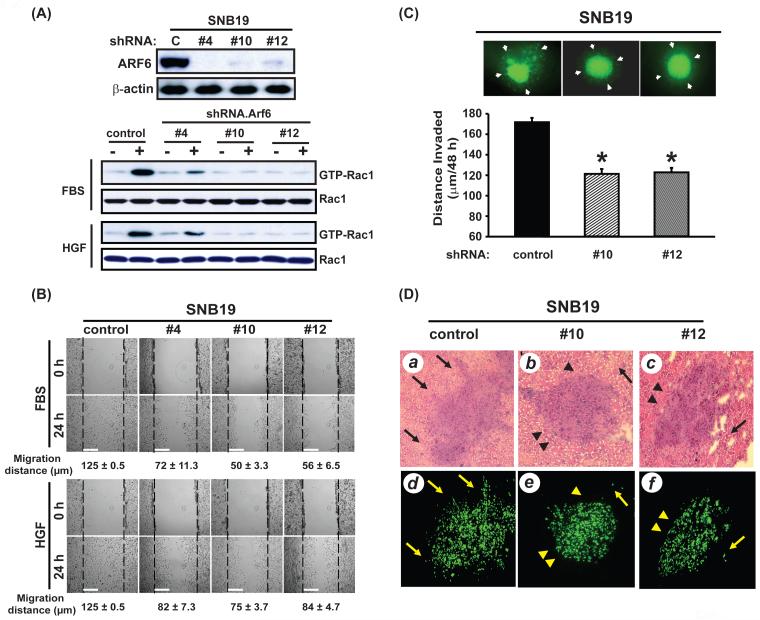
Figure 2. Suppression of ARF6 by siRNA inhibits glioma cell migration in vitro, and invasion ex vivo and in vivo.
(A), SNB19 cells were stably transfected with ARF6 siRNA or control siRNA and cultured in media containing no serum, 10% FBS or 20 ng/ml HGF for 24 hr. Inhibition of ARF6 proteins in the G418-resistent cell clones (control, ARF6-siRNA #4, #10, and #12) and the impact on GTP loading of Rac1 were analyzed by IB with an anti-ARF6 antibody and a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. (B), Cell motility of ARF6-siRNA or control-siRNA cell clones in the presence of 10% FBS or 20 ng/ml HGF was evaluated by an in vitro wound healing assay. Quantification of cell migration (averaging the position of the migrating cells at the wounding edges) in the monolayer wound healing assays are shown under the images as the means values (± SE in μm) of eight measurements at a 24-hr time point for each condition. Bar = 100 μm. (C), Cell invasion of ARF6-siRNA or control-siRNA cell clones was determined by an ex vivo brain slice invasion assay. Top panels, representative epifluorescent images of the GFP-expressing SNB19 ARF6-siRNA or control-siRNA cell clones were captured using a digital camera attached to a stereomicroscope at 40X magnification. White arrows indicate lateral cell migration and invasion 48 hr after initial placement of indicated cells onto the top of brain slices. Bar graph shows invasion depth of ARF6-siRNA or control-siRNA cell clones into a murine brain slice measured by confocal microscopy. Columns, mean distance (μm) invaded in 48 hr from four independent experiments done in six replicates per pair (ARF6-siRNA vs control-siRNA cell clones). Bars, SE. *, P < 0.05, one-way ANOVA followed by Newman-Keuls post hoc. No transfection controls (no siRNA) for SNB19 cells were also done showing no observable effects on cell viability or the invasive ability when comparing the control siRNA and non-transfected cells (data not shown). Data shown in (A) to (C) were from three independent experiments with triplicate (in vitro) and six brain tissue slices (ex vivo) with similar results. (D) Intracranial gliomas established by SNB19 control-siRNA cells (panels a and d), ARF6-siRNA #10 cells (b and e), and ARF6-siRNA #12 cells (c and f) in the murine brains were analyzed by H&E staining (a to c) and epifluorescent images (e to f). Arrows, invasive extensions as well as disseminated tumor clusters. Arrowheads, the clean edge of tumors. The animal experiments were done two independent times with six mice per group with similar results.
