Abstract
microRNAs (miRNAs) are extensively involved in developmental programming. Some miRNAs are highly conserved, while others are lineage specific. All miRNAs maturate through a series of processing steps. Here we review recent progresses in the studies of early steps in miRNA biogenesis, focusing on animal systems. The miRNA maturation pathways are surprisingly diverse, involving transcription by RNA polymerase II or III, cleavage by the Drosha nuclease or the spliceosome, and sometimes modifications by the adenosine deaminase ADAR. The relationship between the diversity in miRNA biogenesis and the apparently rapid evolution of miRNA genes and functions is discussed.
Keywords: microRNA, ribonuclease, RNA-binding proteins, RNA polymerase, spliceosome, adenosine deaminase for RNA
Introduction
miRNAs are a class of non-protein-coding RNAs that are ~22nt in length. They are involved in important biological functions, such as development and cell physiology [1]. Over 700 human miRNAs have been cloned [2]. They regulate the expression of ~30% of protein-coding genes by targeting specific messenger RNAs for cleavage or translational regulation, in a combinatorial fashion [3]. In animals, miRNAs often lead to translational repression, and to some extent mRNA decay, through partially complementary pairing to the 3′ untranslated regions (UTRs) of their targets using the seed region (positions 2 to 7 or 8 from 5′ end). In plants, miRNAs show perfect complementarity to their targets and lead to mRNA degradation. A recent article showed that translational inhibition is also widely effective in gene silencing by plant miRNAs and siRNAs, drawing more similarities between animal and plant miRNAs [4]. Some miRNAs have been shown to be oncogenic, or suppress tumor growth [5]. Several miRNAs are involved in metastasis [6, 7]. Artificial miRNAs that mimic natural pri- or pre-miRNAs (termed small hairpin RNAs, or shRNAs) are being used as a tool for genomic research and for therapeutic purposes [8–10]. The mechanisms of posttranscriptional regulation by miRNAs are nicely reviewed in [3]. In this review, we will focus on recent studies of the early steps in miRNA biogenesis pathways in animals, and discuss their implications in the evolution of miRNA genes and functions.
