Abstract
Aldehyde dehydrogenases (ALDHs) catalyze the irreversible oxidation of a wide range of reactive aldehydes to their corresponding carboxylic acids. Although the proteins have been studied from various organisms and at different growth stages, their roles in seed development have not been well elucidated. We obtained T-DNA insertional mutants in OsALDH7, which is remarkably inducible by oxidative and abiotic stresses. Interestingly, endosperms from the osaldh7 null mutants accumulated brown pigments during desiccation and storage. Extracts from the mutant seeds showed a maximum absorbance peak at 360 nm, the wavelength that melanoidin absorbs. Under UV light, those extracts also exhibited much stronger fluorescence than the wild type, suggesting that the pigments are melanoidin. These pigments started to accumulate in the late seed developmental stage, the time when OsALDH7 expression began to increase significantly. Purified OsALDH7 protein showed enzyme activities to malondialdehyde, acetaldehyde, and glyceraldehyde. These results suggest that OsALDH7 is involved in removing various aldehydes formed by oxidative stress during seed desiccation. The mutant seeds were more sensitive to our accelerated aging treatment and accumulated more malondialdehyde than the wild type. These data imply that OsALDH7 plays an important role in maintaining seed viability by detoxifying the aldehydes generated by lipid peroxidation.
The major regulatory factors that control seed aging are oxidative stress, lipid peroxidation, and respiration (Sun and Leopold, 1995; Bailly et al., 1996; Akimoto et al., 2004). Lipid peroxidation and respiration result in the formation of reactive aldehydes such as malondialdehyde (MDA) and acetaldehyde, which tend to react with proteins and amino acids (Mueller, 1998; Almeras et al., 2003; Weber et al., 2004). Those reactions cause aging and seed damage (Zhang et al., 1995, 1997). Until recently, a physiological approach has been taken in research on seed aging, and molecular and genetic studies have been seldom reported (Clerkx et al., 2004a).
Rice (Oryza sativa) is an important food crop, especially in Asia. Although stress tolerance has been extensively evaluated in an effort to develop advanced cultivars, the aging of seeds is an important economic problem that has been rarely examined (Devaiah et al., 2007). Their deterioration by lipid peroxidation leads to undesirable taste, color, and odor (Robertson et al., 1973; Nakayama et al., 1981; List et al., 1992). This is caused by various reactive aldehydes containing MDA, which are volatile aromatic compounds. Those aldehydes also bind with proteins and amino acids nonenzymatically, resulting in the accumulation of brown pigments during seed storage (Sun and Leopold, 1995).
Aldehydes are intermediates in several fundamental metabolism pathways for carbohydrates, vitamins, steroids, amino acids, and lipids (Yoshida et al., 1998). They are also produced in response to environmental stresses that disturb metabolism, including salinity, dehydration, desiccation, cold, and heat shock (Bartels, 2001). Although indispensable for an organism, excessive amounts threaten seed survival because of their chemically reactive nature and the toxic effect of the molecules (Lindahl, 1992). Therefore, aldehyde levels in cells must be regulated tightly.
Aldehyde dehydrogenases (ALDHs) are represented by a protein super-family that can be categorized into 21 families in eukaryotes (Perozich et al., 1999; Sophos et al., 2001; Sophos and Vasiliou, 2003; Fong et al., 2006). Some ALDHs are substrate specific while others have a highly variable substrate specificity (Sophos and Vasiliou, 2003). ALDHs catalyze the irreversible oxidation of a wide range of reactive aldehydes to their corresponding carboxylic acids (Perozich et al., 1999; Kirch et al., 2005). Therefore, they play a pivotal role in detoxifying the aldehydes generated by environmental stresses. Plant enzymes are represented in 11 ALDH families (Kirch et al., 2004). Despite their importance, the physiological functions of ALDHs in plants have rarely been studied (Liu et al., 2001; Liu and Schnable, 2002; Sunkar et al., 2003; Tsuji et al., 2003).
Family 7 ALDHs (antiquitins) are comparatively distinguishable from other ALDH families, because they show low sequence identity (approximately 30%) to ALDHs from other families (Vaciliou et al., 1999). However, the amino acid sequences among Family 7 members are highly homologous (about 60%–70%; Lee et al., 1994; Fong et al., 2006). In contrast to animal antiquitins that do not exhibit any inducible response to stresses (Lee et al., 1994; Fong et al., 2006), antiquitin in garden pea (Pisum sativum) is inducible by dehydration (Guerrero et al., 1990).
Overexpression of Arabidopsis (Arabidopsis thaliana) and soybean (Glycine max) ALDH7 confers tolerance to osmotic and oxidative stresses in transgenic plants (Kotchoni et al., 2006; Rodrigues et al., 2006). Moreover, their MDA and hydrogen peroxide contents are decreased. This suggests that ALDH7s function not only as aldehyde-detoxifying enzymes but also as efficient scavengers of reactive oxygen species and as lipid peroxidation-inhibiting enzymes.
In this study, we used null mutants in the rice ALDH7 gene to investigate the functional roles of ALDH7 during seed development and storage.
RESULTS
Isolation of a Mutant That Accumulates Brown Pigments in Mature Seeds
Screening T-DNA insertional mutant populations for abnormality in their mature seeds resulted in the identification of a mutant that accumulates brown pigments (Fig. 1A). These pigments were found in the pericarp as well as the inner endosperm (Fig. 1B). This pattern is unusual, because pigments are usually accumulated mainly in the pericarp in color-seed cultivars. The level of pigment increased as the storage time was extended (Fig. 1B). This implies that such accumulation is induced by a factor generated during seed maturation and the storage period.
Figure 1.
Phenotypes of T-DNA-tagged mutant line that accumulates brown pigment in seeds. A, Wild type (WT; left), heterozygous (middle), and homozygous (right) plants. Arrows indicate mutant seeds in heterozygous line. B, Cross sections of WT (left), mutant seeds immediately after harvest (middle), and seeds at 1 year after storage (right). C, Cross sections of WT seeds before (left) and after (right) heat treatment. D, Cross sections of mutant seeds before (left) and after (right) heat treatment. Ho, Homogenic mutant.
During the late stage of seed development and in storage, the water content in rice seeds dropped to <20%, which caused stress to the cells that still survived. Post-harvest heating to dry those seeds was another source of stress for the aleurone and embryo cells. To examine whether pigments were generated by these stresses, we treated wild-type and mutant seeds for 2 months at 60°C. This exposure induced pigment accumulation in the wild-type seeds (Fig. 1C) while enhancing such accumulation in the mutants (Fig. 1D).