Next, we examined the impact of shRNA inhibition of ARF6 on cell motility of SNB19 cells using in vitro and in mouse brain ex vivo assays. As shown in Fig. 2B, in wound healing assays, cell motility of ARF6 siRNA cell clone #4, #10, and #12 was significantly inhibited compared with control siRNA cells. Twenty-four hr after wounding, average distances of the migrating ARF6 siRNA cell clones at the wound edges (mean ± SE) were approximately 72 ± 11.3, 50 ± 3.3 and 56 ± 6.5 μm in 10% FBS; 82 ± 7.3, 75 ± 3.7 and 84 ± 4.7 μm in 20 ng/ml HGF compared to control siRNA cells of 125 ± 0.5 μm in both conditions (the wounds were closed). Similar inhibition of cell migration by ARF6 siRNA was also observed when cell clones were analyzed in Boyden Chamber migration assays (data not shown). Next, we determined whether ARF6 depletion in SNB19 cells abrogated glioma cell lateral migration/invasion and downward invasion into the brain tissues using the ex vivo murine brain tissue slice assay. As shown in Fig. 2C, inhibition of ARF6 by siRNA in SNB19 cells displayed less lateral migration/invasion on the brain tissue slices compared with control siRNA cells (Fig. 2C, white arrows in images at the top panel with green fluorescence). Analyses by confocal laser scanning microscopy showed that glioma cell invasiveness into the brain tissues was significantly attenuated in ARF6 siRNA cell clones (mean ± SE, #10: 121.6 ± 4.5 mm/48h; #12 123.1 ± 4.1 mm/48h) when compared with control siRNA cells (mean ± SE, 172.1 ± 3.9 mm/48h).
Finally, an orthotopic brain glioma model was employed to determine whether inhibition of ARF6 expression by siRNA could suppress glioma cell infiltration into the brain parenchyma of animals. As shown in Fig. 2D, mice that received ARF6 siRNA cell clones developed much less invasive gliomas (Fig. 2D, H&E, panels b and c; GFP: panels e and f) compared with the brains that received control siRNA glioma cells (Fig. 2D, H&E, panel a; GFP, panel d). Taken together, these results suggest that ARF6 is required in the process of glioma cell invasion in the brain.
IQGAP1 is involved in the ARF6-mediated Rac1 activation and glioma cell migration upon stimulation
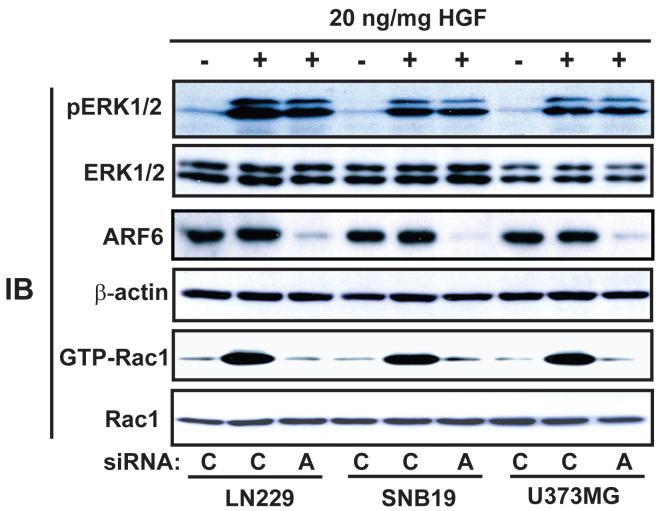
Having demonstrated that the critical role of ARF6 in glioma cell invasion in vitro, ex vivo and in animal brains, we investigated the mechanism by which ARF6 regulates glioma cell migration. Previous studies of ARF6 in melanoma cells, breast cancer cells and Madin-Darby canine kidney (MDCK) cells showed that HGF and EGF induce protein phosphorylation of ERK1/2 through activation of ARF6, leading to an increase in cell migration (7, 33). In glioma cells, EFA6A-enhanced cell motility and invasiveness also require ERK1/2 activation (10). Thus, we examined whether ARF6 is required for HGF stimulation of ERK1/2 in glioma cells. As shown in Fig. 3, 20 ng/ml of HGF strongly stimulated ERK1/2 protein phosphorylation and Rac1 activation in LN229, SNB39 and U373MG glioma cells. Moreover, inhibition of ARF6 protein by siRNA knockdown markedly impaired HGF-stimulated Rac1 activation but had no impact on HGF-stimulated protein phosphorylation of ERK1/2 in these three glioma cell lines. It should be noted that in the experiments shown in Fig. 1 and Fig. 3, same batches of glioma cells were transiently transfected with control and ARF6 siRNA and subjected to analyses of WB for knockdown of ARF6 (Fig. 1A, Fig. 3), in vitro cell migration (Fig. 1B), ex vivo cell invasion (Fig. 1C) and phosphorylation of ERK1/2 (Fig. 3). Together, these data suggest that activation of ERK1/2 by HGF may not play a prominent role in ARF6-mediated glioma cell migration.
Figure 3. ARF6 is not required for HGF-stimulated protein phosphorylation of ERK1/2 in glioma cells.