miRNAs contain 5′-phosphate and 3′-OH, thus they can be cloned using a specific procedure involving size selection and adaptor ligations [11–14]. Most miRNAs are named miR-#, where # represents a number. miRNA sequences are found in either the 5′ or 3′ strand in a hairpin secondary structure of their precursors. If the mature miRNA is located on the 5′ strand, it is called miR-#-5p; if it is located on the 3′ strand it is called miR-#-3p. These common features of miRNAs are defined by miRNA processing factors, as will be discussed later. miRNAs have similar length and chemical structures to another class of small RNAs, called small interfering RNAs (siRNAs) [15]. siRNAs are generated from long double-stranded RNAs, which could be replication intermediates of viruses or transcripts of transposable elements.
The major miRNA maturation pathway
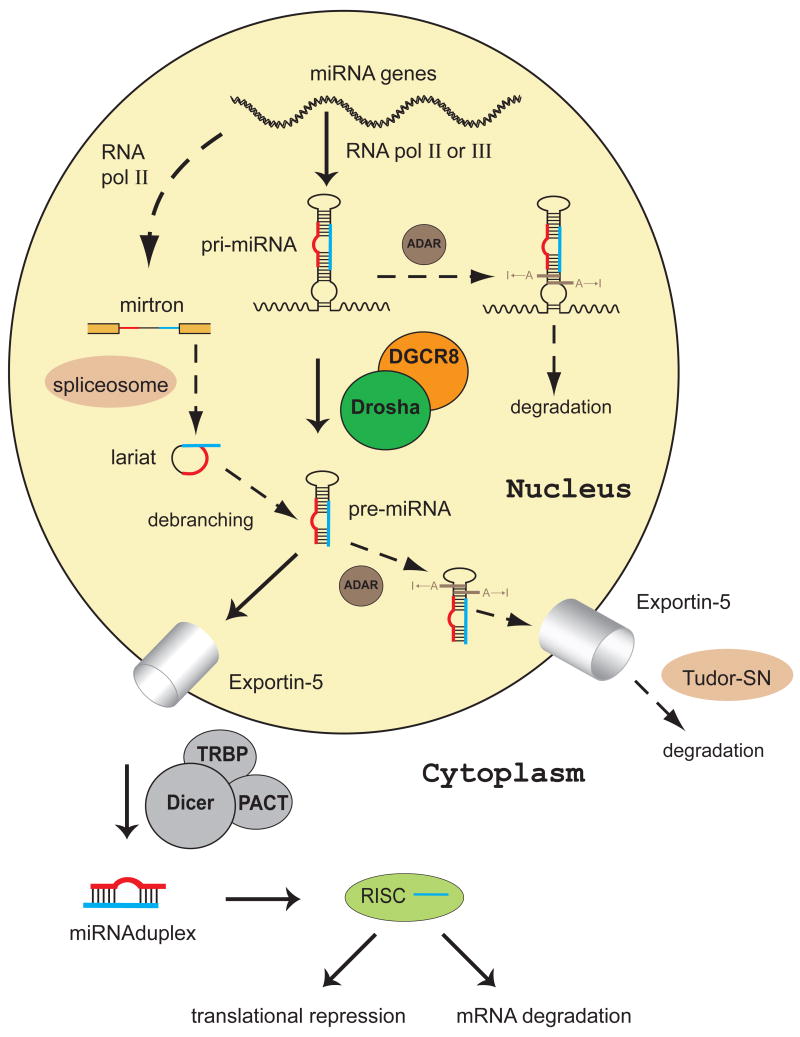
miRNAs are synthesized in cells as long primary transcripts (pri-miRNAs) that often contain thousands of nucleotides [16]. Pri-miRNAs are cleaved by a series of cellular processing factors. The major miRNA processing pathway in animals is illustrated in Fig. 1. A long pri-miRNA is first recognized and cleaved by a ribonuclease III (RNase III), called Drosha, along with an RNA-binding protein DGCR8 or Pasha (Partner of Drosha)[17–21]. Jointly, Drosha and DGCR8 form the Microprocessor complex [18, 19]. The product of the first cleavage event, pre-miRNA with ~70 nt, is then transported across the nuclear membrane by Exportin-5 and Ran-GTP[22, 23].
Fig. 1.
Diverse miRNA maturation pathways in animals. miRNA genes are transcribed by RNA-polymerase II or III into long transcripts termed pri-miRNA. In the major miRNA maturation pathway (solid arrows), a pri-miRNA is cleaved into an intermediate called pre-miRNA by Drosha, an RNase III protein, along with its RNA-binding partner DGCR8. The pre-miRNA, ~ 70 nucleotides in length, is exported by exportin-5 into the cytoplasm, where it is further cleaved by another RNase III enzyme Dicer to yield a miRNA duplex. The miRNA duplex is incorporated into the effector complex RISC and unwound into a mature single strand. The mature miRNA leads to translational repression or mRNA degradation, depending on the complementary to the target mRNAs. Alternative processing pathways have been uncovered (dashed arrows). A class of miRNAs, called mirtrons, is bracketed by splice site sequences. They are processed into pre-miRNAs by the spliceosome and the debranching enzyme. The A-to-I modification of pri-miRNAs and/or pre-miRNAs by the adenosine deaminase ADAR may block their cleavage by Drosha or Dicer, or alter the specificity of the miRNAs. The non-processable RNA species get degraded by nucleases, e.g. Tudor-SN in the cytoplasm.
