Abstract
Tropomyosins are coiled-coil dimers that bind to the major groove of F-actin and regulate its accessibility to actin-modifying proteins. Although approximately 40 tropomyosin isoforms have been identified in mammals, they can broadly be classified into two groups based on protein size, that is, high molecular weight and low molecular weight isoforms. Osteoclasts, which undergo rounds of polarization and depolarization as they progress through the resorptive cycle, possess an unusual and highly dynamic actin cytoskeleton. To further define some of the actin regulatory proteins involved in osteoclast activity, we previously performed a survey of tropomyosin isoforms in resting and resorbing osteoclasts. Osteoclasts were found to express two closely related tropomyosins of the high molecular weight type, which are not expressed in monocytic and macrophage precursors. These isoforms, Tm-2 and Tm-3, are not strongly associated with actin-rich adhesion structures, but are instead distributed diffusely throughout the cell. In this study, we found that Tm-2/3 expression occurs late in osteoclastogenesis and continues to increase as cells mature. Knockdown of these isoforms via RNA interference results in flattening and increased spreading of osteoclasts, accompanied by diminished motility and altered resorptive capacity. In contrast, overexpression of Tm-2, but not Tm-3, caused morphological changes that include decreased spreading of the cells and induction of actin patches or stress-fiber like actin filaments, also with effects on motility and resorption. Suppression of Tm-2/3 or overexpression of Tm-2 resulted in altered distribution of gelsolin and microfilament barbed ends. These data suggest that high molecular weight tropomyosins are expressed in fusing osteoclasts to regulate the cytoskeletal scaffolding of these large cells, due at least in part by moderating accessibility of gelsolin to these microfilaments.
Keywords: Osteoclasts, actin, tropomyosin, cytoskeleton, cell shape
INTRODUCTION
Osteoclasts are formed from fusion of monocyte lineage precursors upon stimulation with macrophage colony stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL). Bone degradation by osteoclasts requires these cells to tightly attach to their substrate and generate the specialized apical ruffled membrane through which protons and proteases, particularly cathepsin K, are secreted. To perform their primary function of bone resorption, osteoclasts must possess a dynamic cytoskeletal organization that permits cell fusion, rapid motility, and cyclical polarization/depolarization. Further, osteoclasts form distinctive actin-based adhesion structures in the form of individual podosomes (when on non-bone substrates) and the sealing zone (when on bone). Therefore, examining the regulatory proteins involved in organization of the osteoclast actin cytoskeleton will aid in our understanding of these remarkable cells’ activity. One class of actin-binding proteins that so far has been poorly studied in osteoclasts is the tropomyosins. Tropomyosins (Tms) are coiled-coil dimers that bind along the length of actin filaments and regulate access of other actin-binding proteins to the microfilament [14]. Cytoskeletal Tms stabilize actin filaments, in part by preventing access of actin severing and depolymerizing proteins such as gelsolin and ADF/cofilin [2, 20, 33]. Tms may work cooperatively with tropomodulin to cap pointed ends of actin filaments and block elongation and depolymerization [42]. Like muscle Tms, cytoskeletal Tms also appear to play a role in actin translocation by regulating recruitment and activity of specific myosins [4]. Mammals express approximately forty isoforms of Tms, which are produced by alternate promoters and exon usage in four genes, termed α, β, γ, and δ [15]. Individual nonmuscle cells express a complement of up to eight or more isoforms [26], and it has become clear in the last decade that individual isoforms segregate to specific actin pools and perform distinct cellular functions [4, 11, 16, 35, 36].
We recently performed a survey of tropomyosins in osteoclasts, and showed that these cells express at least eight isoforms with distinctive intracellular distribution [29]. Individual Tms segregated to various regions within the cell, including podosomes and the sealing zone. For example, we showed that one isoform, Tm-4, is strongly localized to the core of podosomes and the inner face of the actin ring of the sealing zone. More recently, we demonstrated by RNA interference and overexpression studies that Tm-4 is required for proper formation of the podosome core and actin ring, with consequences for cellular motility and bone resorption [30].
Tropomyosins can be segregated into two distinct classes based on their primary sequence. High molecular weight (HMW) tropomyosins are composed of 284 amino acid residues, while the low molecular weight (LMW) forms contain 248 residues. In nonmuscle cells, HMW tropomyosins include Tm-1, a product of the beta tropomyosin gene, and Tm-2 and -3, which are products of the alpha gene. HMW tropomyosins are associated with stress fibers in fibroblast-like cell types. Numerous studies have demonstrated that viral transformation results in suppression of these tropomyosins and disruption of stress fibers [3, 10, 17, 27, 40]. Further, exogenous re-expression of these HMW Tms in virally transformed cells results in reorganization of stress fibers and suppression of neoplastic growth, although the efficacy of each Tm in doing so appears to differ among various cell types studied [3, 12]. Suppression of HMW tropomyosins in transformed cells is believed to be the cause of altered motility and invasive potential. Macrophages, which do not contain extensive stress fibers, were shown to lack expression of the HMW isoforms [32]. Therefore, we were somewhat surprised to find that RAW264.7-derived osteoclasts, which also lack stress fibers, clearly expressed the HMW isoforms Tm-2 and Tm-3 [29]. These isoforms did not segregate with the most clearly defined actin structures in osteoclasts, podosomes and the sealing zone of polarized cells, but were instead distributed rather homogenously throughout the interior of the cell. Given this distribution, we hypothesized that these HMW isoforms may provide structural support for the osteoclast cytoskeleton as they mature from single-nucleus precursors to very large cells containing up to 10 or more nuclei. In this study, we have used loss-of-function and gain-of-function approaches to determine the role of HMW Tms in osteoclasts, and provide evidence that supports this hypothesis.
MATERIALS AND METHODS
Osteoclast Culture
Murine osteoclasts were generated in culture from mouse bone marrow or from the mouse macrophage cell line RAW 264.7 (The American Type Culture Collection, Manassas, VA). To produce bone marrow-derived cells, 4–8 week-old Swiss-Webster mice (Harlan, Indianapolis, IN) were used. Preparation of osteoclasts included incubation of precursor cells overnight in αMEM containing 10% heat inactivated fetal bovine serum and 20 ng/ml M-CSF (R&D Systems, Minneapolis, MN). Non-adherent cells were collected the next day and incubated for an additional 5–7 days in αMEM containing 10% heat inactivated fetal bovine serum, penicillin/streptomycin, 20 ng/ml M-CSF, and 75 ng/ml GST-RANKL, prepared as previously described [24]. The culture medium was replaced every 2–3 days. For preparation of RAW 264.7-derived osteoclasts, macrophage precursors were plated at a density of 20,000 cells/cm2 and cultured in Dulbecco’s Modified Eagle’s medium (DMEM) containing 10% heat inactivated fetal bovine serum, penicillin/streptomycin, and 75 ng/ml GST-RANKL, replacing the culture medium every 2–3 days.
Antibodies and Western analysis
A mouse monoclonal, anti-tropomyosin (TM311) antibody from Sigma-Aldrich (St. Louis, MO) was used to detect Tm-2/3. Mouse monoclonal antibodies against β-actin and gelsolin were from Abcam (Cambridge, MA) and BD Biosciences (San Jose, CA), respectively. Cells were harvested at about 80% confluency for protein analysis and protein was extracted using M-PER Mammalian Protein Extraction Reagent (Pierce Biotechnology, Rockford, IL). Whole-cell lysates were loaded and run in pre-cast PAGE gels (Bio-Rad Laboratories, Hercules, CA), and transferred to Hybond membrane (GE Healthcare Bio-sciences, Piscataway, NJ). Membrane blots were blocked with 5% nonfat dry milk. Primary antibodies were allowed to bind to the membrane overnight, followed by 2–3 washes with 1X PBS. They were then detected using horseradish peroxidase-labeled secondary antibodies coupled with SuperSignal West Pico Chemiluminescent reagents (Pierce Biotechnology, Rockford, IL).