The Pigment Is Likely Melanoidin
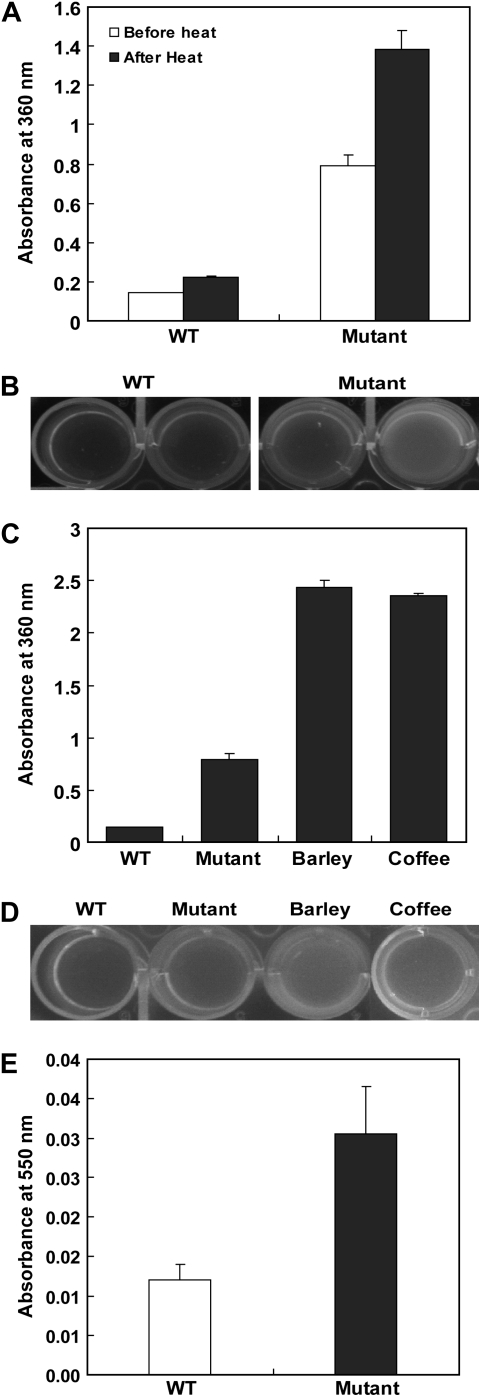
To analyze the components of this accumulated pigment, we scanned the absorption spectra of the aqueous extracts from wild-type and mutant seeds. Extracts from both genotypes peaked at 360 nm, although the mutant extract showed a peak that was up to 4 times higher (Fig. 2A). Heat treatment of seeds for 2 months at 60°C increased the absorbance in both the mutant and wild type (Fig. 2A). Under UV light, the extracts displayed fluorescence, with intensity being much greater from the mutants (Fig. 2B). These results suggest that the pigment is a product of a Maillard reaction, which nonenzymatically produces melanoidin from carbonyl and amino compounds during storage (Adams et al., 2005; Papetti et al., 2006; Adams and Brown, 2007; Niquet and Tessier, 2007). High temperatures and long response times are major factors for melanoidin production. Its absorbing wavelength of approximately 360 nm (Adams et al., 2005) is characteristic of fluorescent materials. Therefore, the pigments accumulated in our mutant seeds are likely melanoidin compounds.
Figure 2.
Absorbance patterns for mature seeds of rice, barley, and coffee. A, Comparison of absorbance patterns for pigment extracts from wild type (WT) or mutant before or after heat stress. B, Fluorescence intensity under UV light for samples in A. C, Comparison of absorbance patterns for extracts from rice, barley, and coffee. D, Fluorescence intensity under UV light for samples in C. E, Measurement of Amadori products of WT and mutant seeds. Seed proteins (200 μg) isolated from stored seeds were used for the test. The protein solution was mixed with nitro blue tetrazolium reagent. After incubation at 42°C for 10 to 20 min, absorbance was measured at 550 nm.
For further verification, we compared our rice extracts with those from the seeds of barley (Hordeum vulgare) and coffee (Coffea arabica), two well-known materials rich in melanoidin. Again, peaks occurred at 360 nm, with their heights being correlated with pigment intensity (Fig. 2C). Those extracts also contained fluorescent materials (Fig. 2D).
Amadori products are intermediates of the Maillard reaction (Sun and Leopold, 1995; Murthy and Sun, 2000). Therefore, measurement of those products has been commonly used for analyzing Maillard reaction products (Chandra et al., 2008). Here, we purified proteins from the seed extracts on 10-DG columns, and the Amadori products were measured by the nitro blue tetrazolium method (Murthy and Sun, 2000). This analysis showed that mutant seeds accumulated up to 3 times more products compared with the wild type (Fig. 2E). These results indicate that the pigments extracted from mutant seeds are likely melanoidin produced by the Maillard reaction.
The Mutant Phenotype Results from a Knockout of OsALDH7 by T-DNA Insertion
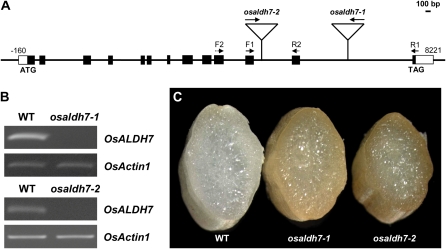
Because the mutant phenotype cosegregated with T-DNA, we determined DNA sequences for the T-DNA flanking region by inverse PCR (An et al., 2003; Jeong et al., 2006). Analysis of the flanking sequence revealed that T-DNA was inserted in the 13th intron of OsALDH7 (Fig. 3A). We designated that mutant as osaldh7-1. We also isolated another allele, osaldh7-2, where T-DNA was located in the 12th intron of OsALDH7 (Fig. 3A). Reverse transcription (RT)-PCR analyses of transcripts accumulated in the seeds showed that OsALDH7 was not expressed in both alleles (Fig. 3B), in which higher levels of brown pigments were accumulated (Fig. 3C).
Figure 3.
Characterization of T-DNA insertional mutants. A, T-DNA insertion positions within OsALDH7. T-DNA is inserted into 12th and 13th introns in osaldh7-1 and osaldh7-2 alleles, respectively. Black boxes indicate exons and white boxes indicate 5′- and 3′-untranslated regions. Solid lines between exons are introns. B, RT-PCR analyses of OsALDH7 expression in osaldh7 mutants, with primers F1, F2, R1, and R2. OsActin1 was used to monitor equal loading. C, Cross sections of mature seeds from WT (left), osaldh7-1 (middle), and osaldh7-2 (right).
