Abstract
Myosin-Va has been shown to have multiple functions in a variety of cell types, including a role in RNA transport in neurons. Using primary cultures of cells from organs of young dilute-lethal (Myo5ad-l/Myo5ad-l) null mutant mice and wild-type controls, we show that in some, but not all, tissues, RNA distribution is dramatically different in the homozygous null mutant cells. The dependence of RNA localization on myosin-Va correlates with the relative abundance of the brain-specific splicing pattern of the myosin-Va tail. We also show that myosin-Va is involved in RNA localization soon after synthesis, because the effects of its absence are diminished for RNAs that are more than 30 min old. Finally, we show that localization of β-actin mRNA is significantly changed by the absence of myosin-Va. These results suggest that myosin-Va is involved in a transient transport or tethering function in the perinuclear region. Cell Motil.
Keywords: Myosin-Va, mRNA transport, β-actin mRNA
INTRODUCTION
Myosin-Va, encoded by the Myo5a (originally dilute) locus [Mercer et al., 1991], is a member of one of the more ancient families within the myosin superfamily [Richards and Cavalier-Smith, 2005; Foth et al., 2006]. Multiple functions have been attributed to this molecular motor, including the transport/tethering of melanosomes, smooth endoplasmic reticulum, recycling endosomes, neurotransmitter vesicles, and mRNA [Reck-Peterson et al., 2000; Langford, 2002]). The absence of myosin-Va in homozygous null mutants causes neurological defects and lethality in dilute-lethal mice [Searle, 1952] and a subset of Griscelli syndrome cases in humans [Pastural et al., 1997]. Both null and hypomorphic Myo5a alleles cause dilutions in pigmentation caused by a mislocalization of melanosomes in skin melanocytes [Provance et al., 1996].
While the molecular basis underlying the pigmentation phenotype of Myo5a mutants is well-characterized [Matesic et al., 2001; Fukuda et al., 2002; Hume et al., 2002; Provance et al., 2002; Wu et al., 2006]), the basis for the mutant neurological phenotype is less clear. Endoplasmic reticulum is missing in dendritic spines of cerebellar Purkinje cells [Dekker-Ohno et al., 1996; Takagishi et al., 1996] from null mutant rats, long-term synaptic depression (LTD) is abolished in null mutant mice [Miyata et al., 2000], and antibody inhibition of myosin Va causes a delay in neurotransmitter release [Rosé et al., 2003]. While these differences can explain the neurological phenotype, a clear pathway has yet to be demonstrated.
A role for myosin-Va in neuronal RNA transport provides an additional mechanism that may explain the neurological phenotype. Myosin-Va is a member of a complex that includes Purα, mStaufen, Fragile X protein, a kinesin, and mRNA [Ohashi et al., 2002]. It also is found in motor axons at periaxoplasmic ribosomal plaques (PARPs) together with kinesin-II [Sotelo-Silveira et al., 2004], zipcode-binding protein 1 (ZBP1) and β-actin mRNA [Sotelo-Silveira et al., 2008], and myosin-Va mRNA [Sotelo-Silveira et al., 2006], suggesting a role in long-range mRNA transport in projection neurons. In more functional experiments, involvement of myosin-Va in movement of mRNA encoding Nd1-L (an F-actin-stabilizing protein) into dendritic spines was shown using null mutants, RNAi, and tail-fragment over-expression [Yoshimura et al., 2006].
Despite the severity of the neurological phenotype, it has not yet been shown that it is responsible for lethality in homozygous null mutant mice and humans. For this reason, and to identify more experimentally tractable systems than neurons for the study of myosin-Va’s role in RNA transport, we assayed RNA localization in primary cells isolated from multiple tissues of null mutant and wild-type control mice. We observed significant differences in RNA localization in fibroblasts cultured from peripheral nervous system, adipose tissue, and lung, while we observed no difference in fibroblasts from spleen. The deficit in null mutant adipose fibroblasts was rescued by transfection with a full-length myosin-Va construct. Using pulse-chase experiments with bromouridine labeling, we show that the differences were most pronounced within 30 min after RNA synthesis. We show a dramatic difference for a specific message known to be transported (β-actin), as well as evidence for the physical association of β-actin mRNA and myosin-Va. These results suggest a general role for myosin-Va in RNA localization in multiple cell types.
RESULTS
Total RNA Distribution Is Altered in Mutant PNS Fibroblasts
To determine if myosin-Va functions in RNA transport or distribution, we stained primary PNS cultures from homozygous null mutant and wild-type control mice with SYTO® RNASelect™, a cell-permeant nucleic acid stain that binds selectively to RNA over DNA. The results were striking; in the mutant cells, it often was difficult to distinguish nucleus from cytoplasm (Fig. 1A), while in wild-type control cells, perinuclear and nuclear fluorescence levels were very different (Fig. 1B). The difference was sufficiently robust that multiple observers blinded to genotype sorted micrographs from each individual experiment for genotype without error. However, we observed considerable variability between experiments; that is, when experiments were combined, observers incorrectly sorted some micrographs.
Fig. 1.
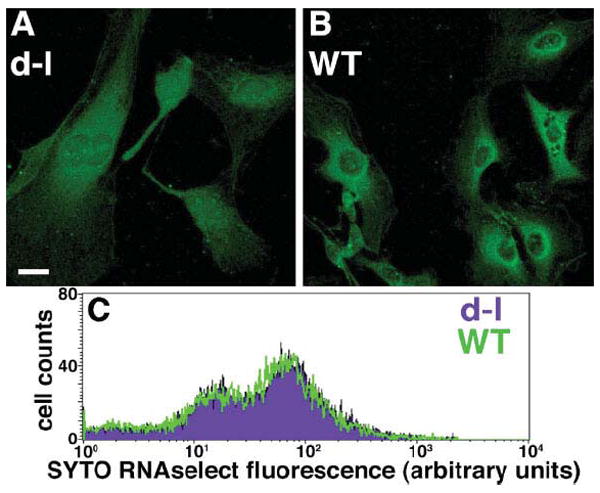
A and B, SYTO® RNASelect™ fluorescent labeling of nucleic acid in homozygous null (dilute-lethal) Myo5a mutant (A) and wild-type control (B) PNS cultures. Bar = 15 μM. (C) flow cytometry analysis of mutant (purple) and wild-type (green) cells labeled with SYTO® RNASelect™.