Competitive RT-PCR of Tm-2/3 mRNA
To determine Tm-2/3 mRNA expression levels by RT-PCR, Tm-2/3 exon-specific primers were created to correspond to sequences within the murine cDNA. The sense primer was of the sequence 5’-GCGGAGAAAAAGGCCACAGATG-3’, and the antisense primer was of the sequence 5’-TCTTCTTTGGCATGGGCCAC-3’. For an internal standard, a cDNA was created that corresponded to the expected RT-PCR product using the primers above, but contained an internal deletion of 17%, a T7 promoter element, and a tail of 15 adenosines, as previously described [21, 25]. This product was transcribed in vitro using the MAXIscript system (Ambion, Austin, TX) and 0.2 pg to 2 pg of the resulting RNA (internal standard) was added to 1 µg of osteoclast total cellular RNA prior to reverse transcription (RT) and PCR. RT-PCR was performed using the Superscript First-strand Synthesis System and TaqDNA Polymerase, both from Invitrogen Corp. (Carlsbad, CA). One-fifth of the resulting RT-PCR products were run in a 2% agarose gel and stained with ethidium bromide to visualize the relative intensities of the sample and internal standard bands. Quantification of bands was performed using a Gel Doc supplied with Quantity One software (Bio-Rad Laboratories, Hercules, CA).
Knockdown and Overexpression of Tm-2/3
For Tm-2/3 RNA interference studies, three siRNAs were designed and synthesized by Ambion (Austin, TX). Two of these siRNAs were used for further studies. SiRNA1 was of the sequence 5’-AACUCAAGGGCACUGAAGAtt-3’ (sense) and 5’-UCUUCAGUGCCCUUGAGUUtc-3’ (antisense). SiRNA2 was of the sequence 5’- GAUGCUGAAGCUCGACAAAtt-3’ (sense) and 5’-UUUGUCGAGCUUCAGCAUCtg-3’ (antisense). These oligonucleotides target sequences within exons 1a and 2b of α-gene tropomyosins. Optimization with different siRNA concentrations (25–100 nM) was performed, and both siRNAs achieved optimal knockdown of Tm-2/3 at 100 nM. Because tropomyosins arise from alternate exon and promoter usage, Tm-2 and Tm-3 differ by only a few amino acids and hence it is not possible to design siRNAs specifically for each isoform. However, none of the other tropomyosins present in osteoclasts (including Tm-5a/5b, Tm-4 and Tm-5NM-1) showed any changes in their levels with Tm-2/3 siRNA knockdown.
To transfect siRNA into RAW 264.7 cells, macrophage precursors were first stimulated by GST-RANKL to induce differentiate into osteoclasts. On day 5 of osteoclast differentiation, siRNA or a non-targeting control dsRNA at the same concentration (Ambion) was transfected with Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA). Transfection efficiency was measured by introducing a commercial fluorescent dsRNA (Sequitur/Invitrogen Corp.) into the cells; efficiency was >95%, as previously described [30]. Highest knockdown was seen on day 2 post transfection and cells were harvested on that day for all experiments. For RNA and protein studies, total cellular RNA was harvested with RNA-Bee (Tel-test Inc., Friendswood, TX), and whole cell lysates for protein assays were harvested using M-PER (Pierce Biotechnology, Rockford, IL). For immunocytochemical studies, cells were either scraped and replated on ivory slices or glass coverslips immediately following transfection or grown on ivory slices or glass coverslips from day 1; both methods worked efficiently and provided similar results.
Osteoclasts derived from mouse bone marrow precursors were transfected by electroporation. On day 4 of differentiation, osteoclasts were scraped, pelleted and resuspended in siPORT buffer (Ambion, Austin, TX). SiRNA or a non-targeting control dsRNA were added to the cell solution and electroporated at 250 V/50 µF, and replated in standard differentiation medium on glass or ivory for immunocytochemistry, or plastic for RNA and protein studies. Transfection efficiencies in marrow-derived osteoclasts were >95% [30].
For overexpression studies, the murine Tm-2 or Tm-3 coding regions were isolated by RT-PCR and subcloned into the eukaryotic expression vector pEF6/V5-His (Invitrogen Corp., Carlsbad, CA). RAW 264.7 macrophages were stably transfected with the tropomyosin plasmids using Lipofectamine and Plus reagents (Invitrogen Corp., Carlsbad, CA). As a control, the empty pEF6/V5-His vector also was stably transfected into the cells using the same method. The transfected cultures were maintained under blasticidin (3 µg/ml) selection.
Immunocytochemistry and microscopy
Osteoclasts on glass coverslips or thinly-cut ivory slices were first briefly fixed in a solution of 1% formaldehyde in pH 6.5 stabilization buffer (127 mM NaCl, 5 mM KCl, 1.1 mM NaH2PO4, 0.4 mM KH2PO4, 2 mM MgCl2, 5.5 mM glucose, 1 mM EGTA, 20 mM Pipes), and then were fixed and permeabilized for 1 hour in a solution of 2% formaldehyde, 0.2% Triton X-100, and 0.5% deoxycholate in the same stabilization buffer [29, 30]. Cells then were probed with primary antibodies diluted in blocking buffer, followed by Alexa-labeled secondary antibodies (Invitrogen Corp., Carlsbad, CA). F-actin was identified using Alexa-labeled phalloidin, also from Invitrogen. Nuclear labeling was performed by incubation of cells with DRAQ5 (Biostatus Limited, San Diego, CA). Cells were visualized using a Zeiss 510 META laser scanning confocal microscope (Campus Microscopy and Imaging Facility, The Ohio State University).
Visualization of free barbed ends
Free barbed ends were labeled essentially as described by Chan et al. [5]. Briefly, osteoclasts were treated for 3 minutes at room temperature in permeabilization buffer (20 mM Hepes, pH 7.5, 138 mM KCl, 2 mM MgCl2, 3 mM EGTA, 0.2 mg/ml saponin, 1 mM ATP, 1% BSA) containing 0.5 µM rhodamine-actin (Cytoskeleton, Inc., Denver, CO). Cells then were fixed for 5 minutes with 3.7% formaldehyde in stabilization buffer, rinsed in PBS, and treated with 10 mM sodium borohydride for 10 minutes, followed by incubation with 488 Alexa phalloidin (Invitrogen Corp.). The specificity of rhodamine-actin binding to barbed ends was established by incubating cells in 100 nM cytochalasin D (which caps barbed ends) for 5 minutes prior to permeabilization and rhodamine-actin addition.
Osteoclast resorption and motility assays
For resorption assays, control- or siRNA-transfected cells were scraped and replated onto BD BioCoat Osteologic Discs (BD Biosciences, San Jose, CA) immediately following transfection and incubated for 3 days. Alternatively, stably transfected tropomyosin-overexpressing and control cells were scraped and replated on Osteologic discs on day 4 of culture and incubated an additional 4 days. The adherent cells were removed by bleach treatment, followed by multiple water rinses. Resorbed areas (clearings) were assayed by photographing the resorbed discs under low magnification light microscopy. The area for each clearing was quantified with SigmaScan Pro 5.0 software (SPSS Science, Chicago, IL) as previously described [30]. Equal numbers of images were compared among test groups.
Migration assays were performed using 8.0 µm pore Transwell migration chambers (Corning Life Sciences, Acton, MA). The bottom side of the membrane was coated with collagen I (3 mg/ml diluted 1:2 with 100% ethanol) and dried overnight. Cells were scraped and replated onto the top side of the membrane immediately following transfection for siRNA treated cells and on day 4 of differentiation for stably transfected cells. After 48 hrs, the cells were stimulated to migrate by the addition of 40 µg/ml osteopontin peptide to the bottom of the wells. The remaining cells on the upper side of the wells were wiped away with a cotton swab and the migrated cells on the bottom side were fixed, and stained for tartrate resistant acid phosphatase using a Leukocyte Acid Phosphatase kit (Sigma, St. Louis, MO).
