Abstract
Radial glia cells are the first distinguishable glial population derived from neural epithelial cells and serve as guides for migrating neurons and as neural progenitor cells in the developing brain. Despite their functional importance during neural development, the determination and differentiation of these cells remains poorly understood at the molecular level. Ets-1 and Ets-2, Ets (E26 transformation-specific) transcription factors, are vertebrate homologues of Drosophila pointed, which is expressed in a subset of glia cells and promotes different aspects of Drosophila glia cell differentiation. However, it remains unsolved that the function of Ets genes is conserved in vertebrate glia development. Here we report that Ets-1 but not Ets-2 is necessary for Xenopus radial glia formation and the activity of Ets-1 is sufficient for radial glia formation prior to neural tube closure. Furthermore, we show that Ras-MAPK (mitogen activated protein kinase) signaling, which acts as an upstream activator of Ets-1 in other biological processes, also regulates radial glia formation. A mutant form of Ets-1, which is not responsive to Ras-MAPK signaling, inhibits radial glia formation promoted by Ras-MAPK signaling. Together, our results show that Ets-1 activated by Ras-MAPK signaling promotes radial glia formation during Xenopus embryogenesis.
Key words: Radial glia, Ets-1, Ets-2, pointed, Ras-MAPK signaling pathway, Xenopus
Introduction
In the developing central nervous system (CNS), most neurons and glia are generated from common neuroepithelial progenitor cells in the ventricular zone.1,2 Radial glia cells are the first glial population that can be distinguished from neuroepithelial cells and are identifiable by morphological and molecular characteristics. They contain a long basal process that extends from the cell body in the ventricular zone through the parenchyma toward the brain surface,3 have endfeet on blood vessels, express glial specific intermediate filaments and contain glycogen granules.4,5 These latter features are shared with cells of the astrocytic lineage. Radial glia cells also express Nestin, RC2, vimentin, glial fibrillary acidic protein (GFAP) and astrocyte-specific glutamate transporter (GLAST)6,7 in common with neuroepithelial cells and astrocytes. In mammals, radial glia cells are observed at early stages of neural tube formation and exist only in the developing CNS.8,9 While Xenopus radial glia cells are also initially detected during the period of neural tube formation, they persist throughout life.10–12
Radial glia in the developing CNS have been shown to guide neuronal migration13,14 and to serve as progenitor cells.7,8,15–20 Importantly, abnormalities in radial glia have been implicated in several distinct human syndromes in which patients often present with epilepsy and mental retardation21 and radial glia have also been suggested as a candidate cancer stem cell for ependymoma.22 Therefore, the molecular characterization of radial glia development is important for understanding the mechanisms of normal neural development as well as a number of neuropathological disorders.
The Ets transcription factor family is characterized by a variant of the winged helix-turn-helix DNA binding motif which recognizes a conserved GGA core sequence. Ets factors are important regulators of development in both invertebrates and vertebrates.23–27 In Drosophila, the pointed gene, an Ets transcription factor, produces two isoforms (P1 and P2) by alternative splicing and directs glia cell differentiation.28,29 In the embryonic CNS, pointedP1 is expressed by lateral glia cells whereas pointedP2 is expressed by midline glia cells.28 In addition to differential expression, these two pointed proteins, P1 and P2, differ with respect to the responsiveness to Ras-MAPK signaling. While pointedP1 behaves as a constitutive activator, pointedP2 seems to be activated by the downstream effecters of Sevenless such as D-Ras1 or ERK-A.30 Indeed, pointedP2 is directly phosphorylated by ERK-A on threonine 151 in the pointed domain in vitro.30,31 The closest vertebrate homologues of pointed gene are Ets-1 and Ets-2 genes28,32,33 and the functions of these Ets genes are conserved between vertebrates and Drosophila.34 pointedP2 efficiently binds to an optimized Ets-binding site and transactivates Ets-l/Ets-2 responsive elements in vertebrate cells. Chicken Ets-1 or Ets-2 is capable of rescuing the Drosophila pointed mutant phenotype in midline glia development. Interestingly, the phosphorylation of the threonine residue homologous to pointedP2 threonine 151 also appears essential for the transactivating function of Ets-l and Ets-2 by the Ras-MAPK signal.35–37 Ets-1 and Ets-2 are widely expressed in many tissues, including the developing nervous system, during murine embryogenesis.38 Ets-1 deficient mice contain decreased the numbers of splenic Natural Killer (NK) cells and displayed severely reduced or absent NK cell function.39 In contrast, Ets-2 deficient mice are lethal before day 8.5 of embryonic development due to defective trophoblast function.40 However, the function of vertebrate Ets genes in radial glia formation still remains unclear.
Here we show that the function of Ets-1 but not Ets-2 is involved in Xenopus radial glia formation. The activation of Ets-1 is sufficient for radial glia formation prior to neural tube closure when radial glia cells appear as reported previously.11,12 Loss-of-function studies by antisense Morpholino Oligonucleotides against Ets genes and a dominant negative form of Ets-1 indicate that only Ets-1 is necessary for radial glia formation. Mutation of threonine 36 of Ets-1, a known target of phosphorylation by Ras-MAPK signaling in other systems, leads to the inhibition of radial glia formation. A constitutively active form of MAPK kinase leads to an increase in the number of radial glia processes and this effect can be reduced by co-injection of this mutant Ets-1. This suggests that Ets-1 is a downstream mediator of Ras-MAPK signaling during radial glia formation. Together, our results suggest that Ets-1 directly regulates radial glia formation as a downstream mediator of Ras-MAPK signaling during Xenopus embryogenesis.
Materials and Methods
Embryo manipulations.
Unfertilized eggs were obtained by subcutaneous injection of gonadotropic hormone (Sigma) into Xenopus laevis females. Eggs were artificially fertilized by using testis homogenate and chemically de-jellied with 3% cysteine-hydrochloride solution (pH 8.0) and cultivated in 0.1x Marc's Modified Ringer's solution (MMR).41 Embryos were staged according to Nieuwkoop and Faber.42
DNA constructs.