To confirm that these were differences in distribution of bulk RNA, and not artifacts caused by different amounts of total RNA, different cell shapes, or different proportions of cell types, we quantified total RNA per cell by flow cytometry of the same cells that were trypsinized, stained with SYTO® RNASelect™ in suspension, and fixed. We observed no difference in RNA content between null mutant and wild-type cells in two independent assays (Fig. 1C), confirming that the micrographs represent differences in total RNA distribution.
The Mutant RNA Distribution Phenotype Is Observed in Primary Fibroblasts From Some, but Not All, Other Tissues
We observed a different pattern of total RNA localization in fibroblasts isolated from adipose tissue and lung than we did in cells from the more heterogeneous PNS cultures described above. In mutant fibroblasts, we observed higher fluorescence in the nucleus than in the cytoplasm (Fig. 2A), while in wild-type control fibroblasts, the difference was less pronounced (Fig. 2B). We also observed concentrated fluorescence just inside the nuclear membrane in mutant fibroblasts (arrows, Fig. 2A). We quantified fluorescence over 20-pixel-long, 5-pixel-wide regions spanning the nuclear/cytoplasmic boundaries, which gave statistically significant differences between wild-type and mutant for PNS, adipose, and lung fibroblasts (Figs. 2D–2F). However, we observed no difference in RNA distribution between wild-type and mutant spleen fibroblasts (Fig. 2G). One difference between tissues is the splicing of the tail domain [Seperack et al., 1995; Huang et al., 1998]); spleen has very little of the brain splicing pattern (Fig. 2C). Together with the results shown in Fig. 1, these results suggest that myosin-Va functions during RNA synthesis or in RNA transport from or immediately outside the nucleus, and that this involves the myosin-Va isoform with the brain-specific splicing pattern.
Fig. 2.
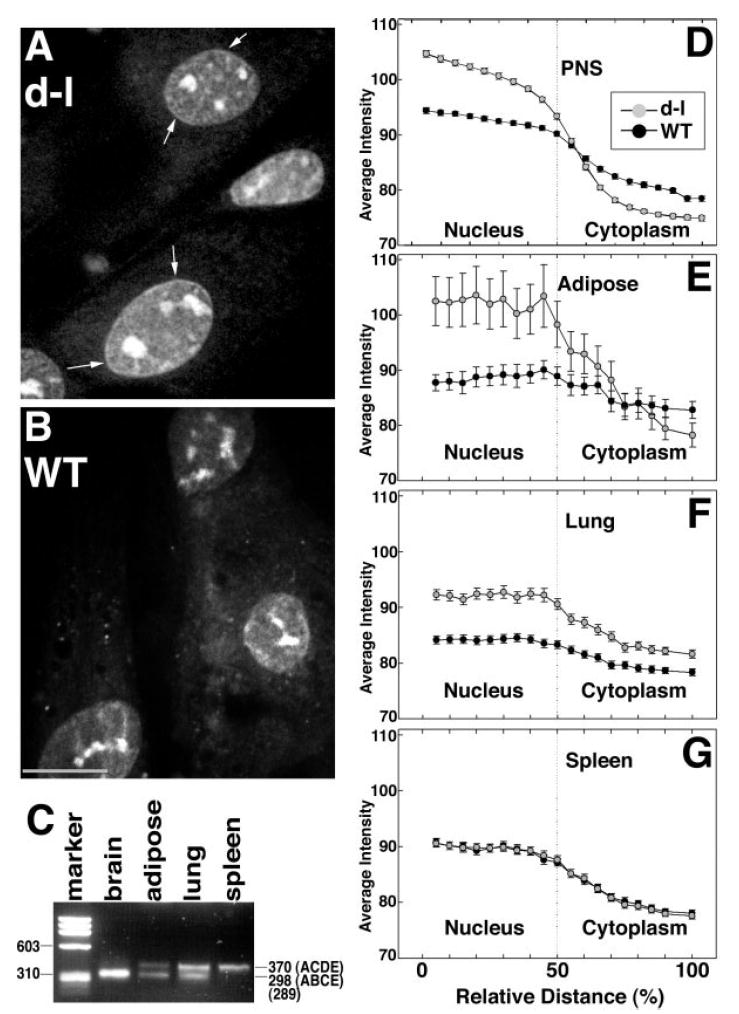
A and B, SYTO® RNASelect™ labeling of adipose fibroblasts from homozygous null mutant (A) and wild-type control (B) mice. Arrows denote the thin band of higher fluorescence at the edge of the nucleus that was more commonly observed in mutant fibroblasts. Bar = 10 μm. (C) RT-PCR illustrating myosin-Va tail splicing in RNA from brain (amount loaded was reduced ten-fold to compensate for its high abundance) and from cultures of adipose, lung, and spleen cells. ACDE and ABDE refer to previously-described splicing patterns [Seperack et al., 1995]. The marker lane is PhiX174 RF DNA cleaved with HaeIII. D-G, quantitation (error bars represent standard error) of SYTO® RNASelect™ fluorescence over 20-pixel-long, 5-pixel-wide regions with their midpoints set at the nuclear/cytoplasmic boundaries of peripheral nervous system (D), adipose (E), lung (F), and spleen (G) cells. Numbers of cells measured for each genotype/cell type combination ranged from 91 to 122, in three independent experiments.
Myosin-Va’s Role in Transport/Tethering Is Early After Transcription
On the assumption that the difference between mutant and wild-type cells observed around the nuclear/ cytoplasmic boundary represented transport or tethering events following soon after transcription, we switched from SYTO® RNASelect™ staining of total, steady-state RNA to bromouridine (BrU) labeling of newly-synthesized RNA.
We tested for myosin-Va function during transport by varying chase times after loading with BrU for 60 min, removing the BrU for 30, 60, or 120 min before fixation. We observed more intense BrU labeling in the nuclei of mutant adipose fibroblasts (relative to cytoplasm) after a 30 min chase (Fig. 3A). By contrast, the ratio of nuclear to cytoplasmic BrU labeling in wild-type control fibroblasts was significantly lower (Fig. 3B).
Fig. 3.