RESULTS
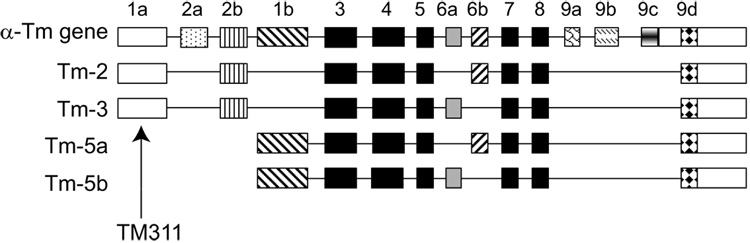
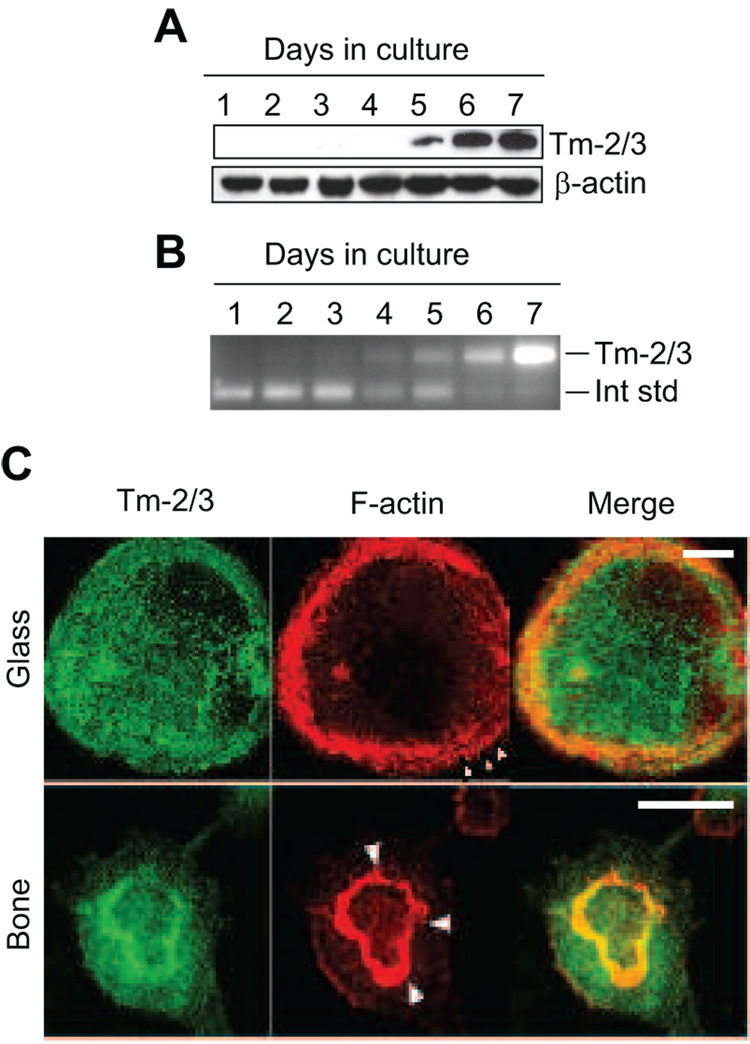
Figure 1 illustrates the α-gene tropomyosins previously demonstrated to be expressed in osteoclasts [29]. Tm-2 and Tm-3 are closely related proteins that differ in primary sequence by only one alternately spliced exon. Tm-2 contains the 76 base-pair exon 6b, while Tm-3 contains exon 6a, also 76 base pairs (Figure 1). Because all exons that compose these proteins are present in other Tm isoforms in osteoclasts, it is not possible to generate reagents specific to either Tm-2 or Tm-3. However, Tm-2 and Tm-3 are unique in containing alpha gene exons 1a and 2b, and antibody and nucleic acid probes can be readily made to these regions. Antibody TM311, which detects an epitope in exon 1a, is used to detect Tm-2 and Tm-3 simultaneously (Figure 1). Through Western analysis and immunocytochemistry, we previously demonstrated that the high molecular weight tropomyosins Tm-2 and Tm-3 were expressed in mature RAW264.7-derived osteoclasts, but showed little to no expression in their macrophage precursors [29], as previously described by others [32]. Based on RT-PCR amplification and random sequencing of their corresponding mRNAs, we found that these tropomyosins were expressed at a ratio of approximately 4:1 (Tm-2: Tm-3) [29]. To extend these findings, we observed expression of Tm-2/3 in mouse bone marrow cultures during a 7 day differentiation period. Figure 2A illustrates that under our conditions, Tm-2/3 levels were undetectable in early stages of culture, but that levels increased dramatically with osteoclast maturation. This increase in expression was paralleled by an increase in Tm-2/3 mRNA, as illustrated in Figure 2B. Occasionally, TM311 also would detect a low abundance, higher molecular band only in mature marrow-derived, but not RAW264.7-derived, cells (not shown). This band potentially could correspond to either Tm-6 (from the alpha gene) or Tm-1 (from the beta gene) since RT-PCR with isoform-specific primers showed the presence of both mRNAs in mature osteoclasts. However, because the presence of this band was not reproducible and because its intensity was significantly less than that of Tm-2/3, our efforts were focused on examination of a role for Tm-2/3 for the purposes of this study.
Figure 1. Schematic of the alpha gene tropomyosins expressed in osteoclasts.
Conserved exons 3–5 and 7–8 are represented in black, while alternately expressed exons are represented in other shades. This diagram was adapted from Goodwin et al. [13]. As indicated, antibody TM311 was derived against sequences in exon 1a.
Figure 2. Expression of high molecular weight tropomyosins in differentiating mouse osteoclast cultures.
A, Osteoclasts were generated from murine bone marrow cultures and assayed by immunoblot for HMW Tm-2/3 and β-actin as a loading control. B, Competitive RT-PCR demonstrates an increase in Tm-2/3 mRNA that parallels regulation of the resulting protein. The upper band corresponds to Tm-2/3 mRNA, while the lower band represents a competitive internal standard. C, Confocal images of osteoclasts on glass or bone demonstrates the diffuse distribution of Tm-2/3 at the cell base and their lack of strong association with individual podosomes and the sealing zone (arrowheads). Scale bars = 20 µm.