Human glioma LN229, SNB19 and U373MG cell lines were transfected with siRNA for ARF6 and control then treated with 20 ng/ml of HGF for 20 min. Inhibition of ARF6 proteins in transfected cells and the impacts on ERK1/2 protein phosphorylation and GTP loading of Rac1 were analyzed with IB using anti-phospho-ERK1/2, anti-ERK1/2 and anti-ARF6 antibodies, and a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. Data are representative of three independent experiments with similar results.
Recent studies showed that Rac1 is a downstream target of ARF6 and several regulatory proteins are likely to be involved in mediating signals between ARF6 and Rac1 (33). Using co-immunoprecipitation (co-IP) methods, we investigated whether ARF6 and Rac1 form a complex upon HGF stimulation, and whether other Rac1-GEFs such as Dock180/ELMO1 (17) and β-PIX (34) or other regulatory/scaffold proteins such as IQGAP1 (20) could be recruited along with ARF6 upon HGF stimulation. As shown in Fig. 4A, ARF6, Rac1, ELMO1 and Dock180 were expressed at high levels whereas β-PIX was found at low levels in SNB19 cells, as well as several other glioma cell lines examined (data not shown). These cells were treated with 20 ng/ml of HGF and total cell lysates were immunoprecipitated with an anti-ARF6 antibody or an isotype-matched IgG control. We found that association of ARF6 with Rac1 became evident in 5 min upon HGF stimulation and further increased with more prolonged HGF treatment. Residual association of ARF6 with IQGAP1 was also observed without stimulation, but the association was induced rapidly upon a 5-min HGF stimulation. In contrast, we could not detect ELMO1, Dock180 or β-PIX proteins in the complex with ARF6 and Rac1 (left panels). These co-immunoprecipitated protein complexes were specific since co-IPs using an isotype-matched IgG control did not result in any pull down of indicated proteins under identical conditions (right panels).
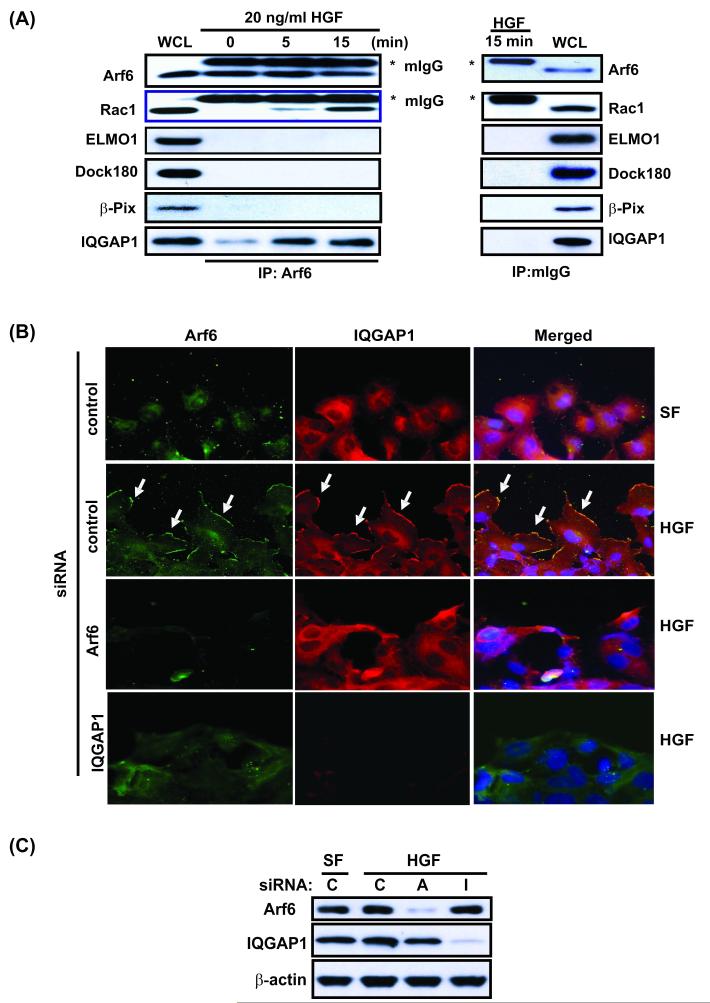
Figure 4. ARF6 is associated with IQGAP1 in glioma cells and controls formation of extended ruffles on plasma membrane.