To examine the subcellular region in which OsALDH7 functions, we conducted a localization experiment using protoplasts purified from Oc suspension cells. OsALDH7-GFP protein was localized in the cytoplasm, which was colocalized with mRFP, a well-known cytosol marker (Supplemental Fig. S1). This result is consistent with a previous report of Arabidopsis ALDH7B4 localization (Kotchoni et al., 2006).
OsALDH7 Expression Is Dramatically Increased under Oxidative Stress
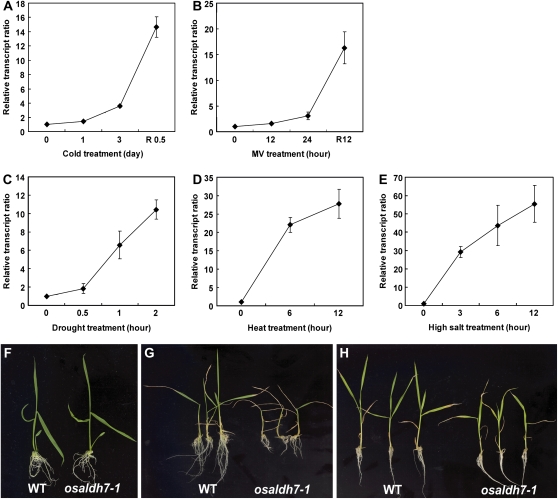
Kotchoni et al. (2006) and Rodrigues et al. (2006) have reported that ALDH7 in Arabidopsis and soybean functions to detoxify aldehydes generated under oxidative stresses. To examine whether rice ALDH7 also plays an important role in abiotic stresses, we investigated OsALDH7 expression patterns after seedlings were treated with cold, heat, drought, paraquat, or high salt. Transcript levels were significantly induced by such stresses (Fig. 4). Interestingly, two expression patterns were revealed. Whereas OsALDH7 transcripts were not increased during cold and paraquat (MV) treatment but increased remarkably during the recovery period following the treatment (Fig. 4, A and B), those transcripts were elevated significantly during the application of drought, heat, or high salt (Fig. 4, C–E). The cold and MV treatments were probably too harsh to sustain most physiological reactions. However, when treated seedlings were returned to normal conditions, their responses such as lipid peroxidation by membrane degradation started to accelerate. We speculate that the OsALDH7 transcription level was increased to remove aldehydes that formed within the lipid peroxidation pathway. By comparison, drought, heat, and high salt stresses did not seriously inhibit cellular operation. Thus, those treated cells could respond to stress without much delay.
Figure 4.
Expression patterns of OsALDH7 during cold (A), MV (B), drought (C), heat (D), or high salt (E) treatments. For cold treatment, seedlings were placed at 4°C under lights for 3 d, then transferred to room temperature for recovery. For MV treatment, seedling roots were submerged in 20 μm MV solution for 1 d, then transferred to distilled water for recovery. For heat stress, seedlings were treated at 50°C. Drought stress was introduced by placing plants on air at 30°C. For high salt stress, seedlings were treated with 250 mm NaCl. Relative transcript ratio implies OsALDH7 transcript levels normalized by OsActin1 mRNA level. One-week-old wild-type (WT) and osaldh7-1 seedlings grown under normal growth condition (F). G and H, Phenotypes of WT (left) and osaldh7-1 (right) seedlings after cold (G) and high salt (H) stresses.
Wild type and osaldh7-1 mutants were germinated and grown on Murashige and Skoog (MS) media. Under the normal growth conditions, the mutant plants were not different from wild type (Fig. 4F). When osaldh7-1 seedlings were treated with cold or high salt, they were more sensitive than the wild type (Fig. 4, G and H). Therefore, we conclude that OsALDH7 is needed for the detoxification of aldehydes that form under various stress conditions.
OsALDH7 Expression Patterns during Seed Development Correspond with Pigment Accumulation Patterns in Seeds
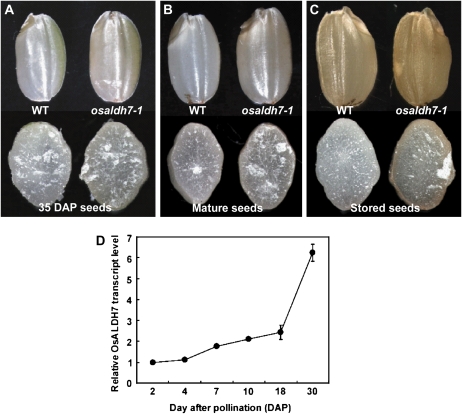
Pigment accumulation started in the late stage of seed development (35 d after pollination), mainly in pericarp tissues (Fig. 5A), with a higher amount of pigment being accumulated at the fully mature stage (Fig. 5B). Stored seeds showed even further accumulation, especially in the mutant (Fig. 5C).
Figure 5.
Pigment accumulation during seed development. A to C, Intact (top) or cross-sectioned (bottom) seeds of 35-d-old (A), mature (B), and stored (C) samples from wild type (WT; left) and osaldh7-1 (right). D, Expression pattern of OsALDH7 during seed development analyzed by real-time PCR.
We looked for correlations between pigment accumulation and ALDH7 transcript levels during seed development. Real-time PCR analyses indicated that those levels were low early on but then significantly increased at the late stage (Fig. 5D). A sudden rise in OsALDH7 expression occurred at the time when pigment accumulation started. This timing coincides with a rapid decline in water content in seeds (Hoshikawa, 1989), suggesting that OsALDH7 is associated with desiccation. Therefore, the phenotypes observed in our osaldh7 mutants were likely due to a failure to remove the aldehydes generated by dehydration stress.
Enzyme Activity of OsALDH7
We examined whether OsALDH7 indeed encodes for an enzyme that catalyzes aldehydes. The OsALDH7 full-length cDNA clone was inserted into the pET-topo vector containing six His residues. This molecule was introduced into Escherichia coli strain BL21 (DE3), and OsALDH7-His tag fusion molecules were induced by isopropylthio-β-galactoside treatment. Using a His-tag column, we purified OsALDH7-His protein and performed immunoblots to demonstrate that the purified protein was OsALDH7 (data not shown).