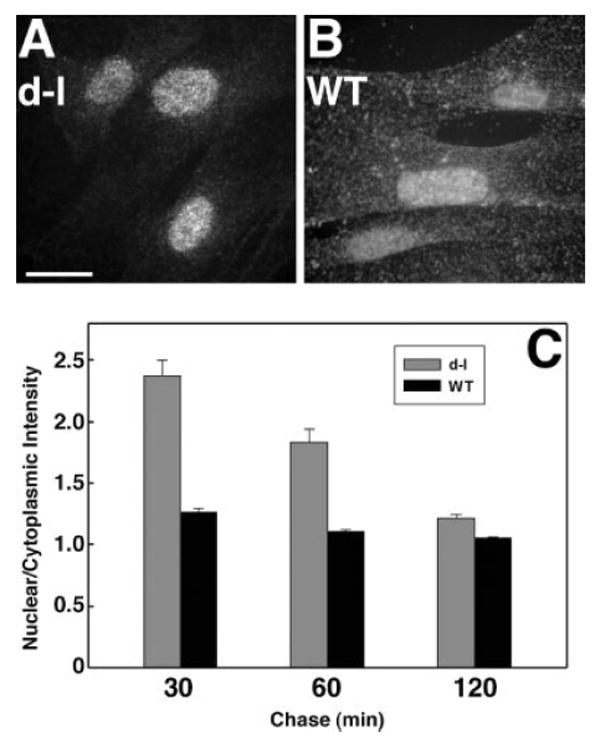
A and B, bromouridine (BrU) incorporation in homozygous null mutant (A) and wild-type control (B) primary adipose fibroblasts incubated with BrU for 60 min and chased with medium lacking BrU for 30 min. Bar = 15 μm. (C) ratio (with standard error) of nuclear to cytoplasmic labeling as a function of genotype and chase duration. Each bar represents measurements from 129 to 156 cells in three independent experiments.
We quantitated RNA transport by measuring the ratio of BrU labeling in nuclei versus cytoplasm for each chase interval (Fig. 3C). We observed statistically-significant differences at all time points, but the differences at shorter chase intervals were much larger, suggesting that myosin-Va is involved in transport early after synthesis, in conjunction with a slower transport mechanism that does not require myosin-Va. These experiments also were performed on the heterogeneous PNS cultures shown in Fig. 1 with similar results (data not shown).
The Absence of Myosin-Va Has No Effect on RNA Synthesis
Since the results described above suggest that myosin-Va functions during or soon after RNA synthesis, we assayed the distribution of newly synthesized RNA by labeling with bromouridine without a chase interval (Fig. 4). Quantitation revealed no differences between mutant and wild-type cells after 10–60 min of BrU loading (Fig. 4G). These data suggest that the absence of myosin-Va affects neither BrU uptake nor RNA synthesis.
Fig. 4.
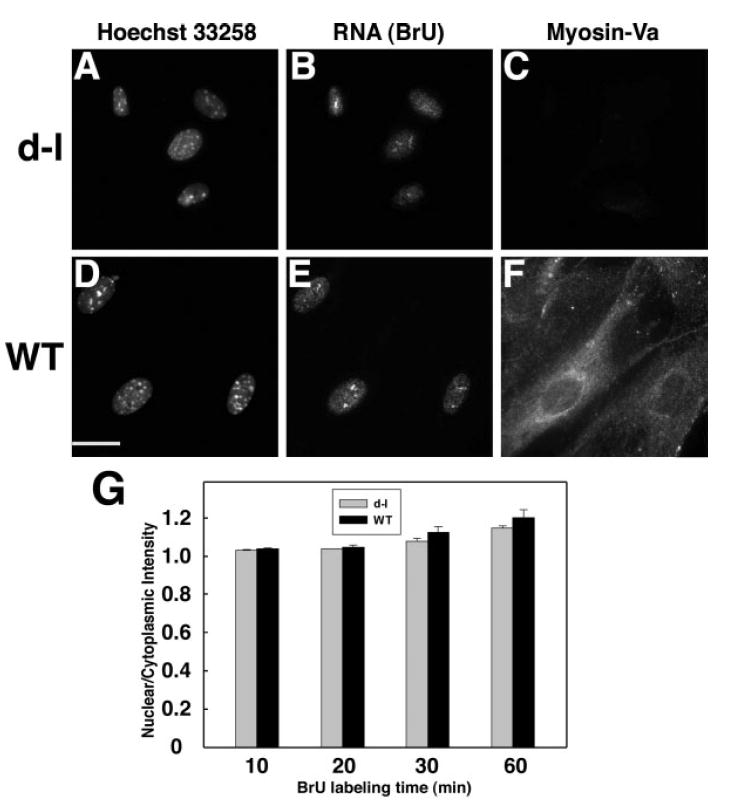
A-F, fluorescence micrographs of homozygous null (A–C) and wild-type control (D–F) adipose fibroblasts stained for DNA with Hoechst 33258 (A,D), newly-synthesized RNA by BrU incorporation followed by anti-BrdU (B,E), and anti-myosin-Va (C,F). Bar = 15μm. (G) distribution of newly-synthesized RNA as a function of time of exposure to BrU in homozygous null mutant (gray bars) and wild-type control (black bars) primary adipose fibroblasts. Each bar represents measurements taken from 99 to 150 cells in three independent experiments.
Rescue of the Mutant Phenotype by Expression of Myosin-Va
To determine if the mutant RNA-distribution phenotype could be rescued by expression of myosin-Va, we transfected homozygous null mutant adipose fibroblasts with a full-length myosin-Va expression construct encoding the brain splicing pattern (ABCE; [Seperack et al., 1995]). On the day after transfection, newly-synthesized RNA was labeled using BrU under the load/ chase conditions described above, nuclear/cytoplasmic fluorescence ratios were measured, and transfected cells were identified by immunofluorescent detection of myosin-Va. The BrU quantitation is shown in Fig. 5A, and newly-synthesized RNA and identification of transfected cells by myosin-Va expression are shown in Figs. 5B and 5C respectively. These data show that mutant cells expressing myosin-Va had nuclear/cytoplasmic fluorescence ratios different from neighboring untransfected cells, and similar to ratios observed in wild-type cells (Fig. 3), with significant differences for 30 and 60 min chase times, but not for 120 min. No difference was observed between transfected and untransfected cells at 10–60 min of BrU loading without a chase period (data not shown). We also observed an overall reduction in the ratios due to the transfection protocol (contrast Y-axis scales of Figs. 3C and 5A); we also observed a similar reduction in labeling of mock-transfected mutant cells in the absence of DNA (data not shown). Transfection of multiple heterologous control DNAs had no effect on the nuclear/cytoplasmic ratio (data not shown).