Xenopus Ets-1 (clone ID: XL200e22) were provided by Dr. Ueno, National Institute for Basic Biology (NIBB) and Ets-2 (IMAGE ID: 6631306) cDNAs were purchased from Open Biosystems. To produce Ets1-GR and Ets2-GR constructs, the human glucocorticoid receptor ligand binding domain (gift from Dr. Sive) was subcloned into the XhoI and XbaI sites of pCS2+43 using PCR with the primers: 5′-GGCGCCGCTCGAGCCTCTGAAAATCCTGGT-3′ and 5′-GGCGGGCATCTAGAACTTTTGATGAAAC-3′ (pCS2+GR), then cDNA of Ets-1 or Ets-2 was subcloned into ClaI and XhoI sites of pCS2+GR using PCR with the primers: 5′-ATTTAGGTGACACTATA-3′ (SP6) and 5′-GAATCTCGAGATTCATCAGTGTCTGGTT-3′ (Ets1GR-D), or 5′-GAATCTCGAGATTCGTCTGTGTCGGGCT-3′ (Ets2GR-D), respectively. To create dn-Ets1-GR construct, cDNA of Ets-1 was subcloned into EcoRI and XhoI sites of pCS2+MT (with six myc-tag) using PCR with the primers: 5′-CTGAATTCAGACTATGTAAGGGAC-3′ and 5′-GGATCTCGAGATCCCAGAAGACTTTG-3′, then the GR domain was subcloned into XhoI and XbaI. To generate the pCS2+HAEts1-GRΔATG (Ets1-GRΔATG in text) and pCS2+HAEts2-GR_ΔATG (Ets2-GRΔATG in text) constructs, the coding sequence of Ets-1 or Ets-2 lacking the translation initiation site was subcloned into the ClaI and XhoI sites of pCS2+GR by PCR with the primers: 5′-GGTAATCGATTACCGACATTAAGCATCA-3′ and Ets1GR-D, or 5′-CGAAATCGATTTCGAAACATGGACCAAG-3′ and Ets2GR-D. The QuikChange Site-Directed Mutagenesis Kit II (Stratagene) was used to create pCS2+Ets1-GRT36A (Ets1-GRT36A in text), using the following primers: 5′-GTCCCGCTTCTGGCACCCAGTAGTAAAG-3′ and 5′-CTTTACTACTGGGTGCCAGAAGCGGGAC-3′, and pCS2+Ets1-GR as a template. The EcoRI/HindIII fragment of cDNA vimentin1 (IMAGE ID: 4888155), which was purchased from Open Biosystems, was subcloned into the same sites of pBlue-scriptKS+ (Stratagene). nrp1a cDNA (IMAGE ID: 6944933) was also obtained from Open Biosystems. pSP64TdnRas (dn-Ras in text, gift from Dr. Whitman) and pSP64T SESEMAPKK (at-MAPKK in text, gift from Dr. Gotoh) have been described previously.44,45
Microinjection of synthetic RNA and morpholino antisense oligonucleotides.
Capped synthetic mRNAs were generated by in vitro transcription with SP6 polymerase, using the mMessage mMachine kit (Ambion, Inc.) as described by the manufacture's instructions. Antisense Morpholino Oligonucleotides (MOs) were designed and produced by Gene Tools, LLC. Ets1MO and Ets2MO were designed to target the following sequences: 5′-GATCTAGCGCAGCTTTCATGGCT-3′ (For Ets-1a), 5′-TTAAGGTCTAGTGCAGCTTTCATG-3′ (For Ets-1b) and 5′-ATTCCAAACTCTGTCATTGGCCCTG-3′ (For Ets-2a and Ets-2b), respectively. The control Morpholino Oligonucleotides (ConMO) has the following sequences: 5′-CCTCTTACCTCAGTTACAATTTATA-3′. For microinjections, 8-cell stage embryos were transferred to 3% Ficoll 400 in 0.1x MMR and injected with 5∼10 nl of the specified amount of RNA and/or MO. After an hour of injection, embryos were transferred into 0.1x MMR and cultured until the desired stage. For the activation of GR-fused protein, dexamethasone (DEX: final concentration 10 µM) was added to the medium. DEX was added at stage 18 except where indicated in the figures.
α-Galactosidase staining and whole-mount in situ hybridization.
Embryos were fixed with MEMFA [0.1 M MOPS, 2 mM EGTA (pH 8.0), 1 mM MgSO4, and 3.7% formaldehyde] for 15 minutes, followed by washing in phosphate-buffered saline (PBS) twice. Then, galactosidase activity was visualized with the RedGal substrate (Research Organics) in 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], 2 mM MgCl2/ PBS. After staining, embryos were re-fixed with MEMFA for several hours. Whole-mount in situ hybridization was performed essentially as described46,47 using DIG (Digoxigenin, Roche Applied Science)-labeled or Fluorescein (Roche Applied Science)-labeled antisense RNA probes and BCIP or BM purple (Roche Applied Science) for chromogenic reaction. Following chromogenic reaction, pigmented embryos were bleached with 10% H2O2 in methanol for several days. After whole mount in situ hybridization, embryos were re-fixed in 4% paraformaldehyde overnight at 4°C and embedded in 3% agarose. One-hundred micrometer sections were prepared on a microtome.
Immunohistochemistry.
RNA or MO was co-injected with green fluorescent protein (GFP) RNA (gift from Drs. Chang and Habas) into one dorsal-animal blastomere of Xenopus embryos at 8-cell stage and embryos, in whose hindbrain GFP was observed, were used for further analysis. Brains isolated from those embryos were fixed with 2% paraformaldehyde in PBS (pH 7.4) for 15 minutes on ice. After rinsing with PBS several times, they were immersed in 30% sucrose in PBS, embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Inc.) and 16 µm sections were prepared on a cryostat at −19 °C. The cryosections were blocked in PBS with 5% goat serum and 0.3% Triton X-100 for one hour to prevent non-specific staining. After incubation with vimentin 14h7 monoclonal antibody (The Developmental Studies Hybridoma Bank, University of Iowa) and Xenopus intermediate filament protein 3 (XIF3) polyclonal antibody (gift from Dr. Szaro)10 or GFAP polyclonal antibody (DAKO) overnight at 4°C, the sections were washed three times in PBS with 0.3% Triton X-100 for 15 minutes each. After treatment with a Texas red-labeled F(ab)2 fragment of goat anti-mouse IgG and FITC-labeled F(ab)2 fragment of goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) for an additional one hour, the sections were washed three times in PBS, for 10 minutes each time. The sections were mounted with GEL/MOUNT™ (Biomedia), and observed on a Leica confocal laser microscopy. We could not observe GFP in the sections after immunostaining. For statistic analysis, data were obtained from 12 sections (three samples each from four animals), stained by anti-vimentin antibody, per each injection experiment. To compare the means of the ratio of radial processes, one-way ANOVA (analysis of variance) was used for statistic analysis and the Newman-Keuls test was followed.