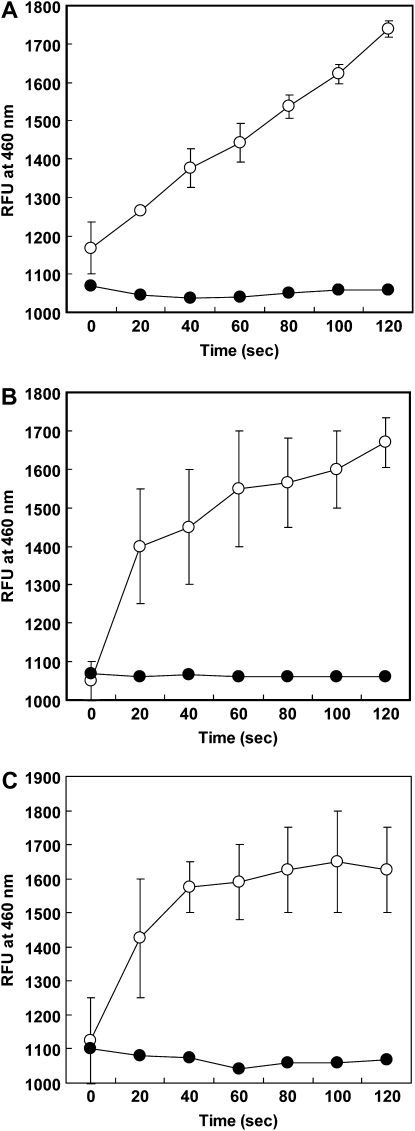
The purified protein had enzyme activity against MDA, a by-product formed in the nonenzymatic lipid peroxidation pathway (Fig. 6A). MDAs are electrophilic, strongly binding to peptides and amino acids. Therefore, the MDA content in cells is evidence of their environmental conditions and can be used as a general index of oxidative stress. Here, OsALDH7 catalyzed other aldehydes as well, including acetaldehyde (Fig. 6B) and glyceraldehyde (Fig. 6C). Acetaldehyde is generated from the alcohol fermentation pathway and glyceraldehyde is related with carbohydrate metabolism. These results suggest that OsALDH7 has broad substrates and may participate in multiple functions such as metabolic pathways and abiotic stresses.
Figure 6.
ALDH enzyme assay. OsALDH7 protein was produced in E. coli, with pET expression vector (pET-OsALDH7). ALDH activity was measured using MDA (A), acetaldehyde (B), and glyceraldehyde (C) as substrates. Extract of E. coli containing pET-U52 vector expressing rice U box gene U52 served as negative control. White circles, pET-ALDH; black circles, pET-U52.
MDA Contents Increase in osaldh7 Seeds
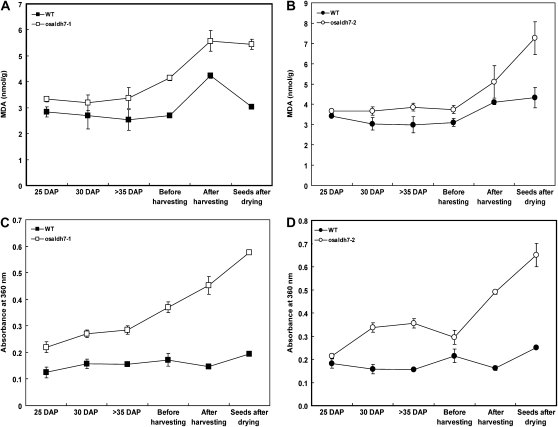
If one of the roles of OsALDH7 is to remove MDA, then MDA contents should be higher in the osaldh7 seeds than in the wild type. Therefore, we measured those contents in developing and mature seeds. During the former stage, seeds are exposed to severe oxidative stress due to desiccation. Such an environment promotes the acceleration of lipid peroxidation and an increase in MDA. Performing a thiobarbituric acid-reactive-substances assay, we found that, as expected, MDA contents were higher in the osaldh7 seeds (Fig. 7, A and B). Although contents in wild-type seeds were somewhat elevated, those in the mutants were more significantly increased (up to 2-fold), especially after seed harvest. Patterns were similar with A360 (Fig. 7, C and D). These data demonstrate that OsALDH7 plays a pivotal role in the removal of MDA that forms during seed desiccation.
Figure 7.
Correlation between MDA and Maillard reaction products during seed development and storage. A and B, MDA levels of wild-type (WT) and osaldh7-1 and osaldh7-2 seeds during late stages of seed development and storage. C and D, Absorbance patterns at 360 nm of the extracts from the mutant seeds (white) and WT (black).
osaldh7 Seeds Show Accelerated Seed Aging
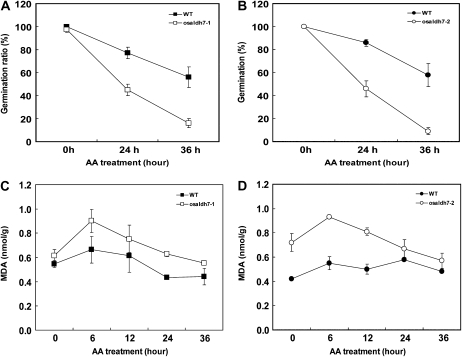
Because lipid peroxidation is involved in seed deterioration (Bailly et al., 1996, 1998), we hypothesized that the viability of osaldh7 mutant seeds would be diminished. Thereupon, we applied an accelerated aging (AA) treatment, a routine experiment (McDonald, 1999). Here, the germination rate was drastically decreased in the osaldh7 seeds to about one-half that of the wild type (Fig. 8, A and B). Mutant seeds also accumulated higher levels of MDA during the AA treatment (Fig. 8, C and D). These results indicate that OsALDH7 is required for the maintenance of seed viability, detoxifying the MDA that is generated during seed storage.
Figure 8.
AA treatment of wild-type (WT) and osaldh7 seeds. A and B, Germination rates of the mutants (white) and WT (black) after AA treatment. After 24- and 36-h treatments, seeds were germinated on MS media. C and D, MDA levels after AA treatment. Seeds were collected at 6-h intervals.