Fig. 5.
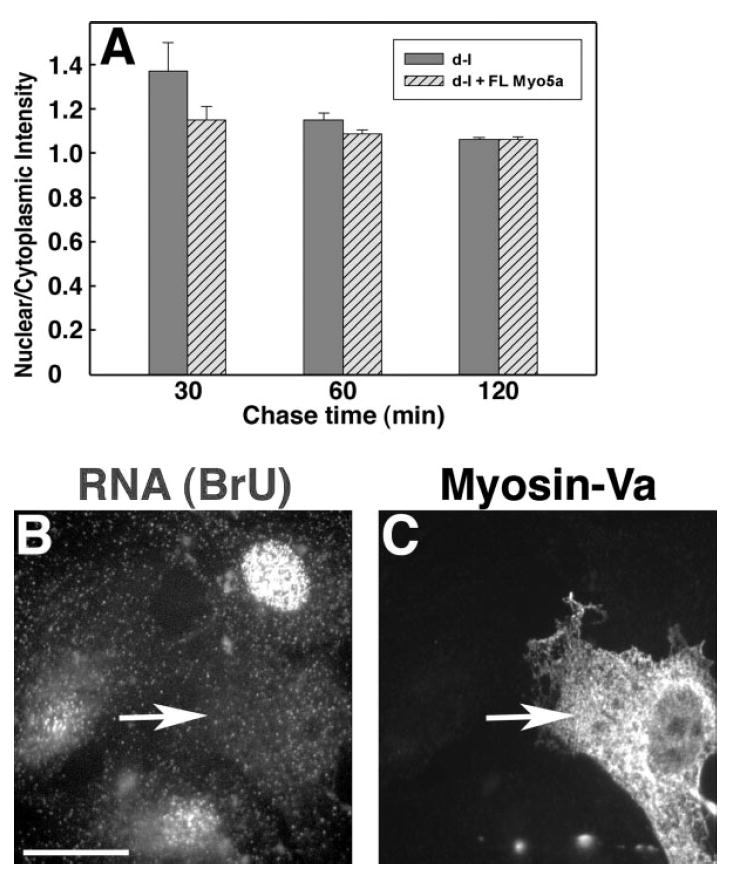
Expression of myosin-Va in null mutant cells rescues the RNA distribution phenotype. (A) nuclear/cytoplasmic intensities of control Myo5a null mutant (d-l, gray bars) and null mutant expressing full-length myosin-Va (d-l + FL Myo5a, hatched bars) adipose fibroblasts. Each bar represents measurements taken from 93 to 130 cells in three independent experiments. (B) representative field after 60′ BrU labeling; the nucleus of the transfected cell (arrow) is indistinguishable. (C) immunofluorescent detection of myosin-Va in transfected cell (arrow), same field as panel B. Bar = 15 μm.
β-Actin Transcript Distribution Is Altered in Mutant Cells
The data described above suggest that myosin-Va functions in the export of newly-synthesized RNA from the nucleus, or transport immediately outside the nucleus. To test the hypothesis that myosin-Va is required for the transport of specific transcripts, we performed in situ hybridization on homozygous null mutant and wild-type cells using a β-actin antisense probe. Homozygous null mutant cells (Fig. 6A and B) had a dispersed distribution, while wild-type control cells (Figs. 6C and 6D) were labeled primarily at the periphery with less-intense perinuclear labeling. Confocal optical sections (not shown) showed that the labeling is not within the nucleus. A negative control probe with the scrambled sequence gave no significant signal (data not shown).
Fig. 6.
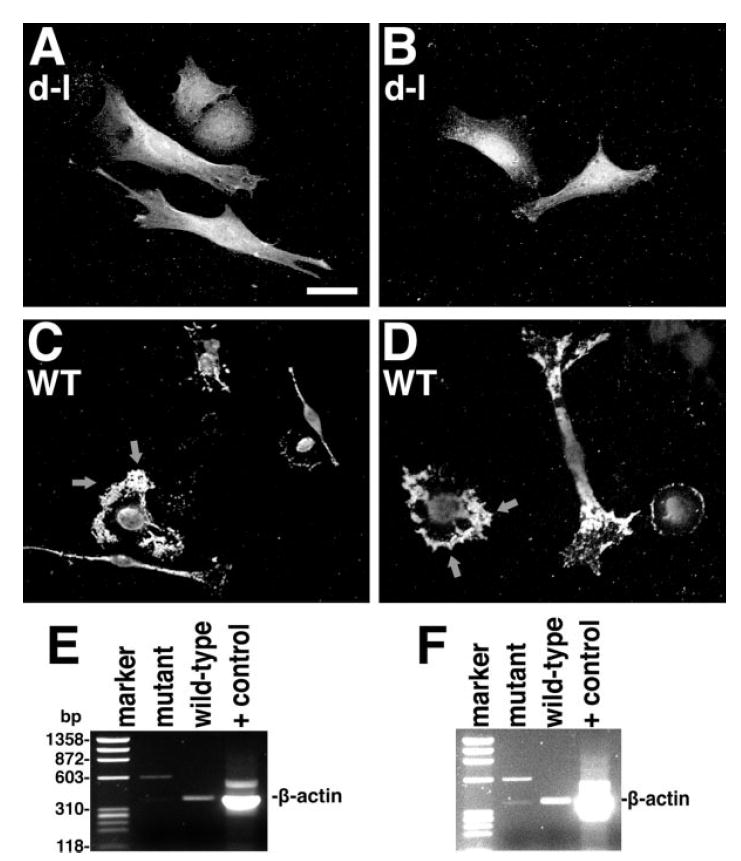
A-D, in situ hybridization of homozygous null mutant (A, B) and wild-type control (C, D) PNS cells with a fluorescent probe for β-actin mRNA; arrows denote the high concentration of β-actin mRNA at the leading edge. Bar = 20μm. (E, F) RT-PCR of anti-myosin-Va immunoprecipitates from homozygous null mutant and wild-type control primary adipose fibroblasts. In F, the levels have been increased to show the faint, but expected band in the mutant control lane, as well as the absence of background bands in the wild-type experimental lane, demonstrating the absence of contamination during gel loading from the positive control lane (RT-PCR from total HeLa cell RNA). The marker lane is PhiX174 RF DNA cleaved with HaeIII.