(A), ARF6 is associated with IQGAP1 in glioma cells. The serum-starved SNB19 cells were treated with 20 ng/ml of HGF for indicated times and lysed. The cell lysates were immunoprecipitated (IP) with an anti-ARF6 antibody (left panels) or an isotype-matched mouse IgG control (right panels) followed by IB separately using anti-ARF6, anti-Rac1, anti-Dock180, anti-ELMO1, anti-β-PIX and anti-IQGAP1 antibodies. IB for whole cell lysates (WCL) was used as a protein input control. (B), SNB19 cells were transiently transfected with ARF6 siRNA, IQGAP1 siRNA or a control siRNA, and then under serum starvation for 48 hr followed by HGF treatment (20 ng/ml) for 30 min. Co-localization of ARF6 and IQGAP1 at the invasion fronts of cells was determined by immunofluorescent staining of various cells using anti-ARF6 and anti-IQGAP1 antibodies. Nuclei were stained by Hoechst (blue). Images were captured using an Olympus BX51 microscope equipped with a digital camera (magnification: 400X). Arrows in the second panels indicate co-localization of ARF6 and IQGAP1 at the invasion fronts. Percentage of cells showing the co-localization was estimated by a blind observer (J.-J. Yiin) in 10 random fields at 10X magnification and is described in the text. (C), IB analyses of siRNA knockdown of ARF6 and IQGAP1 in SNB19 cells. Whole cell lysates of a portion of SNB19 cells that were immunostained in (B) were analyzed by IB for inhibition of ARF6 and IQGAP1 expression by ARF6 siRNA, IQGAP1 siRNA or a control siRNA using anti-ARF6 and anti-IQGAP1 antibodies. IB for β-actin was used as a protein loading control. Data shown in (A) to (C) were from three independent experiments with similar results.
To further examine the connection between ARF6 and IQGAP1, we determined whether ARF6 and IQGAP1 are co-localized in the invasion fronts of HGF-stimulated SNB19 cells by immunofluorescent staining after ARF6 and IQGAP1 were knocked down. As shown in Fig. 4B, in serum-starved control siRNA-treated SNB19 cells, most of ARF6 (green) and IQGAP1 (red) were localized in the cytosol. HGF treatment induced membrane ruffling and formation of cellular protrusions, and triggered redistribution of both ARF6 and IQGAP1 from the cytoplasm to membrane protrusions. Co-localization studies revealed that ARF6 is localized to the leading edge with IQGAP1 (Fig. 4B, arrows in middle panels). Moreover, effective knockdown of ARF6 or IQGAP1 by siRNAs (Fig. 4C) reduced formation of extended ruffles on the plasma membrane and the majority of IQGAP1 or ARF6 stayed in the cytoplasm (Fig. 4B, images in lower two rows). When co-localization of ARF6 and IQGAP1 was quantified , we found that without serum and HGF, approximately 8.7 ± 3.5% (mean ± SE in 10 random fields) control cells showed co-localization of these two proteins, whereas with 20 ng/ml HGF stimulation, 74 ± 3.7% control cells exhibited co-staining of ARF6 and IQGAP1 at lamellipodia. When ARF6 or IQGAP1 was knocked down, only 9.2 ± 3.7% and 7.8 ± 4.6% cells showed overlapping ARF6 and IQGAP labeling. Together, these data suggest that upon HGF stimulation, ARF6 translocation to the membrane triggers redistribution of IQGAP1 from the cytoplasm to the leading edges of membrane ruffling.
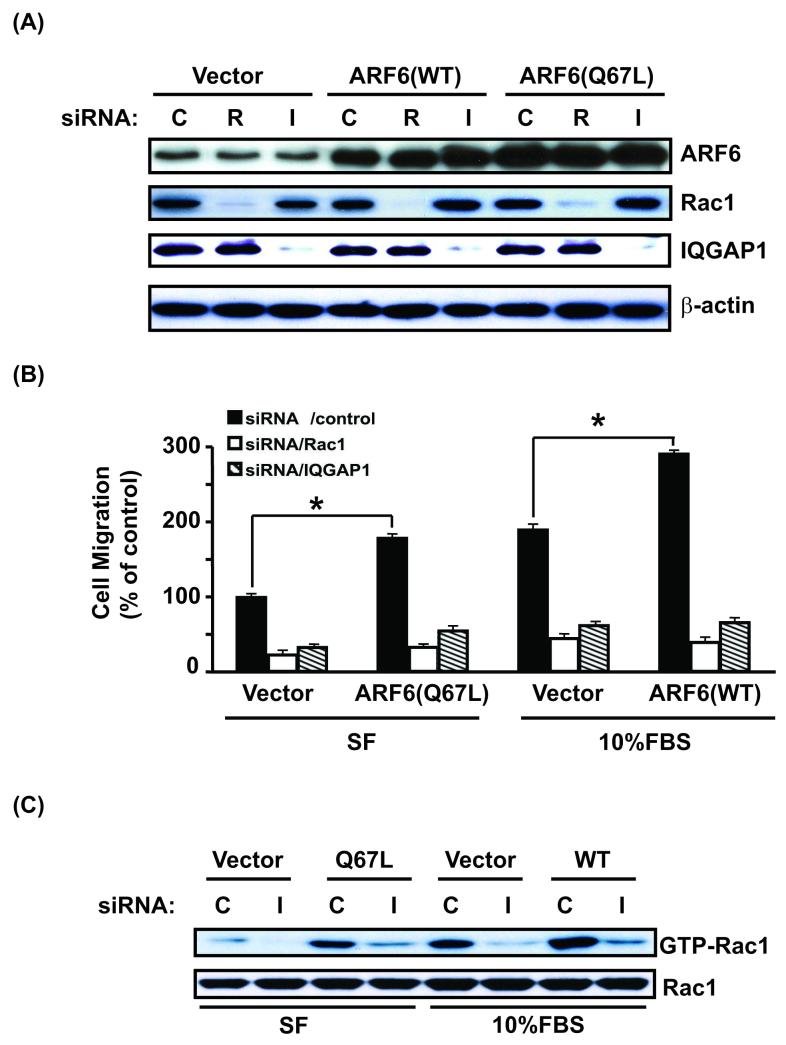