Immunoblotting.
Immunoblotting was performed with anti-Ets-1 antibody (N-276), anti-Ets-1/-2 antibody (C-275), anti-GR antibody (P-20) or anti-α-tubulin antibody (H-300) (Santa Cruz Biotechnology, Inc.) as a primary antibody and HRP-conjugated goat anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Inc.) as a secondary antibody. Embryonic lysates were prepared from 10 embryos for each experiment in 200 µl of 10 mM sodium phosphate pH 7.5, 135 mM KCl, 0.1% Triton X-100, 1 mM DTT, 1 mg/ml Leupeptin, 0.1 mM PMSF, and 5 mM MgCl2.
Results
Ets-1 and Ets-2 are expressed in the central nervous system.
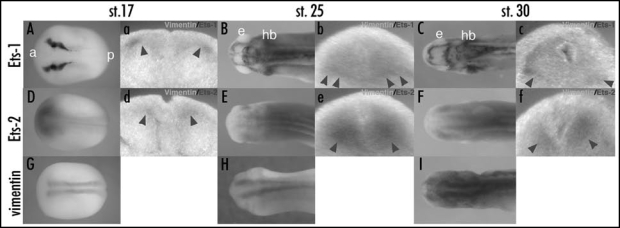
While pointed promotes glia differentiation in Drosophila,28,29 Ets-1 and Ets-2, vertebrate homologues of pointed, have not been previously shown to be involved in radial glia formation. To determine the possible roles of Ets-1 and Ets-2 in Xenopus radial glia formation, we examined the expression of Ets-1, Ets-2 and the radial glial marker, vimentin, at different developmental stages by whole-mount in situ hybridization. The expression of vimentin protein, an intermediate filament protein, is first found in radial glia cells at the time of neural tube closure (st. 19) and is also detected in other tissues such as optic vesicle and mesenchymal cells of the somites and head at later stages during Xenopus embryogenesis.11 Previous data reported that Xenopus Ets-1 and Ets-2 were maternally expressed and their transcripts were localized to the animal pole and the intermediate zone during early embryogenesis.48 At late neurula stage (st. 17), Ets-1 was expressed in neural crest and the lateral edge of neural plate at the level of prospective hindbrain, and Ets-2 was broadly expressed in the neural plate (Fig. 1A and D). The expression of vimentin was observed as two stripes on both sides of midline in neural plate (Fig. 1G). To examine the colocalization of Ets-1 or Ets-2 and vimentin, we performed double whole mount in situ hybridization and embryos were transversally sectioned at the level of prospective hindbrain (Fig. 1a and d). The expression of Ets-1 or Ets-2 overlapped with lateral or inferior edge of vimentin expression indicated by arrow heads in (Fig. 1a and d), respectively. At early tailbud stage (st. 25), both Ets-1 and Ets-2 expression were found in the CNS (Fig. 1B and E). At the level of hindbrain, Ets-1 was expressed in the inferior edge of neural tube and Ets-2 overlapped with vimentin expression in the neural tube (indicated by arrow heads in Fig. 1b and e). At late tailbud stages (st. 30), Ets-1 and Ets-2 were still expressed in the CNS (Fig. 1C and F). At the level of hindbrain, Ets-1 was expressed in lateral and inferior edge of neural tube while the expression of Ets-2 overlapped with that of vimentin (indicated by arrow heads in Fig. 1c and f). These results suggest that Ets-1 may play a spatially and temporally limited role in the formation of radial glia cells while the expression of Ets-2 is more broadly consistent as a candidate for regulating radial glia formation during Xenopus embryogenesis.
Figure 1.
The expression profiles of Ets-1, Ets-2 and radial glia marker, vimentin in Xenopus hindbrain. (A–I) The expressions of Ets-1 (A–C), Ets-2 (D–F), and vimentin (G–I) in Xenopus embryos were shown. All pictures of whole embryos are dorsal views, anterior toward left. (A, D, G) stage 17, late neurula. (B, E and H) stage 25, early tailbud. (C, F and I) stage30, tailbud. (a–f) transversal section cut at the level of prospective hindbrain or hindbrain in (A–F), respectively. Sections were prepared from double whole mount in situ hybridization with Ets-1 or Ets-2 (purple) and vimentin (turquoise blue). Arrow heads in (a–f) indicated the overlapping expression of Ets-1 or Ets-2 and vimentin. a, anterior; p, posterior; e, eye; hb, hindbrain. The original figure contains color pictures.
Overexpression of Ets-1 and Ets-2 promotes radial glia formation in Xenopus hindbrain.
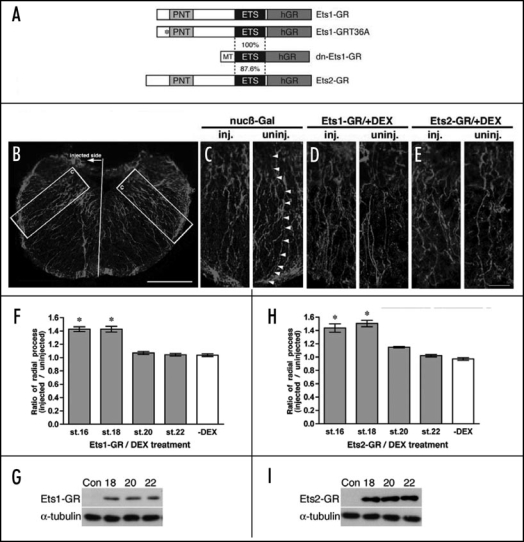
To examine whether Ets-1 and Ets-2 regulates radial glia formation during Xenopus embryogenesis, we generated hormone-inducible forms of Ets-1 (Ets1-GR) and Ets-2 (Ets2-GR), which are fused to the human glucocorticoid receptor ligand binding domain (GR) (Fig. 2A). The activity of these transcription factors can be regulated temporally by the addition of dexamethasone (DEX).49 Preliminary studies revealed that radial glia cells could be induced through stage 18 while similar treatments at later stages had no discernable effect on glial cell numbers (Kiyota et al., unpublished data). We therefore sought to determine the ability of Ets-1 and Ets-2 to effect radial glial formation as well as whether this effect was temporally controlled. In vitro synthesized RNA encoding Ets1-GR or Ets2-GR was injected into one dorsal-animal blastomere of 8-cell stage embryos, which was fated to differentiate into the brain,50 and DEX was added at several different developmental stages. Embryos injected with Ets1-GR or Ets2-GR were cultured until tadpole stage (st. 42), and brains were isolated manually and fixed. Cryosections were prepared and examined by immunostaining with anti-vimentin antibody, and the number of radial processes in the hindbrain was evaluated. We focused on radial glia in the hindbrain at stage 42 (Fig. 2B), as they were well-characterized in a previous study51 and the expression of Ets-1 and Ets-2 overlapped with vimentin expression in the prospective hindbrain at stage 17 and 25 (Fig. 1). As the cell body of each radial glia cell has one radial process,10,19 all vimentin positive radial processes were counted on both the injected and uninjected control half of the hindbrain for each brain section imaged by confocal microscopy. We limited the analysis to radial processes in the image which could be traced from the ventricular zone (the upper side in Fig. 2C) to the brain surface (the lower side of Fig. 2C) while processes which could not be completely traced were excluded. To quantitatively compare different treatments, we determined the ratio of radial processes (RPratio) which is defined as the average ratio of fully traced radial processes on the injected side divided by the number of fully traced processes observed on the uninjected side of the brain. Each value represents the average of 12 independent measurements (three sections each from four animals). Figure 2D and E shows part of the hindbrain of an Ets1-GR-injected or Ets2-GR-injected embryo following DEX treatment at stage 18. The injected side of radial processes was increased by Ets1-GR or Ets2-GR in comparison with the uninjected side, respectively. As shown in Figure 2F and H, overexpression of Ets-1 or Ets-2 could increase the ratio of radial processes when embryos were treated with DEX prior to neural tube closure (st.18) by about 40%. As controls, the protein levels of Ets1-GR and Ets2-GR at stage 18, 20 and 22 were confirmed by immunoblotting with anti-GR antibody (Fig. 2G and I) and DEX itself did not affect radial glia formation (data not shown). These data demonstrate that the activities of both Ets genes are sufficient for radial glia formation prior to neural tube closure and coincided with the period when radial glia cells appear during Xenopus embryogenesis as reported previously.11,12
Figure 2.