In the cytoplasm, the pre-miRNA is further cleaved at a pair of sites close to the hairpin loop by another RNase III enzyme Dicer to give a miRNA duplex[24–29]. The miRNA duplex is then incorporated into the effector complex RISC (RNA induced silencing complex)[30–32], likely through the activity of the RISC loading complex, which contains Dicer, the central RISC component Argonaute and RNA-binding proteins (TRBP and PACT [33–37] in humans, and loquacious [38–40] in flies). A miRNA duplex is unwound into the mature single-stranded form (called the guide strand) [41] and its complementary strand (the passenger strand or miRNA*) is degraded by RISC[33, 42–44]. Mature miRNAs often predominantly originate from one strand of the duplex. Asymmetric incorporation of siRNA strands has been observed [45, 46]. The Drosophila R2D2, the dsRNA binding partner of Dicer-2, binds the end of the duplex with more stable base pairing interaction, providing the biochemical basis for preferential use of siRNA strands [47]. A similar mechanism might be responsible for miRNA strand selection. The stem loop in pre-miRNAs, presumably when bound to the RISC loading complex, likely contributes to the strand selection [48]. The miRNA* also has a chance to be incorporated into RISC and can be actively used in gene regulation [49].
The extensive processing of miRNA transcripts ensures that only RNA molecules with the proper structural features can be used in gene regulation [50]. These structural features are recognized by the miRNA processing factors. For example, it has been shown that Dicer cleaves double-stranded RNA into 21-nt duplexes preferentially from the end of the helix [51, 52]. In addition, the stem and 3′ overhang of pre-miRNAs are important for association with Exportin-5 and Ran-GTP[53]. The recognition of pri-miRNAs by Drosha and DGCR8 is the first step in miRNA maturation, and probably contains the most important constraints for miRNA processing. The RNA structures within an 80-nt hairpin region and the nonstructured regions flanking the hairpin have been shown to be important for processing by Microprocessor [17, 54–56]. The Drosha cleavage sites are about 10-bp from the junction between the miRNA hairpin and the flanking regions [57]. It is agreed that DGCR8 plays a major role in the recognition of pri-miRNAs [57–59]. However, it is far from clear how DGCR8 and Drosha distinguish pri-miRNAs from the large number of other hairpin-containing RNA molecules. In addition, the DEAD-box RNA helicases p68 and p72, which are found in the Microprocessor complex [19], are required for processing of a subset of miRNAs [60]. Their helicase activity is required for the maturation of these miRNAs and the subsequent gene silencing [60, 61]. These helicases may bind pri-miRNAs and modulate the activity of the Microprocessor complex through structural reorganization [60], or facilitate the loading of miRNA duplexes into RISC [19]. siRNAs are also generated through cleavage by Dicer [24]. Mammals only have one Dicer gene: knockout or knockdown of Dicer affects both miRNAs and siRNAs [62, 63], whereas knockdown of Drosha or DGCR8 only affects miRNA maturation [64–67].
Analysis of the genomic locations of miRNA genes indicated that many miRNAs are located in the introns of mRNAs, with some even in the coding regions [68]. A co-transcription of a miRNA with its host gene presumably allows them to be regulated together. Interestingly, a recent report showed that miR-21 is located in an intron of a coding gene, TMEM49; but its transcription is likely driven by its own promoter [69]. Thus, location of a miRNA in an intron does not warrant co-regulation with its host gene. Other miRNAs are in the intergenic regions and are likely transcribed independently. Stems of miRNA hairpins tend to be conserved; the variation of the loop sequences is increased; and there is a sharp decrease in conservation in sequences immediately flanking the hairpin [70]. This type of phylogenetic shadowing profile was successfully used to identify novel miRNAs in primates. There is a tendency in miRNAs to cluster together [11, 12, 31]. Clustering of miRNA hairpins presumably simplified their transcription regulation. Several recent advances in the study of miRNA biogenesis further underscore the diverse origin of miRNAs and will be discussed below.