To determine whether IQGAP1 is a downstream regulator necessary for ARF6 signaling in glioma cell migration, we ectopically expressed wild type ARF6 (ARF6-WT) and a constitutively active, GTPase-deficient mutant of ARF6 (ARF6-Q67L) in U87MG glioma cells that showed low level of ARF6 expression (Fig. S1). G418 drug-resistant cell populations were selected among ARF6-WT-expressing, ARF6-Q67L-expressing, and vector-expressing U87MG cell groups. Various cell groups were then transiently transfected with siRNAs of control, Rac1, and IQGAP, respectively. Exogenous expression of ARF6 or knockdown of Rac1 and IQGAP1 did not significantly affect the expression of other proteins except the targeted molecules (Fig. 5A). We then examined whether knockdown of IQGAP1 blocks the effect of exogenously expressed ARF6-WT and ARF-Q67L, to result in decreased glioma cell migration. As shown in Fig. 5B, in the absence of chemo-attractants (serum-free, SF) in the lower wells of a Boyden Chambers, expression of ARF6-Q67L in U87MG cells results in an increase in cell motility by approximately two-fold compared with the vector-transfected cells (left panel). However, ectopic expression of ARF6-WT did not enhance cell migration under the identical conditions (data not shown). When cells were exposed to 10% FBS in the lower chambers, expression of ARF6-WT in U87MG cells promoted cell migration by 1.5-fold compared to vector-transfected cells (right panel). Furthermore, in the presence or absence 10% FBS in the lower compartments, knockdown of IQGAP1 or Rac1 by siRNA significantly suppressed the motility of U87MG cells enhanced by the ARF6-Q67L expression alone, (left panel) or ARF6-WT expression together with exposure to 10% FBS, (right panel). These data suggest that IQGAP1 plays a critical role downstream of ARF6-mediated glioma cell migration. Lastly, we tested whether knockdown of IQGAP1 affects activation of Rac1 by ARF6 signaling. As shown in Fig. 5C, when the same batches of U87MG glioma cell clones expressing vector, ARF6-WT or ARF6-Q67L that displayed reduced IQGAP expression by IQGAP siRNA knockdown but not control siRNA (Fig. 5A, the third panel) were cultured in media with or without 10% FBS, ectopic expression of ARF6-Q67L or ARF6-WT increased the GTP-bound form of Rac1 compared to vector-transfected U87MG control cells in the absence or presence of 10%FBS, respectively. Depletion of IQGAP1 by IQGAP1 siRNA but not control siRNA significantly decreased GTP-bound forms of Rac1 to minimal levels (Fig. 5C). Taken together, these data suggest a previously unknown connection between ARF6 and IQGAP1 to Rac1 activation.
Figure 5. IQGAP1 is necessary for the signal connection between ARF6 and Rac1, and ARF6-mediated glioma cell migration.
(A), IB analyses of siRNA knockdown of Rac1 or IQGAP1 in various U87MG glioma cells. U87MG cells with low levels of endogenous ARF6 were transiently transfected with ARF6-WT, ARF6-Q67L, or vector only. G418-resistant U87MG, U87MG/ARF6-WT, and U87MG/ARF6-Q67L cell populations were then transiently transfected with control siRNA, Rac1 siRNA, and IQGAP1 siRNA followed by IB with anti-ARF6, anti-Rac1, and anti-IQGAP1 antibodies. IB for β-actin was used as a protein loading control. (B), Impact of knocking down IQGAP1 or Rac1 on cell motility of various indicated cells was determined by in vitro migration assays under conditions of serum-free (SF) and 10% FBS. (C), Effect of inhibition of IQGAP1 on activation of Rac1 in various indicated cells in the absence or presence of 10% FBS was examined using a Rac1 activation assay kit under conditions of serum-free (SF) and 10% FBS. Whole cell lysates were prepared from a portion of various IQGAP1 siRNA-transfected U87MG cells shown in (A) that were cultured in media with or without 10% FBS for an additional 24 hrs after siRNA transient transfection. Expression of endogenous IQGAP1 proteins and the inhibition of IQGAP1 expression by siRNA knockdown in various U87MG cells cultured in media containing 10% FBS are shown in the third panel of IB analysis in (A). IB for Rac1 was used as protein loading controls. Data shown in (A) to (C) are representative from three independent experiments with similar results.