The activities of Ets-1 and Ets-2 are sufficient for promoting radial glia formation prior to neural tube closure. (A) DNA constructs used in this paper. PNT, pointed domain; ETS, Ets-DNA-binding domain; hGR, human glucocorticoid receptor ligand binding domain; MT, myc-tag. (B) nucβ-Gal (nuclear β-Galactosidase)-injected stage 42 Xenopus hindbrain stained by anti-vimentin antibody. All of the high magnification images presented in this paper were taken from a similar part surrounded by a white square in this panel. Scale bar: 100 µm. (C) High magnification view of the hindbrain indicated in (B). Arrow heads indicates a radial process, which can be traced from the ventricular zone to the brain surface. (D and E) High magnification images of the injected and uninjected side of the hindbrain stained by anti-vimentin antibody in 100 pg Ets1-GR-injected (D) and 100 pg Ets2-GR-injected (E) embryos with DEX treatment at stage 18. Scale bar: 25 µm. (F and H) The ratio of radial processes (RPratio) between the injected and uninjected side of the same hindbrain. Gray bars or white bars indicate with DEX treatment or without DEX treatment, respectively. The numbers of X-axis indicate the developmental stages when DEX was added. *p < 0.001 compared with the ration of Ets1-GR-injected (F) or Ets2-GR-injected (H) embryos without DEX treatment, respectively. Each value represents the average of 12 independent measurements. (G and I) The expression of Ets1-GR and Ets2-GR proteins without DEX at 18, 20 and 22. The level of tubulin protein was used for the normalization of samples. The original figure contains color pictures.
Ets-1 is required for radial glia formation in Xenopus hind-brain.
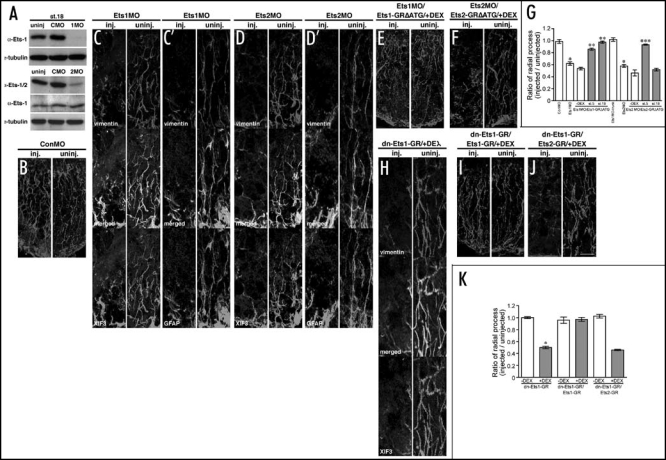
To test whether Ets genes are required for radial glia formation, loss-of-function studies were performed with Morpholino Oligonucleotides (MO), which bind to the translation initiation site of targeted transcripts and inhibits protein translation. Thus the overall levels of endogenous targeted protein are decreased as newly synthesized protein production is blocked.52 To confirm the efficacy of morpholino oligonucleotides targeted against Ets-1 (Ets1MO) and Ets-2 (Ets2MO), we performed immunoblotting analysis on embryonic extracts following microinjection of either Ets1MO or Ets2MO using either anti-Ets-1 antibody or anti-Ets-1/-2 antibody which recognizes both Ets-1 and Ets-2 proteins (Fig. 3A). Ets1MO or Ets2MO was injected into all four blastomeres of four cell stage embryos and embryonic lysates prepared at stage 18. Ets1MO or Ets2MO reduced the level of endogenous Ets proteins significantly, while Control MO (ConMO), which targets a human β-globin intron mutation and doesn't recognize Xenopus Ets transcripts, had no effect on the level of Ets proteins (Fig. 3A). To assess the effect of Ets loss of function on radial glia formation, Ets1MO, Ets2MO or ConMO was injected into one dorsal-animal blastomere of 8-cell stage embryos and the ratio of radial processes (RPratio) was determined as described above. Injection of Ets1MO and Ets2MO but not ConMO reduced the number of radial processes in the injected side of the hindbrain at stage 42 (Fig. 3B–D'). The ratio of radial process (RPratio) in Ets1MO-injected or Ets2MO-injected embryos was determined using the number of vimentin-positive radial glia processes and was 0.62 (p < 0.001) or 0.58 (p < 0.001), respectively (Fig. 3G). Importantly, visual inspection of the injected versus uninjected side in Figure 3C, C', D and D' suggests a significantly greater effect of these constructs on radial process formation. This is due to a non homogeneous level of the injected MO within the hindbrain which leads to an underestimation of the effect on radial process formation when the entire hindbrain region is used to calculate the ratio of radial processes. As the obtained values are statistically significant, the inclusion of the entire hindbrain region avoided the difficulties which might arise in a subjective selection of any sub region of the brain for further analysis. In contrast, Ets1MO injection into the prospective trunk region did not change the number of radial glia processes in the spinal cord (RPratio= 1.02) (Fig. 3D), as Ets-1 is not expressed in the trunk region at stage 17 (Fig. 1A). To examine the specificity of loss-function experiments, we mutated Ets1-GR (Ets1-GRΔATG) or Ets2-GR (Ets2-GRΔATG), and replaced the translation initiation site with sequences encoding the hemaglutanin (HA) epitope tag and which would no longer be recognized by MO. RNA encoding these tagged constructs was co-injected with Ets1MO or Ets2MO respectively and DEX was added soon after the injection (st.5) or prior to neural tube closure (st.18). Radial process numbers were restored to near untreated levels by co-injection of Ets1-GRΔATG and DEX activation at both stage 5 (RPratio = 0.86, p < 0.001) and stage 18 (RPratio=0.98, p < 0.001) (Fig. 3E and G). In addition, Ets1MO-injection into the prospective trunk region did not change radial process numbers in the spinal cord (RPratio = 1.02, Fig. 3G).In contrast, co-injection of Ets2-GRΔATG was able to restore the RPratio to near untreated levels when DEX was added at stage 5 (RPratio = 0.93, p < 0.001) while having no effect at stage 18 (Fig. 3F and G). These results demonstrate that Ets-1 is directly required for radial glia formation and suggest that the depletion of Ets-2 inhibited radial glia formation secondarily.