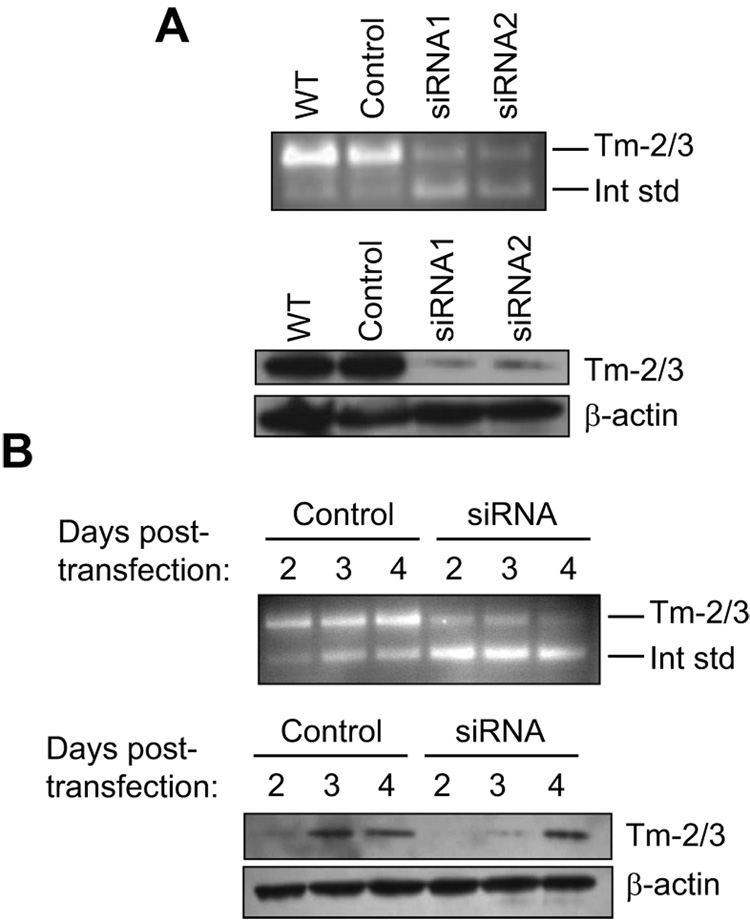
In our previous studies, we found that Tm-2/3 were rather diffusely distributed throughout the interior of osteoclasts, with only modest association with the most predominant F-actin structures of these cells, the podosome belt and the sealing zone of resorbing osteoclasts [29]. In podosome belts and sealing zones, Tm-2/3 strongly associated with neither core filaments nor actin clouds [22], but were only loosely intercalated between these structures. Figure 2C illustrates this pattern; in the merged panels, the juncture between Tm-2/3 and these adhesion structures produces an orange-to-yellow image, indicating imperfect overlap. Rather, these tropomyosins are most prevalent throughout the cell interior. Because tropomyosins are known to stabilize filamentous actin, this distribution led us to speculate that osteoclasts may induce expression of HMW tropomyosins to help support the cytoskeletal scaffolding of these large, highly motile cells [29]. As a first step to determining the role of these molecules in osteoclast function, siRNAs designed to target exon 1a (siRNA2) or 2b (siRNA1) were transfected into mouse bone marrow derived osteoclasts and assayed 2 days later. Figure 3A (top) demonstrates the efficiency of these two siRNAs in suppressing Tm-2/3 mRNA relative to either untreated osteoclasts, or to those treated with a control (non-targeting) siRNA. In multiple experiments, mRNA knockdown by either siRNA was approximately 70%. At the protein level, both siRNAs achieved knockdown levels of 80 – 90% (Figure 3A, bottom). Because Tm-2/3 levels increase during osteoclast maturation, we performed a timed analysis of siRNA knockdown efficiency at days 2, 3, and 4 post-transfection (cells were transfected on day 5 of differentiation, after osteoclast fusion was initiated). As shown in Figure 3B, control-transfected cells demonstrated increased Tm-2/3 protein and mRNA levels during the post-transfection period, and siRNA treatment was able to efficiently suppress protein expression only on days 2 and 3. Therefore, for all following experiments, cells were assayed for morphology and structure on day 2 post siRNA transfection (day 7 of entire culture period). Although only results from marrow-derived osteoclasts are shown, similar results were obtained in RAW264.7 cells.
Figure 3. Suppression of Tm-2/3 by RNA interference.
A, (top) Two siRNAs corresponding to different exons of the Tm-alpha gene efficiently suppress Tm-2/3 mRNA expression in marrow-derived osteoclasts on day 2 post-transfection, as assayed by competitive RT-PCR. (bottom) These same siRNAs efficiently suppress Tm-2/3 protein expression in marrow-derived cells on day 2 post-transfection. B, Competitive RT-PCR (top) and Western analysis (bottom) of marrow-derived osteoclasts demonstrates efficient knockdown of Tm-2/3 mRNA and protein at days 2-3 post-transfection.
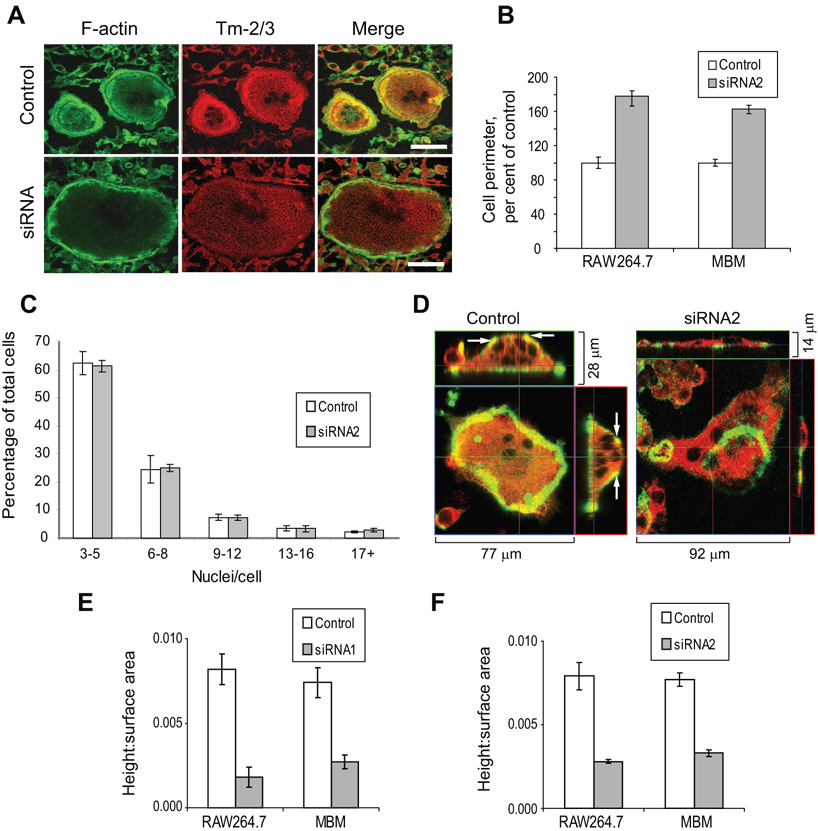
Microscopic imaging of siRNA-treated cells revealed an immediately obvious consequence of Tm-2/3 suppression to be an increase in osteoclast perimeter. Figure 4A demonstrates typical differences in cell perimeter between siRNA and control-transfected cells, as well as the loss of intensity in Tm-2/3 labeling with siRNA treatment. For both RAW264.7 and marrow-derived osteoclasts, siRNA treatment resulted in perimeters that were 60 – 80% larger than control-transfected cells (Figure 4B). Because this change could arise from either increased precursor fusion or increased spreading, numbers of nuclei (in osteoclasts containing 3 or more) were enumerated. In the experiments shown in Figure 4B, RAW264.7 control cells possessed an average of 5.3 ± 1.2 nuclei, while siRNA-treated cells possessed an average of 5.1 ± 1.5 (mean ± std dev). Marrow-derived control osteoclasts contained 3.6 ± 0.7 nuclei, with siRNA-treated cells containing 3.1 ± 0.3. Thus, the change in perimeter derived not from enhanced fusion, but from increased spreading. To extend these findings further, the total distribution of nuclei in RAW264.7 osteoclast cultures were enumerated in three separate experiments. Figure 4C illustrates these results, in which control- and siRNA-treated cells exhibited no statistical differences. These findings suggest that increased osteoclast perimeter in siRNA-treated cells results from enhanced spreading (with resulting flattening of the cells). To further explore this possibility, control- and siRNA-treated cells were examined by confocal microscopy in the Z plane to determine osteoclast height. Because normal osteoclasts plated on bone substrate generally have a more rounded morphology than those plated on glass, we observed cells on bone, in which any differences in cell flattening would be more noticeable. Figure 4D illustrates control- and siRNA-treated cells that both polarize properly, but in which differences in cell flattening are immediately apparent. In these photomicrographs, F-actin is labeled green, allowing visualization of the sealing zone, while Tm-2/3 is in red. (Note that in the siRNA-treated cell, the gain on the Tm-2/3 signal has been enhanced to allow better visualization of the entire cell.) This figure also illustrates a notably consistent difference in F-actin distribution between control and siRNA-treated cells. Control cells demonstrated basolateral F-actin, as demonstrated by yellow co-labeling (arrows), while siRNA-treated cells lacked this actin distribution. Multiple cells treated with control or targeting siRNAs were measured for peak height and surface area, and the ratios between these measurements were calculated. Figure 4E demonstrates that treatment with siRNA1 results in an obvious loss of height relative to surface area (i.e. increased flattening) for both RAW264.7 and marrow-derived osteoclasts. As seen previously in Figure 4B, the control- and siRNA-treated cells assayed possessed similar nuclear numbers (RAW264.7 controls, 6.4 ± 1.1; RAW264.7 siRNA-treated, 6.0 ± 1.2; MBM controls, 3.6 ± 0.9; MBM siRNA-treated, 3.6 ± 0.6). In separate experiments, similar results were obtained for both types of osteoclasts when transfected with siRNA2 (Figure 4F). In these experiments also, nuclear numbers in control- and siRNA-treated cells were similar (RAW264.7 controls, 5.8 ± 1.4; RAW264.7 siRNA-treated, 6.3 ± 1.6; MBM controls, 3.3 ± 0.6; MBM siRNA-treated, 3.4 ± 0.5). Although Tm-2/3 are mildly enriched in the sealing zone of polarized osteoclasts, we saw no effects on actin ring formation or size following siRNA-mediated knockdown. This is in contrast to our previous studies on Tm-4, in which suppression of this tropomyosin caused dramatic decreases in height of the actin ring [30]. Thus, these results are consistent with our previous hypothesis suggesting that natural upregulation of Tm-2/3 during osteoclastogenesis is required to maintain structural integrity of the cell.