To test for a physical interaction between myosin-Va and β-actin mRNA, we immunoprecipitated myosin-Va from mutant and wild-type cytoplasmic fractions and assayed for the presence of β-actin mRNA using RT-PCR. As shown in Figs. 6E and 6F, β-actin transcripts were associated with myosin-Va in wild-type samples, but not in the mutant negative control sample.
DISCUSSION
Multiple functions for members of the myosin-V family have been characterized. Given the ancient origin of this family [Richards and Cavalier-Smith, 2005; Foth et al., 2006], this diversity of function is not surprising. These multiple functions make it more difficult to determine the molecular deficiencies responsible for the neurological and lethal phenotypes observed in homozygous Myo5a null mutant mice and humans homozygous for null mutant MYO5A alleles (Griscelli syndrome) [Pastural et al., 1997].
Directional transport and localization of mRNA has been demonstrated to provide temporal and spatial control of gene expression [Huang and Richter, 2004]. In neurons, the asymmetric distribution of mRNA contributes to synaptic plasticity [Burdwood, 1965; Huang and Richter, 2004; Huang et al., 2006; Schuman et al., 2006]. In yeast, it is well-established that a member of the myosin-V family, Myo4p, transports Ash-1 mRNA [Takizawa et al., 1997; Beach and Bloom, 2001; Muller et al., 2007]. Therefore, an RNA transport/tethering deficit in Myo5a null mutant mice may be sufficient to cause the physiological [Takagishi et al., 1996; Miyata et al., 2000] and morphological [Dekker-Ohno et al., 1996; Takagishi et al., 2007] phenotypes observed in the central nervous system. Circumstantial evidence suggesting a role for myosin-Va in RNA transport in vertebrate neurons was first published in 2002 [Ohashi et al., 2002], but identification of a specific message associated with myosin-Va, Nd1-L, was only recently published [Yoshimura et al., 2006].
We therefore set out to determine if myosin-Va was involved in RNA transport in a wider variety of cell types. Our results from quantitation of newly-synthesized RNA labeled by BrU after variation of pulse and chase intervals (Figs. 3 and 4) suggest that myosin-Va functions as a transporter or tether at a very early stage of RNA transport, within 30 min of RNA synthesis. The prominence of differences in the vicinity of the nuclear membrane suggests that myosin-Va functions either inside the nucleus or just outside the nuclear membrane. There are preliminary reports of myosin-Va in the nucleus [Pranchevicius et al., 2008] to support the former possibility; our future experiments will analyze isolated nuclei to distinguish between them. Our results suggest that another, albeit slower, mechanism exists for transporting RNA out of the nucleus, since newly-synthesized RNA merely leaves the nuclear region more slowly in the absence of myosin-Va.
Since null mutant pups grow more slowly than wild-type controls as soon as the phenotype is evident at 7–8 days of age, a trivial explanation for our observations is that the null mutant cells may not be as healthy as the wild-type cells. However, we observed no differences in total RNA content in PNS fibroblasts (Fig. 1C) or in distribution of RNA in spleen fibroblasts (Fig. 2G) between mutant and wild-type mice. The latter observation correlates with the low levels of Myo5a message and/or the low level of the brain-specific splicing pattern in spleen (Fig. 2C). In combination with recent data demonstrating that interaction of dynein light chain 2 (DLC2) with the myosin-Va tail is dependent on the presence of the brain-specific exon B [Hodi et al., 2006], our results contribute significantly to our understanding of the relevance of alternative splicing of the myosin-Va tail.
Mislocalization of β-actin mRNA in fibroblasts has been shown to change their direction of migration [Shestakova et al., 2001], indicating that it plays a role in establishing cell polarity. Since it has been shown that β-actin mRNA distribution is dependent on actin filaments [Sundell and Singer, 1991], we examined the distribution of β-actin message by in situ hybridization in wild-type and null mutant cells (Fig. 6). Wild-type cells had high concentrations of message in the periphery and just outside the nucleus, while the mutant cells had a more diffuse perinuclear distribution, with no increase at the periphery.
To assay for a physical association between β-actin message and myosin-Va, we immunoprecipitated myosin-Va and used RT-PCR to amplify β-actin mRNA from wild-type, but not null mutant, immunoprecipitates (Figs. 6E and 6F). While this does not demonstrate a direct interaction, it is consistent with the in situ hybridization data. Our data are particularly interesting in the context of the demonstration that myosin-IIB has been shown in fibroblasts to be required for the proper localization of β-actin mRNA in response to growth factors [Latham et al., 1994; Kislauskis et al., 1997; Latham et al., 2001]. From our pulse/chase experiments, myosin-Va appears to be responsible for mRNA transport from the nucleus to the periphery, where myosin-IIB could capture mRNAs.
These data are consistent with multiple models (Fig. 7) that differ primarily in their sites of myosin-Va function. In the first model (Figs. 7A and 7B), nuclear myosin-Va activity is required for the exit of some transcripts from the nucleus to areas from which the mRNAs can be transported anterogradely throughout the cell on microtubules. This model incorporates the preliminary observation of myosin-Va in the nucleus [Pranchevicius et al., 2008], and fits well with the common involvement of microtubule-based motors in RNA transport [Kindler et al., 2005], as well as the coincidence of myosin-V and kinesins on the same cargo [Huang et al., 1999; Stafford et al., 2000]. In mutant cells (Fig. 7B), the absence of myosin-Va would slow exit of transcripts from the nucleus. The second model (Figs. 7C and 7D) incorporates a shuttle (or protective) factor that binds mRNA and reaches equilibrium by moving passively across the nuclear membrane. In mutant cells lacking myosin-Va, transcripts and the shuttle factor would fail to engage microtubule-based transport, and would accumulate in the perinuclear and nuclear regions, decreasing its rate of RNA exit from the nucleus. The granular perinuclear distribution of β-actin message in mutant cells (Fig. 6A) favors the second model (Figs. 7C and 7D). Moreover, treatment of both mutant and wild-type cells with latrunculin A or nocodazole increases the nuclear/cytoplasmic ratio of BrU labeling (data not shown), consistent with both actin and microtubule involvement.