Figure 3.
Ets-1 but not Ets-2 is required for radial glia formation in Xenopus hindbrain. (A) Reduction of endogenous Ets proteins by Morpholino Oligonucleotides (MO) at stage 18. The level of tubulin protein was used for the normalization of samples. (B–F) High magnification images of the injected and uninjected side of the hindbrain in 40 ng ConMO-injected (B) vimentin, 40 ng Ets1MO-injected (C) vimentin and XIF3, (C') vimentin and GFAP, 40 ng Ets2MO-injected (D) vimentin and XIF3, (D') vimentin and GFAP, Ets1MO/100 pg Ets1-GRΔATG-injected (E) vimentin,and Ets2MO/20 pg Ets2-GRΔATG-injected (F) vimentin embryos. DEX was added at stage 18. Scale bar: 25 mm. (G) The ratio of radial processes (RPratio) between the injected and uninjected side of the hindbrain for each injection experiment. Gray bar or white bars indicate with DEX treatment at stage 18 or without DEX treatment, respectively. The numbers of X-axis indicate the developmental stages when DEX was added. *p < 0.001 compared with the ratio of ConMO-injected embryos. **p < 0.001 compared with the ratio of Ets1MO/ Ets1-GRΔATG-injected embryos without DEX treatment. ***p < 0.001 compared with the ratio of Ets2MO/Ets2-GRΔATG-injected embryos without DEX treatment. Each value represents the average of 12 independent measurements. (H–J) High magnification images of the injected and uninjected side of the hindbrain in 50 pg dn-Ets1-GR-injected (H) vimentin and XIF3, dn-Ets1-GR/100 pg Ets1-GR-injected (I) vimentin and dn-Ets1-GR/100 pg Ets2-GR-injected (J) vimentin embryos. DEX was added at stage 18. Scale bar: 25 µm. (K) The ratio of radial processes (RPratio) between the injected and the uninjected side of the hindbrain for each injection experiment. Gray bars or white bars indicate with DEX treatment at stage 18 or without DEX treatment, respectively. *p < 0.001compared with the ratio of dn-Ets1-GR-injected embryos without DEX treatment. **p < 0.001 compared with the ratio of dn-Ets1-GR-injected embryos with DEX treatment. Each value represents the average of 12 independent measurements. The original figure contains color pictures.
The activity of Ets-1 is required for radial glia formation prior to neural tube closure.
To verify whether the activity of Ets-1 is required for radial glia formation prior to neural tube closure, we generated a hormone inducible dominant-negative form of Ets-1 (dn-Ets1-GR), which includes only the Ets-DNA-binding domain53 (Fig. 2A). This dominant negative Ets1-GR, sharing 87.6% similarity with the homologous domain of Ets-2, can also inhibit the activity of Ets-2 (data not shown). Synthetic RNA of dn-Ets1-GR was injected into one dorsal-animal blastomere of 8-cell stage embryos and DEX was added at stage 18. Injected embryos were cultured until tadpole stage (st.42), and immunostaining with anti-vimentin and anti-XIF3 antibodies was performed. The injection of dn-Ets1-GR reduced the number of radial processes in the injected side of the hindbrain (compare the injected and uninjected side in Fig. 3H) and the ratio of radial process number (RPratio) for dn-Ets1-GR-injected embryos was 0.50 (p < 0.001) (Fig. 3J). To demonstrate specificity in this loss-of-function study, both the dominant negative Ets1-GR constructs and Ets1-GR or Ets2-GR were coinjected and treated with DEX at stage 18. Following culture to stage 42, the number of radial processes was determined as before. As shown in Figure 3I and J and quantified in Figure 3K, Ets-1 (RPratio = 0.97, p < 0.001) but not Ets-2 (RPratio = 0.45) could restore the number of radial process to near untreated control levels. These results indicate that Ets-1 but not Ets-2 is required for radial glia formation at the time when radial glial cells appear during Xenopus embryogenesis.11,12
The depletion of Ets-2 but not Ets-1 affected early neural development.