Figure 4. Suppression of Tm-2/3 results in increased osteoclast spreading.
A, siRNA-transfected osteoclasts on glass demonstrate decreased Tm-2/3 labeling and increased perimeter. Scale bars = 50 µm. B, Multiple control- and siRNA-transfected cells like those in panel A were measured for cell perimeter, and the results are presented graphically as mean ± s.e.m.; n = at least 150 cells per group. For both RAW264.7 and MBM cells, P < 1 × 10−16. C, Nuclei were enumerated from control- and siRNA2-transfected RAW264.7-derived osteoclasts from three separate experiments and expressed graphically as mean ± s.e.m. No significant difference in nuclear number was observed between the samples. D, Control- or siRNA-transfected RAW264.7-derived osteoclasts cultured on ivory were imaged by confocal Z-stack analysis to view relative flattening. Arrows indicate regions of basolateral F-actin and Tm-2/3 co-labeling. Green = F-actin; Red = Tm-2/3 E, Multiple control- or siRNA-transfected cells such as those in panel D were measured for height and surface area. The ratio of these measurements are expressed graphically as mean ± s.e.m.; n = 18. For both RAW264.7 and MBM cells, P < 0.002. F, RAW264.7- or marrow-derived osteoclasts were transfected and measured as in panel E, but suppression of Tm-2/3 was achieved by introduction of siRNA2. Bars indicate mean ± s.e.m.; for both RAW264.7 and MBM cells, P < 0.0001.
To determine how cellular flattening might affect osteoclast function, control- and siRNA-treated cells were assayed for their ability to migrate across a membrane in response to osteopontin stimulation. In separate sets of experiments, the motility of control-transfected cells was compared to that of cells transfected with either siRNA1 (Figure 5A) or siRNA2 (Figure 5B). For either siRNA, or either type of osteoclast, motility was diminished by over 50%. Additionally, potential effects on resorptive capacity were measured by quantifying the area of clearance of synthetic bone substrate. As shown in Figure 5C, for either RAW264.7 or marrow-derived cells, total resorption was not altered; however, siRNA-treated cells produced fewer, but larger, resorbed areas. These results are consistent with the siRNA-treated cells as more spread, but less motile, osteoclasts.
Figure 5. Motility, but not total resorptive capacity, was diminished by suppression of Tm-2/3.
A, Control- or siRNA1-transfected osteoclasts were assessed for their ability to migrate across filters in response to osteopontin stimulation. At least 100 cells were counted for each sample. Bars = mean ± s.e.m.; n = 4. For both RAW264.7 and MBM cells, P < 0.01. B, Cells were assessed for migration capacity as in panel A, but were treated with control or siRNA2 oligonucleotides. For both RAW264.7 and MBM cells, P < 0.005. C, siRNA2-treated osteoclasts on synthetic bone substrate demonstrated fewer, but larger clearings than controls (for each pairwise comparison, P < 0.001), while overall resorption was not affected (for both RAW264.7 and MBM cells, P > 0.4). Bars represent mean ± s.e.m. ; n = 10 for each variable. At least 300 clearings were measured for each sample.
As a complement to Tm-2/3 suppression studies, Tm-2 was overexpressed in osteoclasts. RAW264.7 macrophages were stably transfected with Tm-2, and two of the resulting clones (Tm-2.4 and Tm-2.5) were chosen for further study. Figure 6A shows relative expression of Tm-2 in these clones relative to a clone transfected with the empty expression vector. Both overexpressing clones were able to form osteoclasts at the same rate as the control transfectant. In three separate experiments, clone Tm-2.4 formed 98.0 ± 5.8% as many osteoclasts as control transfectants, while clone Tm-2.5 formed 103.0 ± 6.2% as many. Figure 6A shows that these clones express similar levels of Tm-2 in the macrophage form or in the osteoclast form, with the high levels of exogenous Tm-2 apparently swamping out any normally occurring increase in the endogenous form during differentiation. When cultured on glass, the overexpressing clones demonstrated two unusual morphologies, as illustrated in Figure 6B. Unlike control transfectants that generate normal podosome belts (left panel), most Tm-2 overexpressing clones demonstrated patches of F-actin (green) at the base of the cell and a highly rounded morphology (middle panel). Less typically, these cells were of a normal to rounded shape but possessed F-actin stress fibers at the base (right panel). Tm-2 (red) was present in moderate amounts at the coverslip surface, but was very highly expressed in the cell interior. This distribution is not unexpected, given the known localization of Tm-2/3 in wild-type cells. Thus, whereas loss of Tm-2/3 from the cell interior resulted in flattening of osteoclasts, overexpression of Tm-2 in this region produced the converse effect; that is, rounding of the cells.
Figure 6. Overexpression of Tm-2, but not Tm-3, affects osteoclast morphology, motility, and resorptive capacity.
A, Western analysis of stably transfected RAW264.7-derived overexpressing clones demonstrates the high expression levels of Tm-2 relative to control transfectants. B, Confocal imaging demonstrates normal podosome belt formation and cell shape in control-transfected cells, while Tm-2 overexpressing clones are unusual in actin structures and shape. Green = F-actin; Red = Tm-2/3; Blue = nuclei. C, Tm-2 overexpressing clones exhibit a highly rounded morphology with a greater height:surface area ratio than control transfectants. Bars indicate mean ± s.e.m. For comparison of each transfectant to control, P < 0.001. D, Tm-2 overexpressing osteoclast clones demonstrate diminished motility in response to osteopontin stimulation. Bars indicate mean ± s.e.m; n = 4. At least 100 cells were assayed for each sample. For comparison of each Tm-2 transfectant to control, P < 0.01. E, Tm-2 overexpressing osteoclast clones possess diminished resorptive capacity. Bars represent mean ± s.e.m.; n = 8 for each variable. At least 200 clearings were measured for each sample. For all comparisons of Tm-2 transfectants to control, P < 0.005. F, Three clones were generated that overexpressed Tm-3. Unlike Tm-2 overexpressing clones, which possessed similar amounts of the exogenously introduced tropomyosin, cells transfected with Tm-3 showed no alterations in morphology relative to control cells.