Pri-miRNAs can be transcribed by either RNA polymerase II or III
Most pri-miRNAs are likely products of the RNA polymerase II [71, 72]. miRNAs regulate the expression of genes through pairing interactions with mRNAs, especially through their 3′ untranslated regions. Therefore, the expression of miRNAs must be regulated. Since transcription by RNA polymerase II is subjected to the highest degree of regulation out of the three RNA polymerase families, it is not surprising that most pri-miRNAs are transcribed by RNA polymerase II. While it makes sense most pri-miRNAs are transcripts of RNA polymerase II, there is no reason to exclude other RNA polymerases to be used. In fact, RNA polymerase III promoters such as U6 and H1 are often used to transcribe shRNAs [8]. In 2006, it was reported that ~50 human miRNAs are transcribed by RNA polymerase III [73]. In a genomic analysis, the miRNAs in the human chromosome 19 miRNA cluster (C19MC) were found to be dispersed among Alu repeats. Alu sequence is the most abundant transposable element in the human genome. It is derived from the 7SL RNA gene, which encodes the RNA component of the signal recognition particle that functions in protein synthesis. The Alu sequence contains the 7SL promoter, an RNA polymerase III promoter. Subsequent chromatin immunoprecipitation and cell-free transcription assays confirmed that the miRNAs in C19MC are indeed products of RNA polymerase III.
Pre-miRNA can be generated by either Drosha or the spliceosome
Many miRNAs are located within introns. They are in general cleaved by Drosha, parallel to the splicing of their host mRNAs [74]. Deep sequencing of D. melanogaster and C. elegans RNAs allowed the identification of a novel class of intronic miRNAs that do not contain the 10-bp helix at the base of the miRNA hairpin normally required for Drosha cleavage [75, 76]. These pre-miRNAs (mirtrons) turned out to be processed directly by the spliceosome, instead of Drosha (Fig. 1). Like other introns, mirtrons are flanked by 5′ and 3′ splicing sites, and contain branch point sequences. Unlike the pre-miRNAs generated by Drosha, the lariat mirtrons generated by the spliceosome need to be linearized by the debranching enzyme and fold into hairpins, prior to exportation to the cytoplasm by Exportin-5. Both Drosha-processed pre-miRNAs and mirtrons are processed by Dicer. It should be noted that mirtrons are a subset of the miRNAs transcribed by RNA polymerase II.
Pre-miRNAs are typically 60–100 nt in length. This size range coincides with the size of small introns in flies and nematodes. Mammalian introns are usually much longer than pre-miRNAs, thus fewer mirtrons were predicted to exist in mammalian genomes [75]. However, a recent report demonstrated that mirtrons appeared to be present in human and other mammalian genomes [77]. Although several mirtrons are highly conserved within Drosophilids, nematodes and mammals, no mirtron is collectively shared by these animals, suggesting that mirtrons are acquired independently during evolution of different animal clades.
Pri-miRNAs and pre-miRNAs can be modified by RNA editing enzymes
Cleavage by Drosha and Dicer are not the only RNA processing events that miRNAs can go through during maturation. ADAR (adenosine deaminase that acts on RNA) can convert some adenosines (A) to inosine (I) in double stranded RNAs (Fig. 1). I prefers to base-pair with C. The A→I modification disrupts a stable A:U base pair and creates a less stable I:U mismatch. ADAR is the most common type of RNA editing enzyme in metazoans [78]. Editing of pri-miR-142, suppresses its processing by Drosha [79]. Pri-miR-151 and possibly pre-miR-151 are modified by ADARs at two sites close to the Dicer cleavage sites, and these modifications block cleavage by Dicer [80]. Thus, the A-to-I editing could regulate miRNA biogenesis. Interestingly, an A-to-I editing site of miR-376 is located in the “seed” region critical for the recognition of miRNA targets [81]. This modification redirects miR-376 to silence a different set of genes. Therefore, target redirection through ADAR activity can increase the functional diversity of miRNAs. In addition to adenosine deamination, recent deep sequencing of miRNAs extracted from human and rodent showed addition of a single nucleotide at the 3′-end of the miRNAs [82, 83] and cytosine deamination [83]. The functional importance of these modifications remains to be demonstrated.
Possible roles of diverse miRNA biogenesis pathways in evolution
miRNAs can be generated in animals through transcription by either RNA polymerase II or III; as independent transcripts, or together with other genes; from introns or exons. Their precursors can be processed by either Drosha or the spliceosome in the nucleus and be modified by RNA editing enzymes. The diverse pathways to generate functional small non-coding RNAs are further highlighted by miRNAs in plants [84]. Plant genomes do not encode Drosha and DGCR8 homologues; instead miRNAs are processed by the Dicer-like proteins. Unlike their animal counterparts, plant miRNAs are methylated at their 3′ ends through the activity of HEN1. Therefore, the consensus for miRNA biogenesis is: there’s more than one way to skin a cat. As long as the mature miRNAs have the chemical structure and the length required for interaction with RISC and for subsequent gene regulation, it is probably advantageous to have multiple ways to generate them.