DISCUSSION
When ordinary seeds complete their maturation, they must become desiccated if long-term storage is to be successful (Murthy et al., 2003; Gilles et al., 2007). Their tolerance to dry conditions is based on their formation of desiccation-related proteins and materials (osmoprotectants), such as late embryogenesis abundant (LEA) proteins and sugars (Sun et al., 1994; Black et al., 1999; Wise and Tunnacliffe, 2004). LEA genes begin to be expressed in the mid- to late stage of seed development (Bartels et al., 1988; Hartwigsen and Goggi, 2002). Their overexpression enhances tolerance to salt, drought, and osmotic stresses in transgenic plants (Brini et al., 2007; Zhang et al., 2007). Here, we observed that levels of OsALDH7 transcripts started to increase at the late stage of maturation, similar to those of the LEA genes. Expression patterns for OsALDH7 implied that a relationship exists between OsALDH7 and seed desiccation. Kirch et al. (2005) have reported that expression of the Arabidopsis ALDH7B4 gene, in Family 7, is activated by abscisic acid, which regulates the dehydration stress response and the seed desiccation pathway. Other researchers have suggested that osmoprotection may be a major role of the plant antiquitins (Stroeher et al., 1995; Buchanan et al., 2005), compounds that oxidize aldehydes (Fong et al., 2006). Based on those previous studies and our results, we anticipated that OsALDH7 would function during seed desiccation and maturation. We observed that this gene was strongly expressed under abiotic stresses, e.g. drought, cold, and heat, and that osaldh7 mutant seedlings were more sensitive than the wild type. Our data, therefore, suggest that OsALDH7 can be used to confer tolerance to diverse stresses.
ALDHs detoxify the aldehydes generated by various metabolisms (Yoshida et al., 1998; Nakazono et al., 2000; Liu et al., 2001). During seed desiccation, the embryo and aluerone cells must face severe oxidative stress, which increases the level of nonenzymatic lipid peroxidation (Baryla et al., 2000). As a result, MDA is generated, which has a high reactive activity to other molecules such as proteins, amino acids, and DNA (Restow and Obe, 1978; Fraenkel-Conrat and Singer, 1988; Mueller, 1998; Vollenweider et al., 2000; Stintzi et al., 2001; Almeras et al., 2003; Weber et al., 2004). Therefore, MDAs stimulate the expression of genes involved in cell protection/detoxification as well as those implicated in tolerance to stresses such as heat shock and dehydration (Weber et al., 2004). Here, we observed that levels of MDA were higher in osaldh7 seeds during the late developmental stages and while being stored. This suggests that OsALDH7 is needed to reduce the MDA formed during seed dehydration. Our OsALDH7 enzyme assay showed this high reactivity with MDA, thus supporting our assumption.
The brown reaction, i.e. the Maillard reaction, occurs during storage or because of spontaneous heating (Murthy et al., 2003). This reaction is the nonenzymatic response between carbonyl compounds (e.g. reducing sugars and aldehydes) and amino compounds (peptides and amino acids). An increase in aldehyde levels promotes the reaction. During seed desiccation and storage, various aldehydes are formed by metabolisms and lipid peroxidation. The latter in particular is coupled to Maillard reactions during seed storage (Murthy and Sun, 2000; Zamora and Hidalgo, 2005). We observed here that the osaldh7 mutant seeds accumulated more brown pigments, probably due to an increase in MDA contents.
It is difficult to investigate the effect of natural aging, because long-term storage is needed. Here, seed germination rates after short-term storage did not differ significantly between wild type and mutants. Our observation is consistent with that of Sattler et al. (2004), who showed that the germination rate of the vte2 mutant seeds was not much different from that of wild-type seeds, although VTE2 plays an essential role in seed longevity by participating in the tocopherol synthetic pathway. Therefore, we employed the AA treatment, which is broadly used for studying seed aging (Bentsink et al., 2000; Clerkx et al., 2004b).The osaldh7 seeds showed decreased seed viability and generated more MDA after the AA treatment. This indicates that the mutant seeds are more sensitive to oxidative stress and also demonstrates that OsALDH7 is needed to sustain seed viability. Wettlaufer and Leopold (1991) have reported that AA treatment of soybean seeds dramatically increases the Maillard reaction. Furthermore, Sun and Leopold (1995) have suggested that Maillard reactions play a role in seed deterioration. Based on those previous reports and our current results, we postulate that the reduced viability of osaldh7 seeds was caused by brown pigments, the final product of the Maillard reaction.
Elleder (1981) and Uchida (2006) have reported that lipid peroxidation is involved in the formation of lipofuscin, a lipidic brown pigment, with characteristic fluorescence. Pigment accumulations result from binding between MDA and protein. The brown pigments that accumulated in our osaldh7 seeds are similar to lipofuscin in that they are fluorescent, MDA-derivative, and aging related. Therefore, we examined the solubility of the extract in diverse solvents, including water, methanol, ethanol, acetone, and hexan. However, the pigments were soluble only in water, indicating that they are not lipidic material and, thus, not likely to be lipofuscin.
We tried to investigate the chemical nature of this pigment by liquid chromatography-mass spectrometry and gas chromatography-mass spectrometry. However, we were unable to identify any candidate peaks. This may have been because melanoidins are highly polymerized compounds. The structure of those pigments has been poorly defined (Adams et al., 2005), such that no commercial standards are available for structural analysis.
In conclusion, we suggest the following working model. Lipid peroxidation is accelerated by oxidative stress in late-stage seeds, which causes an increase in MDA and other aldehydes. The toxic materials are removed by OsALDH7, inhibiting the Maillard reaction. However, in the osaldh7 mutant, MDA and other aldehydes are not removed, and a Maillard reaction is promoted, forming brown pigments in mature seeds.
MATERIALS AND METHODS
Plant Materials and Growing Conditions
Seeds of osaldh7 and segregant wild-type rice (Oryza sativa ‘Dongjin’) were sterilized with 50% hypochlorite for 30 min, washed three times with sterile distilled water, and placed on an MS medium (Murashige and Skoog, 1962). One-week-old seedlings were then transferred to soil and grown in the greenhouse. After 3 to 4 weeks, they were transplanted to a paddy field and grown to maturity. Their harvested seeds were air-dried for 1 week and stored at room temperature.
Screening of OsALDH7 Mutants from T-DNA Tagging Lines
Defective seeds of the mutants were isolated from T-DNA tagging lines by screening for alterations in their shape or color. The T-DNA insertion position was determined by inverse PCR (An et al., 2003; Jeong et al., 2006). Another allele of osaldh7 was obtained by searching the TES database (http://signal.salk.edu/cgi-bin/RiceGE).