Fig. 7.
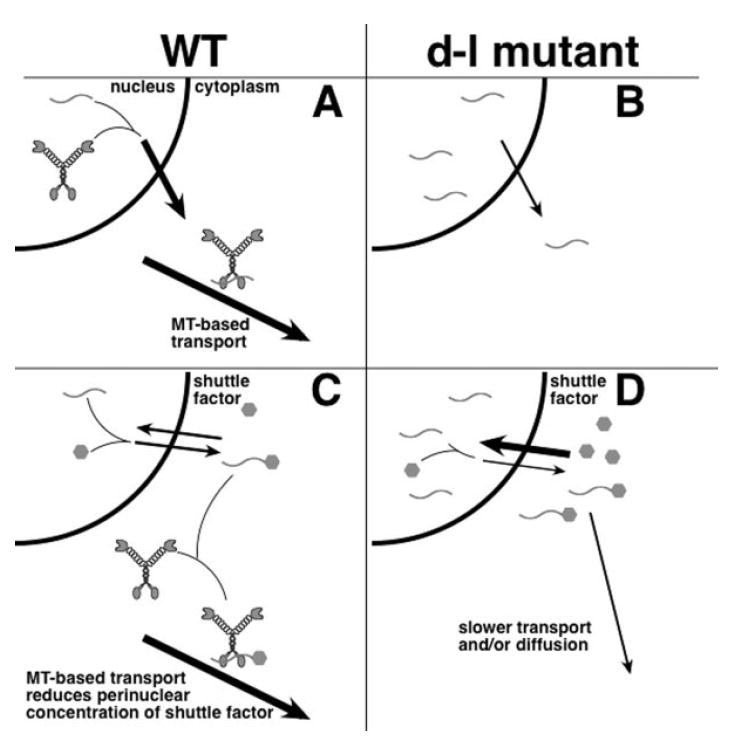
Models for myosin-Va function in RNA transport in wild-type (A, C) and null Myo5a mutant (B, D) cells. (A, B) Nuclear myosin-Va functions in exit of newly-synthesized RNA (wavy lines) from the nucleus to the cytoplasm, also allowing for transport of the complex along microtubules. (B) In the absence of myosin-Va, the ratio of nuclear to cytoplasmic RNA increases. (C, D) Myosin-Va is only present in the cytoplasm, and a shuttle factor (hexagons) binds RNA and is at equilibrium across the nuclear membrane. (D) in null mutant cells, the perinuclear concentrations of the shuttle factor and RNA are higher because transport and/or diffusion to the periphery are retarded.
In summary, these observations parallel observations of myosin-Va function in melanocytes [Provance et al., 1996], the loss of myosin-Va leads to a more granular and dispersed distribution of β-actin mRNA, and a greater nuclear/cytoplasmic ratio for both total and newly-synthesized RNA. Our pulse-chase experiments show that the impact of the absence of myosin-Va is only observed kinetically for newly-synthesized total RNA, but our in situ hybridization data show that myosin-Va has an essential function in β-actin mRNA distribution. We do not know if these functions are mechanistically identical. Any myosin-Va-dependent mechanisms for the proper localization of transcripts in fibroblasts are likely to share components with neuronal mechanisms for mRNA for transport, providing a significantly greater potential for experimental manipulation in fibroblasts.
MATERIALS AND METHODS
Mice
Homozygous dilute-lethal mutant mice (C57BL/6J Myo5ad-l26J/Myo5ad-l26J, C57BL/6J Myo5ad-l20J/Myo5ad-l20J, and DLS/Le Myo5ad-l/Myo5ad-l) mice were maintained in repulsion with short ear (Bmp5se), separated by 0.15 centimorgans; wild-type controls were age-matched C57BL/6J mice. Mice were euthanized at 12–18 days of age according to institutional protocols. Mutant mice were identified by ataxia and normal ears. All procedures were approved by the McLaughlin Research Institute (MRI) Institutional Animal Care and Use Committee. MRI is accredited by AAALAC.
Tissue Culture and Transfection
Primary fibroblasts were cultured from peripheral nervous system (PNS; dorsal root ganglia and sciatic nerve), lung, spleen and visceral adipose tissue of homozygous null mutant and age-matched, wild-type control mice.
PNS fibroblasts were isolated from 12 to 18-day-old mice. Approximately 30 dorsal root ganglia (DRG) or 6 sciatic nerves (~1 cm long), were collected and incubated in collagenase solution (Sigma C9407, 0.2 mg/ml final concentration) in HBSS for 60 min at 37°C with gentle agitation. The fragments were transferred to 0.5 mg/ml trypsin in HBSS and incubated for 15 min. Trypsin activity was neutralized by adding soybean trypsin inhibitor at 0.05 mg/ml, followed by trituration of the cells with a fire-polished Pasteur pipette. The cells were pelleted, washed 2 times with DMEM plus fetal bovine serum (FBS, 10%) and plated on glass coverslips.
Lung, spleen, and visceral adipose tissues were minced and incubated in Dulbecco’s Modified Eagle’s Medium (DMEM) with 1 mg/ml collagenase (Type V, Worthington) at 37°C with shaking at 200 rpm. After 1 h, DMEM with 10% FBS was added and cells were centrifuged at 500g for 3 min. Pelleted cells were plated in DMEM with 10% FCS and 100 units/ml each of penicillin and streptomycin. Cells were maintained in this medium in a humidified 37°C, 10% CO2 incubator for 5–7 passages without allowing the cells to become confluent.
In rescue experiments, the full-length cDNA construct pEF-T7-myosin-Va [Fukuda et al., 2002] with the brain-specific splicing pattern [Seperack et al., 1995] was overexpressed. Primary fibroblasts were transfected with DNA + Lipofectamine 2000 (Invitrogen) or Lipofectamine 2000 alone (mock) in suspension and plated directly onto glass coverslips. After a 1 h incubation, dishes were flooded with complete medium and incubated 12–18 h before their use in experiments.