To ensure that overexpression of Tm-2 did not alter cellular fusion rates, nuclei were enumerated in control transfectants and both overexpressing cell lines. No significant differences were noted (control, 8.0 ± 1.2; Tm-2.4, 7.9 ± 2.2; Tm-2.5, 6.6 ± 1.6). To measure cell spreading, we determined the ratio of cell height to surface area, as previously assessed in siRNA-treated cells. Figure 6C illustrates that control transfectants showed a height:surface area ratio of 0.007, which is identical to cells transfected with control siRNAs (see Figures 4E–F). However, the Tm-2 overexpressing cells demonstrated a notable increase in height:surface area that appeared to correspond with the level of Tm-2 expression (i.e. Tm-2.5 > Tm-2.4). Thus, it appears that overexpression of this high molecular weight tropomyosin results in unusual actin structures at the base of the cell, and can also effect rounding of the entire osteoclast. However, when plated on a bone substrate, no noticeable alterations in sealing zone morphology were apparent (data not shown), suggesting that bone-mediated signaling may play a key role in organization of Tm-mediated actin rearrangements. However, in spite of apparently normal sealing zones, osteopontin-directed motility (Figure 6D) and resorption capacity (Figure 6E) were somewhat compromised. These results indicate that Tm-2’s effects on microfilament organization throughout the cell may have consequences for osteoclast function.
To determine whether any functional differences existed between Tm-2 and Tm-3, Tm-3 overexpressing clones also were generated. Our previous studies showed Tm-3 to be expressed at approximately 20% of the levels of Tm-2 [29]. However, the clones transfected with either exogenous Tm-2 or Tm-3 expressed their respective isoform at approximately equivalent levels (Figure 6F). Curiously, even though the two tropomyosin isoforms differ by only 25 amino acid residues, Tm-3 overexpressing clones demonstrated no difference in morphology from control transfectants (not shown). Thus, in both expression levels and function, Tm-2 appears to the dominant HMW isoform in regulating osteoclast cytoskeletal morphology.
Finally, we performed experiments to obtain a first indication of how alterations in Tm-2/3 levels may affect osteoclast morphology. A protein modulator of microfilament structure, gelsolin, possesses severing activity that is inhibited by tropomyosin. Indeed, it was determined that only HMW tropomyosins such as Tm-2 are capable of inhibiting gelsolin activity [20]. Further, HMW Tms are capable of annealing gelsolin-severed actin filaments, and in doing so, can generate long filaments from small gelsolin-bound actin fragments [19]. LMW tropomyosins are not capable of these functions even at very high concentrations. Therefore, we immunolabeled control, siRNA-treated, and Tm-2 overexpressing cells for F-actin and gelsolin. As shown in Figure 7A, in control cells (rows 1 and 3) gelsolin was distributed primarily in podosomes, consistent with findings from other groups (note the extensive overlap in the merged images) [1, 6]. However, knockdown of Tm-2/3 (row 2) caused redistribution of gelsolin away from podosomes to the cell interior (arrows). This may be best seen by the relative lack of colocalization in the merged image, compared to the control. This finding is consistent with the hypothesis that loss of Tm-2/3, which are highly expressed in the cell interior, permits gelsolin binding to internal actin filaments. In contrast, overexpression of Tm-2 caused a loss of intense gelsolin labeling at the base of the cell (row 4); however, in the same cell imaged several microns off the coverslip and by Z-stack imaging (right of row 4), gelsolin was clearly enhanced in the cortical actin surrounding the entire cell periphery (arrowheads). This is in contrast to the stable control cell above, in which gelsolin was barely detectable in the cell periphery outside of the podosome belt (right of row 3, arrowheads). Thus, it appears that overexpression of Tm-2 (which is highly expressed in the cell interior; see Figure 6B) prevents gelsolin from accumulating in the same area. This finding is consistent with the notion that Tm-2 regulates cell shape by preventing gelsolin access to microfilaments making up the internal scaffolding. In the case of Tm-2/3-depleted cells, the increased presence of gelsolin in these regions could cause severing of these microfilaments, resulting in cell flattening. Conversely, overexpression of Tm-2 may inhibit gelsolin-mediated severing, thus creating the rounded morphology.
Figure 7. Distribution of gelsolin and free barbed microfilament ends in cells expressing altered levels of Tm-2.
A, Control, siRNA-treated, or Tm-2-overexpressing osteoclasts were labeled for gelsolin and F-actin. The most intense areas of gelsolin labeling are indicated by arrows. Stable control cells and Tm-2-overexpressing cells were viewed by Z-stack imaging to illustrate the enhanced distribution of gelsolin in the cortical regions of the overexpressing cells, and its lack in the cell interior (note the intense yellow in the cortical regions of clone Tm-2.5, as indicated by arrowheads). Scale bars = 20 µm. B, Stable control cells and overexpressing clone Tm-2.5 were labeled for F-actin (green) and free barbed ends (reds). In control cells, barbed ends were most abundant at the cell base, while in Tm-2.5, they were most abundant in the cell interior. Scale bars = 20 µm. C, Cells labeled with rhodamine actin following pretreatment with cytochalasin D show specificity of the method for labeling of free barbed ends. Scale bar = 20 µm.
To explore this possibility further, we assayed cells for the prevalence of actin microfilaments containing free barbed (fast-growing) ends. Because gelsolin severs actin filaments and caps the resulting fragments at their barbed ends, decreased gelsolin activity should result in greater numbers of free barbed ends. Therefore, we added rhodamine-labeled monomeric actin to permeabilized osteoclasts under conditions that would allow addition of this actin only to free barbed ends [5]. Figure 7B demonstrates that in control cells, most F-actin structures at the based of the cell contained free barbed ends (row 1). In contrast, Tm-2 overexpressing cells demonstrated the greatest numbers of free barbed ends in the cell interior, where Tm-2 levels were highest and gelsolin was absent (row 2 and accompanying Z-image). This finding is consistent with our hypothesis. We also examined Tm-2/3-suppressed cells for any changes in distribution of free barbed ends, and while the results were suggestive of a loss in their number, our results were somewhat variable (data not shown) and require greater inspection. Finally, to ensure that our method was specific for the detection of free barbed ends, rhodamine-actin was incubated with permeabilized cells that previously had been treated with cytochalasin D, which caps these ends. Virtually no labeling with rhodamine-actin was detectable, indicating the specificity of this method (Figure 7C). Taken together, these findings are consistent with a role for HMW tropomyosins in osteoclasts regulating cell morphology by controlling access of gelsolin to structural microfilaments.
DISCUSSION
The results presented here are consistent with our hypothesis that expression of high molecular weight tropomyosin Tm-2 is strongly induced during osteoclastogenesis to provide structural support for osteoclasts as they increase in size to diameters of 50–100 microns or more. This increased expression is dramatic; by Western analysis and competitive RT-PCR, Tm-2/3 protein and mRNA are essentially undetectable in osteoclast precursors but notably abundant in mature cultures. Consistent with our findings, proteomic analysis of mice subjected to ovariectomy-induced estrogen withdrawal (which stimulates direct differentiation of osteoclast precursors, among other effects [39]) resulted in the finding that two HMW tropomyosins from the alpha and beta genes were among the proteins most strongly upregulated by this process [34]. Notably, the upregulated alpha gene product (accession number NP_077745) was identical to Tm-2. In addition, a beta gene product was identified (accession number NP_033442); however, this tropomyosin was identical to the skeletal muscle β-TM [28], rather than the expected related nonmuscle isoform Tm-1. It is possible that this protein was misidentified due to the extensive sequence identity between Tm-1 and β-TM. However, isoform-specific RT-PCR revealed that mRNAs corresponding to both isoforms were present in our osteoclast cultures (data not shown). Even so, detection of a protein band potentially corresponding to the beta gene isoforms was not entirely reproducible. Therefore, it is unclear whether beta gene products play a necessary function in osteoclast activity. In our in vitro cultures, it appears that Tm-2 is the dominant HMW tropomyosin isoform, both in abundance and in function. It is possible that osteoclasts in vivo are subjected to triggers that may more potently induce expression of beta gene tropomyosins; nonetheless, our findings provide evidence to demonstrate that natural upregulation of Tm-2 plays a key role in regulation of osteoclast motility and function.