Discussion
In this study, we show that ARF6 mediates glioma cell migration upon HGF and FBS stimulation in vitro and is critical for glioma cell invasion in the brain of animals. We demonstrate that ARF6 is expressed in various human glioma cell lines examined. When ARF6 is suppressed by siRNA in invasive LN229, U373MG and SNB19 glioma cells, HGF- and FBS-stimulated Rac1 activation and cell motility was substantially attenuated in vitro. When ARF6 was knocked down by siRNA in invasive SNB19 gliomas ex vivo and in the brain, glioma cell invasion into brain parenchyma was also significantly inhibited. Our mechanistic studies reveal that ARF6 mediates HGF stimulation of glioma cell motility through Rac1 and IQGAP1. Inhibition of Rac1 and IQGAP1 by siRNA abrogates HGF-stimulated cell migration while knockdown of IQGAP1 by siRNA suppressed FBS-stimulated cell migration and Rac1 activation. Our data provide convincing evidence that ARF6 regulates HGF- and FBS-stimulated glioma cell migration through formation of a complex with IQGAP1 and Rac1 in vitro and is critical for invasive behaviors of malignant glioma cells in the brain.
The role of ARF6 in mediating cell motility has been studied in several experimental model systems in vitro. Ectopic expression of ARF6-WT enhanced in MDA-MB-231 breast cancer cell migration and GTPase-deficient mutant ARF6-Q67L inhibited cell migration (8). In LOX melanoma cells and MDCK cells, expression of ARF6-Q67L sustained HGF-stimulated cell migration while a dominant negative ARF6-T27N inhibition HGF stimulation (7, 33). In U373MG glioma cells, activation of ARF6 by one of its own GEF, EFA6A also stimulated cell migration (10). In the later three studies, it was shown that ARF6 mediates HGF stimulation of cell migration through activation of ERK1/2 and Rac1 activities. Additionally, co-expression of another GEF of ARF6, GEP100 in MCF-7 cells evoke EGF-dependent cell invasion in vitro while inhibition of GEF100 blocked lung metastasis of mouse 4T1 mammary tumor cells in mice (9). Our results corroborated with these studies. We show that ARF6 is critical for HGF-stimulated glioma cell migration in vitro, cell invasion ex vivo and in the brain of animals. However, as shown in Fig. 3, HGF stimulation of 4 glioma cell lines, LN229, U373MG, SNB19 and D54MG (not shown), induced significant protein phosphorylation of ERK1/2. Moreover, effective knockdown of ARF6 in these glioma cells impaired HGF-stimulated Rac1 activity but did not affect HGF-stimulated phosphorylation of ERK1/2. Additionally, in U87MG/ARF6-Q67L cells, expression of constitutively active ARF6-Q67L mutant had minimal impact on phosphorylation of ERK1/2, Akt and JNK (data not shown). Although we cannot rule out the existence of compensatory pathways that lead to ERK activation upon depletion of ARF6 in glioma cells, our data suggest that ARF6-mediated HGF or FBS stimulation of cell migration differs from that in breast cancer cells, melanoma cells and MDCK cells (7, 33). The discrepancy of our results in the involvement of ERK1/2 activation in HGF-stimulated cell migration with the aforementioned studies probably reflects a cell-type difference. Moreover, in addition to Rac1 activation, ERK1/2 activation could also be required for HGF-stimulated glioma cell migration. A study of the impact of inhibition of ERK1/2 activation by an MEK inhibition U0126 on HGF-stimulated cell motility is warranted to test this hypothesis. Nonetheless, our results provide the first in vivo evidence that inhibition of ARF6 by siRNA impaired glioma cell infiltration into brain parenchyma in animals, underscoring the critical role of ARF6 in modulation of glioma invasion in the brain.