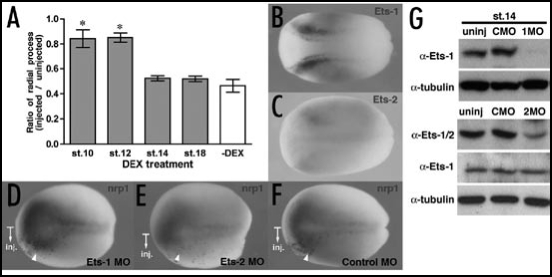
As noted above, the ability to rescue the Ets2MO phenotype was dependent on the time of Ets-2GR activation and was distinct from the results obtained for Ets-1. To examine more closely this potential temporal control, co-injection of Ets2MO and Ets2-GRΔATG was performed followed by DEX addition at different stages (Fig. 4A). The ratio of radial processes (RPratio) was calculated as described above. The reduction of radial process number by Ets2MO was only rescued by Ets2-GRΔATG when DEX was added prior to early neurula stage (st.14) but not at later stages (Fig. 4A), suggesting that the depletion of Ets-2 affected early neural development rather than radial glia formation. As Ets-2 is weakly expressed in the part of the neural plate and in neural crest at stage 12 (Fig. 4C), we performed loss-of-function studies using Ets2MO at an early developmental stage. To assess the hypothesis that the observed effects of Ets-2 correspond to defects in neural tissue formation, Ets2MO, Ets1MO or ConMO were injected into one dorsal-animal blastomere of 8-cell stage embryos and the expression of nrp1, a pan-neural marker,54 was examined by whole-mount in situ hybridization. As predicted, the expression of nrp1 was inhibited by Ets2MO but not Ets1MO and ConMO (Fig. 4D–F). As a control, we confirmed that the level of both endogenous Ets proteins are significantly reduced at stage 14 (Fig. 4G). These data support that Ets-2 affects early neural development and is not directly involved in radial glia formation.
Figure 4.
The depletion of Ets-2 but not Ets-1 affected early neural development. (A) The ratio of radial processes (RPratio) between the injected and uninjected side of the hindbrain of Ets2MO/Ets2-GRΔATG-injected embryos. Gray bars or a white bar indicate with DEX treatment or without DEX treatment, respectively. The numbers of X-axis indicate the developmental stages when DEX was added. *p < 0.001 compared with the ratio of Ets2MO/Ets2-GRΔATG-injected embryos without DEX treatment. Each value represents the average of 12 independent measurements. (B and C) The expression of Et1 (B) and Ets2 (C) genes in stage 12 embryos. All panels are dorsal views, anterior toward left. (D–F) Whole-mount in situ hybridization analysis of nrp1 expression at stage 14. (D) n = 29 (no change)/29, (E) n = 20 (reduced)/20, (F) n = 21(no change)/21. The injected side in each embryo is indicated by b-Gal staining (red color). (G) Reduction of endogenous Ets proteins by Morpholino Oligonucleotides (MO) at stage 14. The level of tubulin protein was used for the normalization of samples. The original figure contains color pictures.
Ras-MAPK signaling is upstream of Ets-1 during radial glia formation.
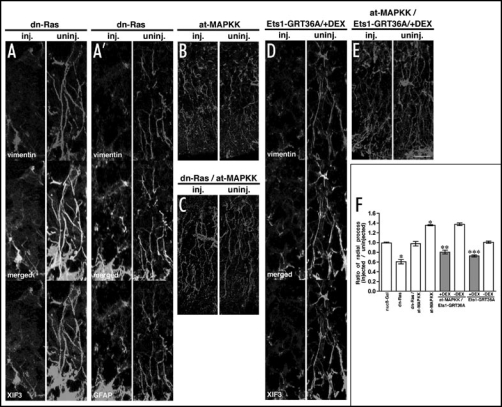
In other biological processes, Ets-1 is phosphorylated and activated by Ras-MAPK signaling.35,55 We therefore tested whether Ras-MAPK signaling was also involved in the process of radial glia formation. We used a constitutive active form of MAPKK (at-MAPKK)45 for gain-of-function studies which we predicted would increase the number of radial glia processes and a dominant negative form of Ras (dn-Ras)44 for loss-of-function study that we predicted would decrease radial glia process numbers. All injections and analysis were performed as previously described. As expected, the number of radial processes was reduced in the dn-Ras-injected side of the hindbrain when compared with the uninjected side (Fig. 5A and A'), and the ratio of radial process number (RPratio) was 0.61 (p < 0.001) (Fig. 5F). In contrast, injection of at-MAPKK increased the number of radial processes in the injected side of the hindbrain when compared with the uninjected side (Fig. 5B) and the ratio of radial process number (RPratio) in at-MAPKK-injected embryos was 1.35 (p < 0.001) (Fig. 5F). Furthermore, co-injection with at-MAPK restored the number of observed radial processes and RPratio in dn-Ras-injected embryos to levels found in untreated hindbrain (Fig. 5C and F). These results suggest that Ras-MAPK signaling is both necessary and sufficient for radial glia formation.
Figure 5.
Ras-MAPK signaling is upstream of Ets-1 and promotes radial glia formation. (A–E) High magnification images of the injected and uninjected side of the hindbrain in 250 pg dn-Ras-injected (A) vimentin and XIF3, (A') vimentin and GFAP, 100 pg at-MAPKK-injected, (B) vimentin, dn-Ras at-MAPKK-injected (C) vimentin, 100 pg Ets1-GRT36A-injected, (D) vimentin and XIF3, and at-MAPKK/Ets1-GRT36A-injected, (E) vimentin embryos. DEX was added at stage 18. Scale bar: 25 µm. (F) The ratio of radial processes (RPratio) between the injected and uninjected side of the hindbrain for each injection experiment. Gray bars or white bars indicate with DEX treatment at stage 18 or without DEX treatment, respectively. *p < 0.001 compared with the ratio of nucβ-Gal-injected embryos. **p < 0.001 compared with the ratio of at-MAPKK/Ets1-GRT36A-injected embryos without DEX treatment. ***p < 0.001 compared with the ratio of Ets1-GRT36A-injected embryos without DEX treatment. Each value represents the average of 12 independent measurements. The original figure contains color pictures.
Previous studies not only predicted the involvement of Ras-MAPK signaling in radial glia formation through Ets-1, these studies suggested that threonine 36 would play a critical and essential role during Ets-1 mediated radial glia formation.35 To address this possibility, we prepared a mutant Ets1-GR (Ets1-GRT36A) whose threonine in the position 36 was changed to alanine (Fig. 2A). Synthesized RNA of Ets1-GRT36A was injected and analyzed as described above. The number of radial processes in Ets1-GRT36A-injected hindbrain was reduced when compared with the uninjected side (Fig. 5D) and the ratio of radial process number (RPratio) was 0.73 (p < 0.001) (Fig. 5F), indicating 36-threonine of Ets-1 is crucial for the function of Ets-1 in radial glia formation and that loss of this critical residue is inhibitory. We co-injected at-MAPKK with Ets1-GRT36A and the number of radial processes was determined as described above. The increase of RPratio by at-MAPKK itself (RPratio=1.35, p <0.001) was reduced to 0.81 (p < 0.001) by co-injection of Ets1-GRT36A (Fig. 5E and F). Therefore, the effect of radial glia formation observed in at-MAPKK-injected embryos seems not to be secondary to mesoderm induction by at-MAPKK at early developmental stage. All together, our results suggest that Ets-1 is a downstream effector of Ras-MAPK signaling and promotes radial glia formation.