HMW tropomyosins Tm-2 and Tm-3 differ from other alpha gene tropomyosins expressed in osteoclasts by virtue of their length (284 versus 248 amino acid residues) and upstream exons. Tm-5a and Tm-5b are identical to Tm-2 and Tm-3, respectively, with the exception that the former pair do not contain exons 1a and 2b, but instead the single exon 1b (see Figure 1). This difference of about 80 amino acid residues is sufficient to target each pair to distinct actin pools within osteoclasts. While Tm-2/3 are distributed in internal actin pools, Tm-5a/b are found in actin clouds that surround podosome cores [29]. However, Tm-5a/b also are upregulated during osteoclastogenesis, like their high molecular weight counterparts. The kinetics of this increased Tm-5a/b expression differs from that of Tm-2/3, in that the rise of Tm-5a/b is of smaller magnitude and begins earlier in osteoclastogenesis (data not shown). The use of alternate promoters by the alpha gene has not been explored in nonmuscle cells, but it will be of interest to determine the factors involved in expression of these gene products, particularly for Tm-2/3, which are not expressed in osteoclast precursor cells. By virtue of their amplified expression during osteoclastogenesis, alpha gene tropomyosins differ from other osteoclast Tms, such as Tm-4 and 5NM-1, which remain at constant levels, or other gamma gene Tms that are downregulated [29].
The derivation of Tm-2 and Tm-3 from alternate exons that are shared with other alpha gene tropomyosins prohibited our ability to specifically suppress expression of one isoform without the other. Further, we were unable to differentiate between the native Tm-2 and Tm-3 by immunolocalization studies. However, overexpression of Tm-2 induced formation of cells with altered morphologies, while overexpression of Tm-3 had no discernable effect. These results are consistent with previous comparisons of these isoforms. In fibroblast-like cells, both Tm-2 and Tm-3 have been shown to localize to stress fibers [37]. However, when these isoforms were individually transfected into a ras-transformed cell line that lacked Tm-2 and Tm-3, both tropomyosins were able to induce stress fiber formation and spreading, although Tm-2 was much more potent in doing so [12]. Further, cells expressing Tm-2 were no longer able to grow in low serum, while those expressing Tm-3 maintained this ability. Additional HMW isoform-specific differences have been noted in other cell models (rev. in [14]), and these tropomyosins appear to maintain these differences in regulating the osteoclast cytoskeleton. In vitro binding assays of bacterially-expressed Tm-2 and Tm-3 demonstrated these isoforms to possess similar actin binding qualities [37], so the reasons for differences in function between the isoforms are unclear. Nonetheless, like many other tissue and cell types [38], osteoclasts express more Tm-2 than Tm-3; this difference in expression, coupled with its potency in altering cell morphology, suggests Tm-2 to be dominant over Tm-3 in activity.
The mechanisms by which cytoskeletal Tms regulate actin dynamics have only recently begun to be assessed, but several studies and our own data suggest potential means by which Tm-2 may help regulate the actin infrastructure of osteoclasts. We demonstrated the potential role of Tm-2 as a modulator of gelsolin accessibility in Figure 7. Under normal circumstances, gelsolin is distributed primarily in adhesion structures, where its activity is regulated by the tyrosine kinase PYK2 and binding to phosphatidylinositol lipids [7, 8, 41]. In Tm-2/3-suppressed cells, its redistribution away from podosomes may contribute to the loss of osteopontin-mediated cell motility seen in Figure 5, given that osteoclasts from mice completely deficient in gelsolin can not assemble podosomes nor become motile in response to osteopontin [6]. Further, gelsolin’s redistribution to the cell interior appears to cause a corresponding redistribution in microfilament barbed ends, which are indicative of microfilament turnover and may account for altered cell morphology. In addition, tropomodulins bind cooperatively with tropomyosin to cap actin filaments at their pointed ends [42], thus preventing both microfilament elongation and depolymerization. Two isoforms, Tmod1 and Tmod3, have been shown by mRNA dot-blot to be ubiquitously expressed in all human tissues tested, including bone marrow [9]. Our preliminary studies reveal mRNAs for both isoforms to be present in osteoclasts, as assessed by RT-PCR and immunocytochemistry (data not shown). It was recently shown that reduction of Tmod3 in polarized intestinal epithelial cells resulted in disruption of the spectrin membrane skeleton, which has been shown to control the height of polarized epithelia [23, 43]. Tropomyosin-tropomodulin binding may similarly play a role in stabilization of the cytoskeleton in osteoclasts in addition to modulation of gelsolin distribution. Experiments to define the roles of these proteins in stabilization of osteoclast morphology are underway.
Finally, regulated phosphorylation of HMW tropomyosin in other cell models is known to play a role in membrane/cytoskeletal dynamics. Tm-2 was shown to be a target of PI3K protein kinase activity that was activated during ligand-stimulated endocytosis of the β-adrenergic receptor [31]. Phosphorylation of tropomyosin at Ser 61 was shown to be necessary for endocytosis of the receptor-ligand complex. In an earlier study, phosphorylation of the HMW tropomyosin Tm-1 downstream of ERK signaling was shown to regulate stress fiber formation [18]. In preliminary studies, we tested osteoclast cultures on glass and bone for the presence of phosphoserine residues in Tm-2, but were unable to detect these residues. However, phosphorylation may be a transient event that requires specific signaling pathways that we have not yet identified. More thorough examination of the role of Tm-2 phosphorylation in osteoclasts is required.
In summary, high molecular weight tropomyosins, while not specific to the osteoclast lineage, are a marker of osteoclast maturity due to their pronounced upregulation during differentiation and fusion. These actin-binding proteins, especially Tm-2, play a role in maintaining osteoclast morphology, motility, and resorptive capacity. This study, coupled with our previous work on the low molecular weight isoform Tm-4 [30], demonstrates the clear segregation of tropomyosin distribution and function in osteoclasts, and provides clues about the extent of actin cytoskeletal diversity in these cells.