Abiotic Stress Treatments
One-week-old seedlings were grown in an MS medium. To induce cold stress, the seedlings were transferred to 4°C and incubated for 3 d. During this treatment, they were harvested daily (beginning at d 0) for RNA isolation and real-time PCR analysis. After the 3-d test period, the seedlings were allowed to recover at room temperature for 12 h before sampling. For the heat treatment, seedlings were held at 50°C and sampled at 0, 6, and 12 h. For drought experiments, water was blotted from the seedling roots with paper towels before the seedlings were placed on a paper towel at 30°C. These treated seedlings were sampled at 0.0, 0.5, 1.0, and 2.0 h. The paraquat treatment involved submerging the seedling roots in a 20 μm MV solution, then sampling them at 12-h intervals. After 1 d, the seedlings were transferred to distilled water and sampled after 12 h. For high salt treatment, seedling roots were submerged in 250 mm NaCl for 12 h. Afterward, they were allowed to recover at room temperature.
Pigment Extraction and Analysis
Hulled-whole seeds (about 100 mg) of the wild type and osaldh7 were ground in a milling machine, and the powder was dissolved in 1 mL of distilled water. After vigorous vortexing, the solution was mixed at 4°C for 24 h on a rotator. This solution was then centrifuged at 13,000 rpm for 15 min, and the supernatant was retrieved into a new Eppendorf tube. The solution was filtered using a 0.2-μm syringe filter (Sartorius). The extracts were scanned with a UV spectrophotometer (Shimadzu) over a spectral range of 200 to 700 nm. The extracts were also observed under UV light to determine whether the materials were fluorescent.
Measurement of Amadori Products
Stored seeds (10 mg) were ground with a milling machine before 1.2 mL of 50 mm phosphate buffer, pH 7.2, was added to the powder. Nucleic acids were removed by adding 200 μL of 10% streptomycin sulfate, then centrifuging at 15,000 rpm for 15 min. The supernatant was transferred to a new tube, and proteins were precipitated with ammonium sulfate (0.55 g mL−1). The proteins precipitated by centrifugation were dissolved in 50 mm phosphate buffer, pH 7.2, and the protein solution was further purified on 10-DG columns (Bio-Rad). Those purified proteins were then used for measurement of Amadori products by the nitro blue tetrazolium method, as described previously (Murthy and Sun, 2000).
RT-PCR and Quantitative Real-Time PCR
Samples were homogenized in a milling machine (Retsch), and total RNA was extracted using TRIzol reagent (Invitrogen). For first-strand cDNA synthesis, 2 μg of total RNA was reverse-transcribed in a total volume of 25 μL that contained 10 ng of oligo(dT)12-18 primer, 2.5 mm dNTPs, and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) in a reaction buffer. After RT at 37°C for 90 min, RT-PCR was conducted for 33 cycles in a 25-μL solution containing 20 pmol of gene-specific primers, 0.2 mm dNTPs, 1 unit of Taq DNA polymerase (Enzynomics), and 1× reaction buffer. Primers were designed at different exons, allowing the differentiation of cDNA products from genomic DNA contamination. Real-time PCR was performed using a Roche LightCycler II as previously described (Han et al., 2006). The procedure utilized a 20-μL solution containing 1 μL of cDNA solution, 20 pmol of gene-specific primers, 1× SYBR premix Ex Taq (TakaRa Shuzo). For RT-PCR analyses in osaldh7-1 and osaldh7-2, F1 (5′-ccaaaatttattccttgtgc-3′)/R1 (5′-aatctgaaggaagcagttga-3′) and F2 (5′-gtactgcttccacgaatcac-3′)/R2 (5′-cggttgtgagagaagaactc-3′) primer sets were used, respectively (Fig. 3A). The mRNA level of OsActin1 served to normalize the relative expression level of OsALDH7.
Vector Construction
To construct our localization vectors, we performed PCR with the following primers: 5′-gcACTAGTatggggagcttcgcgaggaa-3′ and 5′-gcACTAGTgccaaaatttattccttgt-3′ (underlined parts indicate SpeI-recognized sequences). Full-length cDNA of OsALDH7 was inserted into the pBluescript SK+ vector (Stratagene), which was digested with EcoRV. The subclone was digested with SpeI and then introduced into the SpeI site in pGA3452, which contains the sGFP gene driven by the maize (Zea mays) ubiquitin promoter. Protoplasts prepared from the Oc cell line of rice were transformed with the localization vectors by methods described previously (Han et al., 2006; Woo et al., 2007).
Enzyme Assays
Full-length OsALDH7 cDNA was isolated by the primers 5′-caccatggggagcttcgcgagg-3′ (forward primer) and 5′-ctagccaaaatttattcctt-3′ (reverse primer). The cDNA clone was inserted into pET100/d-TOPO, and the resulting plasmid was transformed into Escherichia coli strain BL21 (DE3). Transformed cells were incubated at 37°C overnight, then transferred to 250 mL of a fresh medium (1.6% tryptone, 1.0% yeast extract, and 0.5% NaCl) and incubated in a 37°C shaker. Once the OD600 reached 0.5 to 0.8, 1 mm isopropylthio-β-galactoside was added, and incubation was continued at 30°C for another 8 h. For OsALDH7 assays, cells were harvested and resuspended in 10 mL of B-PER protein extraction reagent (Thermo Fisher Scientific). This solution was incubated on ice for 20 min and sonicated three times, for 10 s each, using a Branson Sonifier model 450 at maximum output. The lysate was centrifuged at 12,000 rpm for 20 min. After the supernatant was transferred to a new tube, 6× His-tagged OsALDH7 proteins were purified on a Ni-NTA Spin column (Qiagen). For our enzyme assay, 10 μg of purified protein was added to a reaction mixture containing 1.5 mm NAD (Sigma) and 0.1 m sodium pyrophosphate buffer, pH 8.5. The total volume was adjusted to 300 μL with water before 20 μg of an aldehyde (MDA, acetaldehyde, or glyceraldehyde) was added to the mixture. Finally, the emission fluorescence of NADH was recorded for up to 2 min, at 20-s intervals, on a SPECTRAMAX Gemini (Elsevier Biosoft).
Lipid Peroxidation Assay
Levels of lipid peroxidation were assayed according to the thiobarbituric acid test, which determines the amount of MDA as an end product of the reaction (Cakmak and Horst, 1991; Loreto and Velikova, 2001). Seeds (0.2 g) were homogenized in 5 mL of 0.1% (w/v) TCA solution on ice. After the homogenate was centrifuged at 3,000 rpm for 10 min, the supernatant (0.5 mL) was transferred to a new tube, and 1 mL of 20% (w/v) TCA solution containing 0.5% thiobarbituric acid was added. The mixture was kept in a boiling water bath for 30 min, then quickly cooled in ice. Following centrifugation at 2,000 rpm for 15 min, absorbance of the supernatant was measured at 532 nm and 600 nm.