Discussion
In the developing CNS, radial glia cells function as scaffolding cells for neuronal migration,13,14 and as progenitor cells for neurons and glia cells.7,8,15–20 While pointed, an Ets transcription factor, is required for glia cell differentiation in Drosophila,28,29 the function of vertebrate homologues of pointed, Ets-1 and Ets-2, in vertebrate radial glia formation remains poorly understood. We have demonstrated that the overexpression of Ets-1 or Ets-2 increases the number of radial processes in the hindbrain and that either activity is sufficient for radial glia formation prior to neural tube closure during Xenopus embryonic development. In contrast, although Morpholino Oligonucleotides (MO) against Ets-1 and Ets-2 both inhibited radial glia formation, the reduction of radial processes by Ets1-MO could be rescued by mutant Ets-1 activated at stage 18 while rescue of the Ets-2-MO injected embryos required the activation of Ets-2 by stage 14. In addition, the inhibition of radial glia formation by a dominant negative form of Ets-1, which can inhibit the activity of both Ets-1 and Ets-2, was rescued by only co-injection of Ets-1. These results have shown that Ets-1 but not Ets-2 regulates radial glia formation, suggesting that Ets-2 affects radial glia formation indirectly. In fact, the formation of neural tissues from neural ectoderm represented by nrp1 expression was inhibited by MO against Ets-2 but not Ets-1 (Fig. 4). These data support that the function of Ets-2 has an effect on early neural development and Ets-2 indirectly regulates radial glia formation. However, it is not known how Ets-2 affects the expression of nrp1 and neural development, while previous studies showed that Ets-2 plays an essential role in mesoderm patterning during early Xenopus development.56 The further analysis is necessary to investigate the function of Ets-2 during neural development.
Radial glia appear right after neural tube closure during Xenopus embryogenesis11,12 and we have shown that the activity of Ets-1 is required for radial glia formation prior to neural tube closure (Figs. 2 and 3). This indicates that Ets-1 may play a significant role in the process of radial glia cell fate determination. On the other hand, the expression of Ets-1 at late neurula stage overlaps with the lateral edge of vimentin expression in the prospective hindbrain but not other region of the central nervous system (Fig. 1 and data not shown). Thus, Ets-1 may be involved in radial glia cell fate determination in a limited region of developing Xenopus hindbrain. Interestingly, all radial glia cells in mammalian brain are not homogeneous, as evidenced by the expression of Pax-6 in radial glia of the developing cortex but not of the basal telencephalon.57 Xenopus radial glia cells may also have heterogeneous characteristics and Ets-1 may regulate the cell fate determination of some sort of radial glia cells. While Ets-1 is also expressed in the inferior or lateral edge of neural tube at stage 25 or 30 and share borders with vimentin expression (Fig. 1), it remains unresolved whether Ets-1 is involved in radial glia development in hindbrain at later stages.
Ets-1 is phosphorylated at threonine 38, which is identical to Xenopus Ets-1 threonine 36, and activated by Ras-MAPK signaling to activate the transcription of downstream target genes.35–37 Our data have shown that changing the threonine 36 to alanine in Ets-1 inhibits radial glia formation, suggesting that the phosphorylation of this threonine residue must be important for activating Ets-1 in radial glia formation. Consistent with the role of Ets-1 and the importance of threonine 36, we have shown that a dominant negative form of Ras inhibits radial glia formation and a constitutive active form of MAPK kinase (MAPKK) increases the number of radial processes. This enhancement of radial glia formation by a constitutive active form of MAPKK was inhibited by threonine 36 mutated Ets-1 (Ets1-GRT36A in Results). These data suggest that Ets-1 lies downstream of Ras-MAPK signaling in the promotion of radial glia cell fates. FGF and Neuregulin-ErbB signalings have been shown to be involved in mammalian radial glia development.58–62 While FGF signal promotes radial glial character in vivo, it lacks the signaling capacity to promote proliferation in vitro.58 Neuregulin signal promotes the maintenance and elongation of radial glia cells.59–62 Since FGF and Neuregulin signaling activate Ras-MAPK signaling,63–65Ets-1 may be a downstream component for FGF or Neuregulin during radial glia development. However, how Ras-MAPK-Ets-1 signaling is activated in radial glia formation is not understood completely.
Ets-1 is known to form complexes with other transcription factors and co-factors, and activate the transactivation of target genes.66—68 Those partners interact with Ets-1 through Ets-1 DNA binding domain.67,68 On the other hand, the PNT domain of Ets-1 regulates Ets-1 phosphorylation by serving as a MAPK (ERK2) docking site69 and enhances the activity of the Ets-1 transactivation domain.70 The phosphorylation of threonine 38 of mammalian Ets-1 is required for the interaction with CBP/p300, a co-activator, and this interaction enhances the transactivation of target genes.71 In our study, threonine 36 mutated Xenopus Ets-1 inhibited radial glia formation promoted by a constitutive active form of MAPKK. Therefore, overexpressed mutated Ets-1 may replace endogenous Ets-1 on the promoter of target genes and block recruitment of transcriptional partners and co-activators such as CBP/p300.
Taken together, Ets-1 but not Ets-2 is required for radial glia formation in the hindbrain during Xenopus embryogenesis and Ets-1 is also a downstream effector of Ras-MAPK signaling in radial glia formation. Our data suggest that Ets-1 is phosphorylated and activated by Ras-MAPK signaling prior to neural tube closure.
Acknowledgements
We thank N. Ueno (NIBB), M. Whitman, H. Sive, C. Chang, R. Habas and Y. Gotoh for kindly gifts of plasmids, and B.G. Szaro and The Developmental Studies Hybridoma Bank for antibody. We also thank C. Altmann and G. Corfas for critical suggestions to the manuscript.