RNA Labeling and Detection
Cells were plated at subconfluent densities on glass coverslips a day before experiments. Total RNA was detected with SYTO® RNASelect™ (Molecular Probes) according to manufacturer’s instructions. For flow cytometry, 2 × 106 cells were trypsinized, stained (10μM, 20 min), washed 3× with PBS, fixed in 4% paraformaldehyde, pelleted and resuspended in PBS with 3% BSA. Fluorescence and scatter was quantitated using a Becton Dickinson FACScan. For BrU incorporation, modified from published methods [Jackson et al., 1998; Kysela et al., 2005], cells were incubated in 2.5 mM BrU (made fresh in cold, complete medium) for the indicated times at 37°C. To chase, labeling medium was exchanged for cold, complete medium and incubated for indicated times at 37°C. Experiments were ended by fixing cells in fresh 4% paraformaldehyde in PBS. BrU was detected with an Alexa488-conjugated anti-BrdU monoclonal antibody (Molecular Probes), followed by an Alexa488-conjugated goat anti-mouse secondary antibody (Molecular Probes). For myosin-Va detection, a rabbit polyclonal anti-myosin-Va (DIL2, gift of V. Gelfand) was used with an Alexa 546-conjugated goat anti-rabbit secondary. Nuclei were stained with Hoechst 33258.
Microscopy and Image Analysis
Coverslips with BrU incorporation were imaged by a 20× PlanApo lens (N.A. 0.75) on a TE2000E microscope (Nikon) with a Quantix-57 camera (Photometrics) controlled by MetaMorph software (Molecular Dynamics). Acquisition parameters were constant within all data sets. SYTO® RNASelect™ staining was imaged with a 40× PlanApo lens (N.A. 0.95) on the same system. In MetaMorph, a 5-pixel-wide, 20-pixel-long line was drawn from the inside of the nucleus to the cytoplasm, avoiding nucleoli, and with the edge of the nucleus approximately centered in each line. Average intensity along the line was exported to Excel. Because diagonal lines had fewer than 20 pixels, each pixel position was converted to a percentage by dividing by the number of pixels in the line. Data were binned for 18 relative positions along the line. In PNS cultures, Schwann cells were identified by their spindle morphology and excluded from analysis. For the BrU-labeled cells, nuclear regions were identified and selected by MetaMorph using the Hoechst image without access to the BrU image, transferred to the BrU image, and the regions were manually moved to an adjacent cytoplasmic area. Images were obtained with a 60× WI PlanApo (N.A. 1.2) objective.
In situ Hybridization
Cells on coverslips were washed in Krebs-Ringer buffer and fixed in Krebs-Ringer with 2% paraformaldehyde for 2 h at 37°C. After fixation, coverslips were washed in PBS with 5 mM MgCl2 and 0.1% triton for = min, followed by two washes with PBS-MgCl2, then equilibrated (15 min, 42°C) in hybridization solution (4× SSC, 50% formamide, 0.5 mg/ml sheared single stranded salmon sperm DNA, 0.25 mg/ml yeast tRNA, 1× Denhardt’s and 10% Dextran sulfate). After equilibration, the coverslips were incubated in hybridization solution containing the probe (1 ng/μl of each oligonucleotide) at 42°C overnight. Coverslips were washed in 4× SSC with shaking for 20 min, twice in 2× SSC for 15 min, twice in 1× SSC for 15 min, and then incubated with a monoclonal anti-Cy3/Cy5 antibody (Sigma, C0992) followed by an Alexa 546-conjugated goat anti-mouse antibody. The probe for β-actin was a combination of two oligonucleotides complementary to the 5′ untranslated region of the mRNA, labeled with Cy3 at both 3′ and 5′ ends (Laboratory of Molecular Technology, NCI-SAIC, Frederick) oligo 1, GCTGTCGCCTTCACCGTTCCAGTTTTTAAATCCTGAGTCAAAAGCGCCAA; oligo 2, TAGGAGTGGGGGTGGCTTTTGGGAGGGTGAGGGACTTCCTGTAACCACTT; oligo3 (control, scrambled), AATCGAGGTACTCCAGTTCCAGTCCACCTGATCGACTGGGACCTTTCCGG.
Immunoprecipitation and RT-PCR
1 × 107 cells were collected, resuspended in hypotonic buffer (10 mM Tris pH 7.8, 10 mM β-mercaptoethanol (Sigma), Complete™ protease inhibitor (Roche), 0.5 mM PMSF with 1U/μl RNAsin (Invitrogen)), incubated in ice for 20 min and passed through a cell cracker (clearance 37μm) 20 times. Nuclei were pelleted at 1000 g, 15 min, 4°C. The supernatant was diluted with 2× Ripa Buffer (50 mM Tris pH8.0, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 0.5 % DOC (10%)), Complete™, and 1:1000 PMSF, DIL2 anti-Va antiserum (1:1000) was added and rocked overnight at 4°C. The next day, 30 μl of Protein A agarose was added for 30 min at 4°C. The agarose was collected by pelleting and washed with Rippa Buffer, Rippa Buffer plus 500 mM NaCl, and 50 mM Tris pH 7.0. The beads were resuspended in water and reverse transcribed using random primers (Invitrogen). β-actin was detected by PCR (30 cycles, 60°C annealing) using the primers GGAGAAGAGCTATGAGCTGC and CCTGCTTGCTGATCCACATC (predicted to produce a product of 376 bp, separated on an agarose gel, stained with ethidium bromide, and imaged using a VersDoc gel documentation system (BioRad).
Acknowledgments
The authors thank Nancy Jenkins for Myo5ad-l26J and Myo5ad-l20J mutant mice, Mitsunori Fukuda for the myosin-Va cDNA construct, David Munroe for supplying the β-actin probes, Rajeev Kumar and George Carlson for assistance with flow cytometry, Ryan Karcher and Deborah Cabin for helpful discussions, and Colleen Silan for technical support.
Contract grant sponsor: NIH; Contract grant number: R01 GM066901; Contract grant sponsors: A Coordenação de Aperfeiçoamento de Pessoal de Nível Superior fellowship; Comisión Sectorial de Investigación Cienítfica-UDELAR and PEDECIBA; Contract grant sponsor: The Pew Latin American Fellows Program in the Biomedical Sciences and ALSA; Contract grant number: 963.
Footnotes
Published online 20 March 2008 in Wiley InterScience (www. interscience.wiley.com).