Activation of Rac1 has been linked with ARF6-mediated cell migration in response to various stimuli (4). In LOX cells, MDCK cells and U373MG cells, stimulation of ARF6 activates Rac1 through protein phosphorylation of ERK1/2 (7, 10, 33). Alternatively, in CHO and MDCK cells, activation of ARF6 stimulates Rac1 through interaction with Rac1 coupled with the Dock180/ELMO1 complex (13, 17), perhaps also through binding to β-PIX (18). β-PIX was shown to be upstream of ARF6 and modulate ARF6 activity through a G protein-coupled receptor kinase-interacting protein 1 (GIT1) (34). Our data show that Rac1 could be effectively co-precipitated with ARF6 upon HGF stimulation in a time-dependent manner. However, we failed to detect Dock180, ELMO1 and β-PIX in the complex of ARF6/Rac1 in our glioma cell models (Fig. 4A). Additionally, knockdown of DOCK180/ELMO1 complex by siRNA (16) only partially blocked the ARF6-mediated activation of Rac1 and cell migration in LN229, U373MG and SNB19 cells (data not shown). Thus mechanisms that result in Rac1 activation downstream of ARF6 likely vary depending on cell types.
IQGAP1 is an effector of Rac1 and Cdc42 that regulates cell adhesion and migration by modulating actin filaments and microtubule polarization. In response to EGF and VEGF, IQGAP1 forms complexes with various proteins including Rac1 enhancing cell migration (20, 22, 35). In NIH3T3 cells, ARF6 GTPase activity is required to regulate IQGAP1-dependent recruitment of a protein complex at membrane ruffles, suggesting coordinated modulation of cell motility by ARF6 and IQGAP1 at a close proximity near membrane ruffles (19). We show that upon HGF stimulation IQGAP1 was rapidly recruited into a complex of ARF6 and Rac1 in glioma cells. IQGAP1 is also co-localized with ARF6 at HGF-induced membrane ruffles and cell protrusions. When ARF6 or IQGAP1 was knocked down by siRNA, recruitment of IQGAP1 into the leading edges was diminished. Furthermore, inhibition of IQGAP1 or Rac1 by siRNA in U87MG cells with ectopically expressed ARF6-WT or ARF6-Q67L attenuated ARF6-mediated HGF stimulation of cell migration and serum-stimulated Rac1 activities. Thus, these results establish for the first time a link between ARF6 and Rac1 by IQGAP1 and demonstrate an association of ARF6, Rac1 and IQGAP1 in a complex within membrane ruffles. Moreover, in breast and ovarian cancer cells, IQGAP1 directly binds to ERK2 and induces ERK2 phosphorylation. Knockdown of IQGAP1 reduced CD44-, IGF- and EGF-stimulated ERK1/2 phosphorylation inhibiting cell migration (28, 36). In our study, we found that inhibition of ARF6 did not affect HGF-stimulated ERK1/2 phosphorylation, thus suggesting that ERK1/2 activation might not have a prominent role in ARF6-mediated stimulation of IQGAP1 and Rac1. Whether IQGAP1 still interacts with phosphorylated ERK1/2 contributing HGF-stimulated cell migration warrants further investigation.
In summary, the key findings of this study are that ARF6 mediates HGF- and serum-stimulated glioma cell migration through IQGAP1 and Rac1. Inhibition of ARF6 significantly inhibited glioma cell infiltration ex vivo and in the brain. Our results demonstrate for the first time a critical role of ARF6 in regulating the invasive phenotype of glioma cells in vivo and place IQGAP1 between ARF6 and Rac1 in HGF-stimulated signaling in glioma cell invasion. Further investigation of ARF6-IQGAP1-Rac1-mediated glioma cell migration could establish molecules involved in this pathway as potential therapeutic targets for treatment of patients inflicted with malignant gliomas.
Supplementary Material
Acknowledgement
We would like to thank D. Bigner for D54MG and A172 cells and Y-H. Zhou for SNB19 cells, G. Borisy for a pG-SUPER/GFP vector, K. Ravichandran for the anti-ELMO1 antibody, G. Wang for assistance using the laser confocal microscope, Y. Wan for using an inverted microscope and N. Balass for proofreading of this manuscript. This work was supported by grants from the National Institutes of Health (CA102011, CA130966), American Cancer Society (RSG CSM-107144) to S.-Y. Cheng, the National Institutes of Health (CA115316) to C. D-S, and the Hillman Fellows Program to S.-Y. Cheng and B. Hu.