Obviously, the distinct pathways allow miRNAs to be controlled through different mechanisms [85]. For example, the Lin-28 RNA-binding protein specifically associates with the let-7 family pri-miRNAs and blocks their processing by Drosha and Dicer [86]. The SMAD proteins, a TGF-β and BMP signal transducers, promote the processing of pri-miR-21 by Drosha [87]. hnRNP A1, a nucleocytoplasmic shuttling protein, binds specifically to human pri-miR-18a and facilitates its Drosha-mediated processing[88]. We found that the DGCR8 protein binds heme and this interaction may be part of a molecular mechanism that regulates miRNA maturation [58]. ADAR editing of pri-and/or pre-miRNAs allows tissue-specific regulation of their processing and target specificity [79–81]. The presence of multiple miRNA maturation pathways predicts that altering the expression or activity of miRNA processing factors changes the abundance of only a subset of miRNAs.
It is possible that distinct pathways used to generate miRNAs allow these small gene regulators to evolve more readily, thus conferring fitness to their hosts. It was suggested that the mirtron pathway may have helped the emergence of miRNAs before the advent of Drosha [75]. When deep sequencing of miRNAs from three species of Drosophila was performed, a large class of evolutionarily young miRNAs was observed [89]. The estimated birth rate of new miRNA genes is 12 per million years. Among the new genes, 96% disappeared quickly in the course of evolution, 4% were retained, and only 2.5% became modestly or highly expressed. Furthermore, sequencing and comparative analysis of the C19MC in nine diverse primate species suggested an Alu-mediated rapid expansion of this miRNA family [90]. Single nucleotide polymorphisms (SNPs), the most abundant form of DNA variation in the human genome, are found in miRNA genes and may provide a snapshot of miRNAs during evolution [91]. Some miRNA SNPs have been shown to alter miRNA processing and target specificity [92, 93]. Editing by ADAR allows miRNA isoforms with different target specificity to be generated from a single miRNA gene [81]. The emergence of new miRNAs correlates with the introduction of developmental complexity during evolution, suggesting important functional contributions from the expansion of miRNA genes, as well as other non-protein-coding RNAs [94, 95]. Large numbers of miRNA-like hairpin structures [96, 97] and mirtron-sized introns [75–77] found in animal genomes could provide potential novel miRNA candidates to facilitate the fast evolution of miRNAs.
Acknowledgments
This work is supported by NIH grant GM080563-01A1 to F.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bushati N, Cohen SM. microRNA Functions. Annu Rev Cell Dev Biol. 2007 doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 7.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 9.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 12.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 19.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 25.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 26.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 27.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 30.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 31.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 33.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. Embo J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 38.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 42.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 43.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 44.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 47.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 48.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb Symp Quant Biol. 2006;71:73–80. doi: 10.1101/sqb.2006.71.042. [DOI] [PubMed] [Google Scholar]
- 53.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 56.Gottwein E, Cai X, Cullen BR. A Novel Assay for Viral MicroRNA Function Identifies a Single Nucleotide Polymorphism That Affects Drosha Processing. J Virol. 2006;80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 58.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 59.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat Struct Mol Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 60.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O’Malley BW, Kato S. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 61.Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282:32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- 62.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 70.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 71.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 74.Kim YK, Kim VN. Processing of intronic microRNAs. Embo J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 79.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X. MicroRNA metabolism in plants. Curr Top Microbiol Immunol. 2008;320:117–136. doi: 10.1007/978-3-540-75157-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smalheiser NR. Regulation of mammalian microRNA processing and function by cellular signaling and subcellular localization. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbagrm.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 89.Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 90.Zhang R, Wang YQ, Su B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 2008;25:1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- 91.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 93.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 95.Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]