References
- Beach DL, Bloom K. ASH1 mRNA localization in three acts. Mol Biol Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdwood W. Bidirectional particle movement in neurons. J Cell Biol. 1965;27:115a. [Google Scholar]
- Dekker-Ohno K, Hayasaka S, Takagishi Y, Oda S, Wakasugi N, Mikoshiba K, Inouye M, Yamamura H. Endoplasmic reticulum is missing in dendritic spines of Purkinje cells of the ataxic mutant rat. Brain Res. 1996;714:226–230. doi: 10.1016/0006-8993(95)01560-4. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: Implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- Hodi Z, Nemeth AL, Radnai L, Hetenyi C, Schlett K, Bodor A, Perczel A, Nyitray L. Alternatively spliced exon B of myosin Va is essential for binding the tail-associated light chain shared by dynein. Biochemistry. 2006;45:12582–12595. doi: 10.1021/bi060991e. [DOI] [PubMed] [Google Scholar]
- Huang JD, Mermall V, Strobel MC, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. Molecular genetic dissection of mouse unconventional myosin-VA: Tail region mutations. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JD, Brady ST, Richards BW, Stenolen D, Resau JH, Copeland NG, Jenkins NA. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD. Regulation of local mRNA translation. Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AN, Collinson Lm, Hopkins CR, Strom M, Barral DC, Bossi G, Griffiths GM, Seabra MC. The leaden gene product is required with Rab27a to recruit myosin Va to melanosomes in melanocytes. Traffic. 2002;3:193–202. doi: 10.1034/j.1600-0854.2002.030305.x. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. β-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kysela K, Philimonenko AA, Philimonenko VV, Janacek J, Kahle M, Hozak P. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol. 2005;124:347–358. doi: 10.1007/s00418-005-0042-8. [DOI] [PubMed] [Google Scholar]
- Langford GM. Myosin-V, a versatile motor for short-range vesicle transport. Traffic. 2002;3:859–865. doi: 10.1034/j.1600-0854.2002.31202.x. [DOI] [PubMed] [Google Scholar]
- Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. β-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH. A Rho-dependent signaling pathway operating through myosin localizes β-actin mRNA in fibroblasts. Curr Biol. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- Matesic LE, Yip R, Reuss AE, Swing DA, O’Sullivan TN, Fletcher CF, Copeland NG, Jenkins NA. Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci USA. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Miyata M, Finch EA, Khiroug L, Hashimoto K, Hayasaka S, Oda SI, Inouye M, Takagishi Y, Augustine GJ, Kano M. Local calcium release in dendritic spines required for long-term synaptic depression. Neuron. 2000;28:233–244. doi: 10.1016/s0896-6273(00)00099-4. [DOI] [PubMed] [Google Scholar]
- Muller M, Heuck A, Niessing D. Directional mRNA transport in eukaryotes: Lessons from yeast. Cell Mol Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Pastural E, Barrat FJ, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, Seger R, Griscelli C, Fischer A, de Saint Basile G. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat Genet. 1997;16:289–292. doi: 10.1038/ng0797-289. [DOI] [PubMed] [Google Scholar]
- Pranchevicius MCS, Baqui MMA, Ishikawa-Ankerhold HC, Lourenco EV, Leao RM, Banzi SR, Tavares dos Santos C, Barreira MCR, Espreafico EM, Larson RE. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motility and the Cytoskeleton. 2008 doi: 10.1002/cm.20269. in press. [DOI] [PubMed] [Google Scholar]
- Provance DW, Wei M, Ipe V, Mercer JA. Cultured melanocytes from dilute mutant mice exhibit dendritic morphology and altered melanosome distribution. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provance DW, James TL, Mercer JA. Melanophilin, the product of the leaden locus, is required for targeting of myosin-Va to melanosomes. Traffic. 2002;3:124–132. doi: 10.1034/j.1600-0854.2002.030205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Provance DW, Mooseker MS, Mercer JA. Class V myosins. Biochim Biophys Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rosé SD, Lejen T, Casaletti L, Larson RE, Pene TD, Trifaro JM. Myosins II and V in chromaffin cells: Myosin V is a chromaffin vesicle molecular motor involved in secretion. J Neurochem. 2003;85:287–298. doi: 10.1046/j.1471-4159.2003.01649.x. [DOI] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle AG. A lethal allele of dilute in the house mouse. Heredity. 1952;6:395–401. [Google Scholar]
- Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci USA. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Cárdenas M, Koenig E, Sotelo JR. Myosin Va and kinesin II motor proteins are concentrated in ribosomal domains (periaxoplasmic ribosomal plaques) of myelinated axons. J Neurobiol. 2004;60:187–196. doi: 10.1002/neu.20015. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Crispino M, Puppo A, Sotelo JR, Koenig E. Myelinated axons contain β-actin mRNA and ZBP-1 in periaxoplasmic ribosomal plaques and depend on cyclic AMP and F-actin integrity for in vitro translation. J Neurochem. 2008;104:545–557. doi: 10.1111/j.1471-4159.2007.04999.x. [DOI] [PubMed] [Google Scholar]
- Stafford P, Brown J, Langford GM. Interaction of actin- and microtubule-based motors in squid axoplasm probed with antibodies to myosin V and kinesin. Biol Bull. 2000;199:203–205. doi: 10.2307/1542899. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Takagishi Y, Oda S, Hayasaka S, Dekkerohno K, Shikata T, Inouye M, Yamamura H. The dilute-lethal gene attacks a Ca2+ store in the dendritic spine of Purkinje cells in mice. Neurosci Lett. 1996;215:169–172. doi: 10.1016/0304-3940(96)12967-0. [DOI] [PubMed] [Google Scholar]
- Takagishi Y, Hashimoto K, Kayahara T, Watanabe M, Otsuka H, Mizoguchi A, Kano M, Murata Y. Diminished climbing fiber innervation of Purkinje cells in the cerebellum of myosin Va mutant mice and rats. Dev Neurobiol. 2007;67:909–923. doi: 10.1002/dneu.20375. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Wu X, Sakamoto T, Zhang F, Sellers JR, Hammer JA. In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va. FEBS Lett. 2006;580:5863–5868. doi: 10.1016/j.febslet.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Myosin-Va facilitates the accumulation of mRNA/ protein complex in dendritic spines. Curr Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]