References
- 1.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Nakada M, Nakada S, Demuth T, et al. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–78. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giese A, Bjerkvig R, Berens ME, et al. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–36. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 4.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–6. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- 6.Sabe H. Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. J Biochem. 2003;134:485–9. doi: 10.1093/jb/mvg181. [DOI] [PubMed] [Google Scholar]
- 7.Tague SE, Muralidharan V, D’Souza-Schorey C. ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proc Natl Acad Sci USA. 2004;101:9671–6. doi: 10.1073/pnas.0403531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto S, Onodera Y, Hashimoto A, et al. Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci USA. 2004;101:6647–52. doi: 10.1073/pnas.0401753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishige M, Hashimoto S, Ogawa E, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat Cell Biol. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Ng SS, Wang J, et al. EFA6A enhances glioma cell invasion through ADP ribosylation factor 6/extracellular signal-regulated kinase signaling. Cancer Res. 2006;66:1583–90. doi: 10.1158/0008-5472.CAN-05-2424. [DOI] [PubMed] [Google Scholar]
- 11.Palacios F, D’Souza-Schorey C. Modulation of Rac1 and ARF6 activation during epithelial cell scattering. J Biol Chem. 2003;278:17395–400. doi: 10.1074/jbc.M300998200. [DOI] [PubMed] [Google Scholar]
- 12.Nishiya N, Kiosses WB, Han J, et al. An alpha4 integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat Cell Biol. 2005;7:343–52. doi: 10.1038/ncb1234. [DOI] [PubMed] [Google Scholar]
- 13.Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 15.Chan AY, Coniglio SJ, Chuang YY, et al. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene. 2005;24:7821–9. doi: 10.1038/sj.onc.1208909. [DOI] [PubMed] [Google Scholar]
- 16.Jarzynka MJ, Hu B, Hui KM, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–11. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15:1749–54. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 18.Cotton M, Boulay PL, Houndolo T, et al. Endogenous ARF6 interacts with Rac1 upon angiotensin II stimulation to regulate membrane ruffling and cell migration. Mol Biol Cell. 2007;18:501–11. doi: 10.1091/mbc.E06-06-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma M, Henderson BR. IQ-domain GTPase-activating protein 1 regulates beta-catenin at membrane ruffles and its role in macropinocytosis of N-cadherin and adenomatous polyposis coli. J Biol Chem. 2007;282:8545–56. doi: 10.1074/jbc.M610272200. [DOI] [PubMed] [Google Scholar]
- 20.Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16:242–9. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Mateer SC, Wang N, Bloom GS. IQGAPs: integrators of the cytoskeleton, cell adhesion machinery, and signaling networks. Cell Motil Cytoskeleton. 2003;55:147–55. doi: 10.1002/cm.10118. [DOI] [PubMed] [Google Scholar]
- 22.Noritake J, Watanabe T, Sato K, et al. IQGAP1: a key regulator of adhesion and migration. J Cell Sci. 2005;118:2085–92. doi: 10.1242/jcs.02379. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda S, Fukata M, Kobayashi K, et al. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–7. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 24.Mataraza JM, Briggs MW, Li Z, et al. IQGAP1 promotes cell motility and invasion. J Biol Chem. 2003;278:41237–45. doi: 10.1074/jbc.M304838200. [DOI] [PubMed] [Google Scholar]
- 25.Nabeshima K, Shimao Y, Inoue T, et al. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002;176:101–9. doi: 10.1016/s0304-3835(01)00742-x. [DOI] [PubMed] [Google Scholar]
- 26.Furnari FB, Lin H, Huang HS, et al. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci USA. 1997;94:12479–84. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brugnera E, Haney L, Grimsley C, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–82. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 28.Bourguignon LY, Gilad E, Rothman K, et al. Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J Biol Chem. 2005;280:11961–72. doi: 10.1074/jbc.M411985200. [DOI] [PubMed] [Google Scholar]
- 29.Kojima S, Vignjevic D, Borisy GG. Improved silencing vector co-expressing GFP and small hairpin RNA. Biotechniques. 2004;36:74–9. doi: 10.2144/04361ST02. [DOI] [PubMed] [Google Scholar]
- 30.Hu B, Jarzynka MJ, Guo P, et al. Angiopoietin 2 induces glioma cell invasion by stimulating matrix metalloprotease 2 expression through the alphavbeta1 integrin and focal adhesion kinase signaling pathway. Cancer Res. 2006;66:775–83. doi: 10.1158/0008-5472.CAN-05-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valster A, Tran NL, Nakada M, et al. Cell migration and invasion assays. Methods. 2005;37:208–15. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Kondraganti S, Mohanam S, Chintala SK, et al. Selective suppression of matrix metalloproteinase-9 in human glioblastoma cells by antisense gene transfer impairs glioblastoma cell invasion. Cancer Res. 2000;60:6851–5. [PubMed] [Google Scholar]
- 33.Tushir JS, D’Souza-Schorey C. ARF6-dependent activation of ERK and Rac1 modulates epithelial tubule development. Embo J. 2007;26:1806–19. doi: 10.1038/sj.emboj.7601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahuna O, Quellari M, Achard C, et al. Thyrotropin receptor trafficking relies on the hScrib-betaPIX-GIT1-ARF6 pathway. Embo J. 2005;24:1364–74. doi: 10.1038/sj.emboj.7600616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species--dependent endothelial migration and proliferation. Circ Res. 2004;95:276–83. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
- 36.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem. 2004;279:17329–37. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.