ACKNOWLEDGMENTS
The authors would like to thank Tejdeep Singh for generation of preliminary data and the Campus Microscopy and Imaging Facility at The Ohio State University for aid and technical advice. This work was supported by grants RO1 AR051515 and RO1 DK052131 (to B.S.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Akisaka T, Yoshida H, Inoue S, Shimizu K. Organization of cytoskeletal F-actin, G-actin, and gelsolin in the adhesion structures in cultured osteoclast. J. Bone Miner. Res. 2001;16:1248–1255. doi: 10.1359/jbmr.2001.16.7.1248. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 3.Braverman RH, Cooper HL, Lee HS, Prasad GL. Anti-oncogenic effects of tropomyosin: isoform specificity and importance of protein coding sequences. Oncogene. 1996;13:537–545. [PubMed] [Google Scholar]
- 4.Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci. 1998;111(Pt 2):199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- 6.Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol. 2000;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellaiah MA. Regulation of podosomes by integrin alphavbeta3 and Rho GTPase-facilitated phosphoinositide signaling. Eur J Cell Biol. 2006;85:311–317. doi: 10.1016/j.ejcb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Chellaiah MA, Biswas RS, Yuen D, Alvarez UM, Hruska KA. Phosphatidylinositol 3,4,5-trisphosphate directs association of Src homology 2-containing signaling proteins with gelsolin. J Biol Chem. 2001;276:47434–47444. doi: 10.1074/jbc.M107494200. [DOI] [PubMed] [Google Scholar]
- 9.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 10.Cooper HL, Bhattacharya B, Bassin RH, Salomon DS. Suppression of synthesis and utilization of tropomyosin in mouse and rat fibroblasts by transforming growth factor alpha: a pathway in oncogene action. Cancer Res. 1987;47:4493–4500. [PubMed] [Google Scholar]
- 11.Dalby-Payne JR, O'Loughlin EV, Gunning P. Polarization of specific tropomyosin isoforms in gastrointestinal epithelial cells and their impact on CFTR at the apical surface. Mol Biol Cell. 2003;14:4365–4375. doi: 10.1091/mbc.E03-03-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimona M, Kazzaz JA, Helfman DM. Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc Natl Acad Sci U S A. 1996;93:9618–9623. doi: 10.1073/pnas.93.18.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin LO, Lees-Miller JP, Leonard MA, Cheley SB, Helfman DM. Four fibroblast tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene via alternative RNA splicing and the use of two promoters. J Biol Chem. 1991;266:8408–8415. [PubMed] [Google Scholar]
- 14.Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 15.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Had L, Faivre-Sarrailh C, Legrand C, Mery J, Brugidou J, Rabie A. Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. J Cell Sci. 1994;107(Pt 10):2961–2973. doi: 10.1242/jcs.107.10.2961. [DOI] [PubMed] [Google Scholar]
- 17.Hendricks M, Weintraub H. Tropomyosin is decreased in transformed cells. Proc Natl Acad Sci U S A. 1981;78:5633–5637. doi: 10.1073/pnas.78.9.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houle F, Rousseau S, Morrice N, Luc M, Mongrain S, Turner CE, Tanaka S, Moreau P, Huot J. Extracellular signal-regulated kinase mediates phosphorylation of tropomyosin-1 to promote cytoskeleton remodeling in response to oxidative stress: impact on membrane blebbing. Mol Biol Cell. 2003;14:1418–1432. doi: 10.1091/mbc.E02-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa R, Yamashiro S, Matsumura F. Annealing of gelsolin-severed actin fragments by tropomyosin in the presence of Ca2+. Potentiation of the annealing process by caldesmon. J Biol Chem. 1989;264:16764–16770. [PubMed] [Google Scholar]
- 20.Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7497. [PubMed] [Google Scholar]
- 21.Jeyaraj S, Dakhlallah D, Hill SR, Lee BS. HuR stabilizes vacuolar H+-translocating ATPase mRNA during cellular energy depletion. J Biol Chem. 2005;280:37957–37964. doi: 10.1074/jbc.M502883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurdic P, Saltel F, Chabadel A, Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur J Cell Biol. 2006;85:195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Kizhatil K, Yoon W, Mohler PJ, Davis LH, Hoffman JA, Bennett V. Ankyrin-G and beta2-spectrin collaborate in biogenesis of lateral membrane of human bronchial epithelial cells. J Biol Chem. 2007;282:2029–2037. doi: 10.1074/jbc.M608921200. [DOI] [PubMed] [Google Scholar]
- 24.Krits I, Wysolmerski RB, Holliday LS, Lee BS. Differential localization of myosin II isoforms in resting and activated osteoclasts. Calcif Tissue Int. 2002;71:530–538. doi: 10.1007/s00223-001-1112-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee BS, Holliday LS, Krits I, Gluck SL. Vacuolar H+-ATPase activity and expression in mouse bone marrow cultures. J. Bone Miner. Res. 1999;14:2127–2136. doi: 10.1359/jbmr.1999.14.12.2127. [DOI] [PubMed] [Google Scholar]
- 26.Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol. 1997;170:1–38. doi: 10.1016/s0074-7696(08)61619-8. [DOI] [PubMed] [Google Scholar]
- 27.Masuda A, Takenaga K, Kondoh F, Fukami H, Utsumi K, Okayama H. Role of a signal transduction pathway which controls disassembly of microfilament bundles and suppression of high-molecular-weight tropomyosin expression in oncogenic transformation of NRK cells. Oncogene. 1996;12:2081–2088. [PubMed] [Google Scholar]
- 28.McInnes C, Leader DP. The tropomyosin mRNAs of mouse striated muscles: molecular cloning of beta-tropomyosin. Biochim Biophys Acta. 1988;951:117–122. doi: 10.1016/0167-4781(88)90031-0. [DOI] [PubMed] [Google Scholar]
- 29.McMichael BK, Kotadiya P, Singh T, Holliday LS, Lee BS. Tropomyosin isoforms localize to distinct microfilament populations in osteoclasts. Bone. 2006;39:694–705. doi: 10.1016/j.bone.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 30.McMichael BK, Lee BS. Tropomyosin 4 regulates adhesion structures and resorptive capacity in osteoclasts. Exp Cell Res. 2008;314:564–573. doi: 10.1016/j.yexcr.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naga Prasad SV, Jayatilleke A, Madamanchi A, Rockman HA. Protein kinase activity of phosphoinositide 3-kinase regulates beta-adrenergic receptor endocytosis. Nat Cell Biol. 2005;7:785–796. doi: 10.1038/ncb1278. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Sakiyama S, Takenaga K. Suppression of syntheses of high molecular weight nonmuscle tropomyosins in macrophages. Cell Motil Cytoskeleton. 1995;31:273–282. doi: 10.1002/cm.970310404. [DOI] [PubMed] [Google Scholar]
- 33.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastorelli R, Carpi D, Airoldi L, Chiabrando C, Bagnati R, Fanelli R, Moverare S, Ohlsson C. Proteome analysis for the identification of in vivo estrogen-regulated proteins in bone. Proteomics. 2005;5:4936–4945. doi: 10.1002/pmic.200401325. [DOI] [PubMed] [Google Scholar]
- 35.Percival JM, Hughes JA, Brown DL, Schevzov G, Heimann K, Vrhovski B, Bryce N, Stow JL, Gunning PW. Targeting of a tropomyosin isoform to short microfilaments associated with the Golgi complex. Mol Biol Cell. 2004;15:268–280. doi: 10.1091/mbc.E03-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Percival JM, Thomas G, Cock TA, Gardiner EM, Jeffrey PL, Lin JJ, Weinberger RP, Gunning P. Sorting of tropomyosin isoforms in synchronised NIH 3T3 fibroblasts: evidence for distinct microfilament populations. Cell Motil Cytoskeleton. 2000;47:189–208. doi: 10.1002/1097-0169(200011)47:3<189::AID-CM3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 37.Pittenger MF, Helfman DM. In vitro and in vivo characterization of four fibroblast tropomyosins produced in bacteria: TM-2, TM-3, TM-5a, and TM-5b are co-localized in interphase fibroblasts. J Cell Biol. 1992;118:841–858. doi: 10.1083/jcb.118.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schevzov G, Vrhovski B, Bryce NS, Elmir S, Qiu MR, O'Neill GM, Yang N, Verrills NM, Kavallaris M, Gunning PW. Tissue-specific tropomyosin isoform composition. J Histochem Cytochem. 2005;53:557–570. doi: 10.1369/jhc.4A6505.2005. [DOI] [PubMed] [Google Scholar]
- 39.Shevde NK, Bendixen AC, Dienger KM, Pike JW. Estrogens suppress RANK ligand-induced osteoclast differentiation via a stromal cell independent mechanism involving c-Jun repression. Proc Natl Acad Sci U S A. 2000;97:7829–7834. doi: 10.1073/pnas.130200197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takenaga K, Masuda A. Restoration of microfilament bundle organization in v-raf-transformed NRK cells after transduction with tropomyosin 2 cDNA. Cancer Lett. 1994;87:47–53. doi: 10.1016/0304-3835(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Xie Y, Du QS, Wu XJ, Feng X, Mei L, McDonald JM, Xiong WC. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J Cell Biol. 2003;160:565–575. doi: 10.1083/jcb.200207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber KL, Fischer RS, Fowler VM. Tmod3 regulates polarized epithelial cell morphology. J Cell Sci. 2007;120:3625–3632. doi: 10.1242/jcs.011445. [DOI] [PubMed] [Google Scholar]