AA Test of Seeds
To promote AA, the hulls from 50 seeds each of the wild type and osaldh7-1 and osaldh7-2 mutants were removed. The seeds were transferred to a high-humidity chamber (100% relative humidity) and incubated at 50°C for 24 and 36 h. These AA-treated seeds were then surface-sterilized with 50% hypochlorite and placed on an MS medium. Germination rates were scored 7 d after sowing. This experiment was repeated three times.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF323586.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of OsALDH7∷sGFP fusion protein.
Supplementary Material
Acknowledgments
We thank Hea-kyung Jung, Hee-Jung Woo, Hyunsook Lee, Jeonghi Lee, Kyoungmi Han, and Kyung-Mi Kim for assisting in sequencing the T-DNA insertion sites; In-Soon Park and Kyungsook An for generating the T-DNA insertional lines; Nam-In Baek and Jong-Soo Yoo for analyses of pigment components; Yang-Seok Lee, Yongmin Woo, Sunok Moon, and Jong-jin Park for valuable discussion; and Priscilla Licht for critical reading of the manuscript.
This work was supported in part by the Crop Functional Genomic Center, the 21st Century Frontier Program (grant no. CG1111), by the Biogreen 21 Program (grant no. 20070401–034–001–007–03–00), Rural Development Administration, by the Korea Science and Engineering Foundation through the National Research Laboratory Program funded by the Ministry of Science and Technology (grant no. M10600000270–06J0000–27010), and by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund; grant no. KRF–2007–341–C00028).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gynheung An (genean@postech.ac.kr).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Adams A, Borrelli RC, Fogliano V, Kimpe ND (2005) Thermal degradation studies of food melanoidins. J Agric Food Chem 53 4136–4142 [DOI] [PubMed] [Google Scholar]
- Adams JB, Brown HM (2007) Discoloration in raw and processed fruits and vegetables. Crit Rev Food Sci Nutr 47 319–333 [DOI] [PubMed] [Google Scholar]
- Akimoto T, Cho S, Yoshida H, Furuta H, Esashi Y (2004) Involvement of acetaldehyde in seed deterioration of some recalcitrant woody species through the acceleration of aerobic respiration. Plant Cell Physiol 45 201–210 [DOI] [PubMed] [Google Scholar]
- Almeras E, Stolz S, Vollenweider S, Reymond P, Mene-Saffrane L, Farmer EE (2003) Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J 34 202–216 [DOI] [PubMed] [Google Scholar]
- An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, et al (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D (1996) Changes in malondialdehyde content and in superoxide dismutase, catalase and glutathione reductase activities in sunflower seeds as related to deterioration during accelerated aging. Physiol Plant 97 104–110 [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D (1998) Free radical scavenging as affected by accelerated aging and subsequent priming in sunflower seeds. Physiol Plant 104 646–652 [Google Scholar]
- Bartels D (2001) Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci 6 284–286 [DOI] [PubMed] [Google Scholar]
- Bartels D, Singh M, Salamini F (1988) Onset of desiccation tolerance during development of the barley embryo. Planta 175 485–492 [DOI] [PubMed] [Google Scholar]
- Baryla A, Laborde C, Montillet JL, Triantaphylides C, Chagvardieff P (2000) Evaluation of lipid peroxidation as a toxicity bioassay for plants exposed to copper. Environ Pollut 109 131–135 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M, Corbineau F, Gee H, Come D (1999) Water content, raffinose, and dehydrins in the induction of desiccation tolerance in immature wheat embryos. Plant Physiol 120 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A, Pages M, Masmoudi K (2007) Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep 26 2017–2026 [DOI] [PubMed] [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, Klein RR, Pratt LH, Cordonnier-Pratt MM, Klein PE, et al (2005) Sorghum bicolor's transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol 58 699–720 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Horst WJ (1991) Effect of aluminium on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol Plant 83 463–468 [Google Scholar]
- Chandra R, Bharagava RN, Rai V (2008) Melanoidins as major colourant in sugarcane molasses based distillery effluent and its degradation. Bioresour Technol 99 4648–4660 [DOI] [PubMed] [Google Scholar]
- Clerkx EJ, Blankestijn-De Vries H, Ruys GJ, Groot SP, Koornneef M (2004. a) Genetic differences in seed longevity of various Arabidopsis mutants. Physiol Plant 121 448–461 [Google Scholar]
- Clerkx EJ, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M (2004. b) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol 135 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang X (2007) Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J 50 950–957 [DOI] [PubMed] [Google Scholar]
- Elleder M (1981) Chemical characterization of age pigments. In RS Sohal, ed, Age Pigments. Elsevier/North-Holland Biomedical Press, Amsterdam, pp 204–241
- Fong WP, Cheng CHK, Tang WK (2006) Antiquitin, a relatively unexplored member in the superfamily of aldehyde dehydrogenases with diversified physiological functions. Cell Mol Life Sci 63 2881–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Singer B (1988) Nucleoside adducts are formed by cooperative reaction of acetaldehyde and alcohols: possible mechanism for the role of ethanol in carcinogenesis. Proc Natl Acad Sci USA 85 3758–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles GJ, Hines KM, Manfre AJ, Marcotte WR Jr (2007) A predicted N-terminal helical domain of a Group 1 LEA protein is required for protection of enzyme activity from drying. Plant Physiol Biochem 45 389–399 [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Jones JT, Mullet JE (1990) Turgor-responsive gene transcription and RNA levels increase rapidly when pea shoots are wilted. Sequence and expression of three inducible genes. Plant Mol Biol 15 11–26 [DOI] [PubMed] [Google Scholar]
- Han MJ, Jung KH, Yi G, Lee DY, An G (2006) Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol 47 1457–1472 [DOI] [PubMed] [Google Scholar]
- Hartwigsen JA, Goggi SA (2002) Expression of a Dehydrin-like protein in maize seedlings germinated from seed exposed to freezing. J Plant Biol 45 225–229 [Google Scholar]
- Hoshikawa K (1989) The Growing Rice Plant: An Anatomical Monograph. Nobunkyo, Tokyo, pp 256–257
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ (2004) The ALDH gene superfamily of Arabidopsis. Trends Plant Sci 9 371–377 [DOI] [PubMed] [Google Scholar]
- Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol 57 315–332 [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29 1033–1048 [DOI] [PubMed] [Google Scholar]
- Lee P, Kuhl W, Gelbart T, Kamimura T, West C, Beutler E (1994) Homology between a human protein and a protein of the green garden pea. Genomics 21 371–378 [DOI] [PubMed] [Google Scholar]
- Lindahl R (1992) Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol 27 283–335 [DOI] [PubMed] [Google Scholar]
- List GR, Mounts TL, Lanser AC (1992) Factors promoting the formation of nonhydratable soybean phosphatides. J Am Oil Chem Soc 69 443–446 [Google Scholar]
- Liu F, Cui X, Horner HT, Weiner H, Schnable PS (2001) Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Schnable PS (2002) Functional specialization of mitochondrial aldehyde dehydrogenases. Plant Physiol 130 1657–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto F, Velikova V (2001) Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol 127 1781–1787 [PMC free article] [PubMed] [Google Scholar]
- McDonald MB (1999) Seed deterioration: physiology, repair and assessment. Seed Sci Technol 27 177–237 [Google Scholar]
- Mueller MJ (1998) Radically novel prostaglandins in animals and plants: the isoprostanes. Chem Biol 5 R323–333 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Murthy UMN, Kumar PP, Sun WQ (2003) Mechanisms of seed aging under different storage conditions for Vigna radiata (L.) Wilczek: Lipid peroxidation, sugar hydrolysis, Maillard reactions and their relationship to glass state transition. J Exp Bot 54 1057–1067 [DOI] [PubMed] [Google Scholar]
- Murthy UMN, Sun WQ (2000) Protein modification by Amadori and Maillard reactions during seed storage: roles of sugar hydrolysis and lipid peroxidation. J Exp Bot 51 1221–1228 [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Sayo K, Kito M (1981) Decomposition of phospholipids in soybean during storage. Cereal Chem 58 260–264 [Google Scholar]
- Nakazono M, Tsuji H, Li Y, Saisho D, Arimura SI, Tsutsumi N, Hirai A (2000) Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol 124 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niquet C, Tessier FJ (2007) Free glutamine as a major precursor of brown products and fluorophores in Maillard reaction systems. Amino Acids 33 165–171 [DOI] [PubMed] [Google Scholar]
- Papetti A, Daglia M, Aceti C, Quaglia M, Gregotti C, Gazzani G (2006) Isolation of an in vitro and ex vivo antiradical melanoidin from roasted barley. J Agric Food Chem 54 1209–1216 [DOI] [PubMed] [Google Scholar]
- Perozich J, Nicholas HB, Wang BC, Lindahl R, Hempel J (1999) Relationships within the aldehyde dehydrogenase extended family. Protein Sci 8 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restow H, Obe G (1978) Acetaldehyde induces cross-links in DNA and cause sister chromatide exchanges in human cells. Mutat Res 58 115–119 [DOI] [PubMed] [Google Scholar]
- Robertson JA, Morrison WH III, Burdick D (1973) Chemical evaluation of oil from field- and storage-damaged soybeans. J Am Oil Chem Soc 50 443–446 [Google Scholar]
- Rodrigues SM, Andrade MO, Gomes APS, Damatta FM, Baracat-Pereira MC, Fontes EPB (2006) Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J Exp Bot 57 1909–1918 [DOI] [PubMed] [Google Scholar]
- Sattler SC, Gilliland LU, Magallanes-Lundback M, Pollard M, Dellapenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophos NA, Pappa A, Ziegler TL, Vasiliou V (2001) Aldehyde dehydrogenase gene superfamily: the 2000 update. Chem Biol Interact 130–132 323–337 [DOI] [PubMed] [Google Scholar]
- Sophos NA, Vasiliou V (2003) Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact 143–144 5–22 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeher VL, Boothe JG, Good AG (1995) Molecular cloning and expression of a turgor-responsive gene in Brassica napus. Plant Mol Biol 27 541–551 [DOI] [PubMed] [Google Scholar]
- Sun WQ, Irving TC, Leopold AC (1994) The role of sugar, vitrification and membrane phase-transition in seed desiccation tolerance. Physiol Plant 90 621–628 [Google Scholar]
- Sun WQ, Leopold AC (1995) The Maillard reaction and oxidative stress during aging of soybean seeds. Physiol Plant 94 94–104 [Google Scholar]
- Sunkar R, Bartels D, Kirch HH (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 35 452–464 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Tsutsumi N, Sasaki T, Hirai A, Nakazono M (2003) Organ-specific expressions and chromosomal locations of two mitochondrial aldehyde dehydrogenase genes from rice (Oryza sativa L.), ALDH2a and ALDH2b. Gene 305 195–204 [DOI] [PubMed] [Google Scholar]
- Uchida K (2006) Lipofuscin-like fluorophores originated from malondialdehyde. Free Radic Res 40 1335–1338 [DOI] [PubMed] [Google Scholar]
- Vaciliou V, Bairoch A, Tipton KF, Nebert DW (1999) Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9 421–434 [PubMed] [Google Scholar]
- Vollenweider S, Weber H, Stolz S, Chetelat A, Farmer EE (2000) Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J 24 467–476 [DOI] [PubMed] [Google Scholar]
- Weber H, Chetelat A, Reymond P, Farmer EE (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J 37 877–888 [DOI] [PubMed] [Google Scholar]
- Wettlaufer SH, Leopold AC (1991) Relevance of Amadori and Maillard products to seed deterioration. Plant Physiol 97 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A (2004) POPP the question: what do LEA proteins do? Trends Plant Sci 9 13–17 [DOI] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su'udi M, Yang JI, Park JJ, Back K, Park YM, An G (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65 125–136 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Rzhetsky A, Hsu LC, Chang C (1998) Human aldehyde dehydrogenase gene family. Eur J Biochem 251 549–557 [DOI] [PubMed] [Google Scholar]
- Zamora R, Hidalgo FJ (2005) Coordinate contribution of lipid oxidation and Maillard reaction to the nonenzymatic food browning. Crit Rev Food Sci Nutr 45 49–59 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou RX, Zhang LJ, Wang RY, An LZ (2007) CbLEA, a novel LEA gene from Chorispora bungeana, confers cold tolerance in transgenic tobacco. J Plant Biol 50 336–343 [Google Scholar]
- Zhang M, Nagata S, Miyazawa K, Kikuchi H, Esashi Y (1997) A competitive enzyme-linked immunosorbent assay to quantify acetaldehyde-protein adducts that accumulate in dry seeds during aging. Plant Physiol 113 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Nakamura Y, Tsuda S, Nagashima T, Esashi Y (1995) Enzymatic conversion of volatile metabolites in dry seeds during storage. Plant Cell Physiol 36 157–164 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.