Abbreviations
- Ets
E26 transformation-specific
- MAPK
mitogen activated protein kinase
- CNS
central nervous system
- GFAP
glial fibrillary acidic protein
- MO
morpholino oligonucleotides
Note
Color figures may be found online at: http://www.landesbioscience.com/journals/organogenesis/article/5171
Footnotes
Previously published online as an Organogenesis E-publication: http://www.landesbioscience.com/journals/organogenesis/article/5171
References
- 1.Anderson DJ. The neural crest cell lineage problem: Neuropoiesis? Neuron. 1989;3:1–12. doi: 10.1016/0896-6273(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 2.Lillien L. Neural development: Instructions for neural diversity. Curr Biol. 1997;7:R168–R171. doi: 10.1016/s0960-9822(97)70080-0. [DOI] [PubMed] [Google Scholar]
- 3.Rakic P. Radial versus tangential migration of neuronal clones in the developing cerebral cortex. Proc Natl Acad Sci USA. 1995;92:11323–11327. doi: 10.1073/pnas.92.25.11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi BH. Radial glia of developing human fetal spinal cord: Golgi, immunohistochemical and electron microscopic study. Brain Res. 1981;227:249–267. doi: 10.1016/0165-3806(81)90112-7. [DOI] [PubMed] [Google Scholar]
- 5.Levitt P, Rakic P. Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J Comp Neurol. 1980;193:815–840. doi: 10.1002/cne.901930316. [DOI] [PubMed] [Google Scholar]
- 6.Götz M, Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Götz M. Neuronal or glial progeny: Regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 8.Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 9.Hartfuss E, Galli R, Heins N, Götz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 10.Gervasi C, Stewart CB, Szaro BG. Xenopus laevis peripherin (XIF3) is expressed in radial glia and proliferating neural epithelial cells as well as in neurons. J Comp Neurol. 2000;423:512–531. [PubMed] [Google Scholar]
- 11.Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Messenger NJ, Warner AE. The appearance of neural and glial cell markers during early development of the nervous system in the amphibian embryo. Development. 1989;107:43–54. doi: 10.1242/dev.107.1.43. [DOI] [PubMed] [Google Scholar]
- 13.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 14.Hatten ME, Mason CA. Mechanisms of glial-guided neuronal migration in vitro and in vivo. Experientia. 1990;46:907–916. doi: 10.1007/BF01939383. [DOI] [PubMed] [Google Scholar]
- 15.Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 16.Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 18.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 19.Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamaki S, Eckert K, He D, Sutton R, Doshe M, Jain G, Tushinski R, Reitsma M, Harris B, Tsukamoto A, Gage F, Weissman I, Uchida N. Engraftment of sorted/expanded human central nervous system stem cells from fetal brain. J Neurosci Res. 2002;69:976–986. doi: 10.1002/jnr.10412. [DOI] [PubMed] [Google Scholar]
- 21.McManus MF, Golden JA. Neuronal migration in developmental disorders. J Child Neurol. 2005;20:280–286. doi: 10.1177/08830738050200040301. [DOI] [PubMed] [Google Scholar]
- 22.Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J, Macdonald T, Rutka J, Guha A, Gajjar A, Curran T, Gilbertson RJ. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Karim FD, Urness LD, Thummel CS, Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA, Gunther CV, Nye JA. The ETS-domain: A new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes and Development. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- 24.Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994;33:13509–13516. doi: 10.1021/bi00250a001. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Mao X, Olejniczak ET, Nettesheim DG, Yu L, Meadows RP, Thompson CB, Fesik SW. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994;1:871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- 26.Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 27.Bassuk AG, Leiden JM. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64:65–104. doi: 10.1016/s0065-2776(08)60887-1. [DOI] [PubMed] [Google Scholar]
- 28.Klambt C. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117:163–176. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- 29.Klaes A, Menne T, Stollewerk A, Scholz H, Klambt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 31.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 32.Pribyl LJ, Watson DK, McWilliams MJ, Ascione R, Papas TS. The Drosophila ets-2 gene: Molecular structure, chromosomal localization, and developmental expression. Dev Biol. 1988;127:45–53. doi: 10.1016/0012-1606(88)90187-x. [DOI] [PubMed] [Google Scholar]
- 33.Albagli O, Soudant N, Ferreira E, Dhordain P, Dewitte F, Begue A, Flourens A, Stehelin D, Leprince D. A model for gene evolution of the ets-1/ets-2 transcription factors based on structural and functional homologies. Oncogene. 1994;9:3259–3271. [PubMed] [Google Scholar]
- 34.Albagli O, Klaes A, Ferreira E, Leprince D, Klambt C. Function of ets genes is conserved between vertebrates and Drosophila. Mech Dev. 1996;59:29–40. doi: 10.1016/0925-4773(96)00568-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffer P, de Jonge M, Mettouchi A, Binetruy B, Ghysdael J, Kruijer W. JunB promoter regulation: Ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene. 1994;9:911–921. [PubMed] [Google Scholar]
- 37.Bradford AP, Conrad KE, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Functional interaction of c-Ets-1 and GHF-1/Pit-1 mediates Ras activation of pituitary-specific gene expression: Mapping of the essential c-Ets-1 domain. Mol Cell Biol. 1995;15:2849–2857. doi: 10.1128/mcb.15.5.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–1565. [PubMed] [Google Scholar]
- 39.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes and Development. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng HB. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 1991;36:657–662. [PubMed] [Google Scholar]
- 42.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Garland Publishing, Inc; 1994. [Google Scholar]
- 43.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes and Development. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 44.Whitman M, Melton DA. Involvement of p21ras in Xenopus mesoderm induction. Nature. 1992;357:252–254. doi: 10.1038/357252a0. [DOI] [PubMed] [Google Scholar]
- 45.Gotoh Y, Masuyama N, Suzuki A, Ueno N, Nishida E. Involvement of the MAP kinase cascade in Xenopus mesoderm induction. EMBO J. 1995;14:2491–2498. doi: 10.1002/j.1460-2075.1995.tb07246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harland RM. In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 47.Takada H, Hattori D, Kitayama A, Ueno N, Taira M. Identification of target genes for the Xenopus Hes-related protein XHR1, a prepattern factor specifying the midbrain-hindbrain boundary. Dev Biol. 2005;283:253–267. doi: 10.1016/j.ydbio.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 48.Meyer D, Durliat M, Senan F, Wolff M, Andre M, Hourdry J, Remy P. Ets-1 and Ets-2 proto-oncogenes exhibit differential and restricted expression patterns during Xenopus laevis oogenesis and embryogenesis. Int J Dev Biol. 1997;41:607–620. [PubMed] [Google Scholar]
- 49.Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura O, Yamada K. Differences in the fine structure and chemical constitution of nucleolar bodies and the true nucleolus in Xenopus laevis embryos. Dev Growth Differ. 1971;13:303–321. doi: 10.1111/j.1440-169x.1971.00303.x. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida M, Colman DR. Glial-defined rhombomere boundaries in developing Xenopus hindbrain. J Comp Neurol. 2000;424:47–57. [PubMed] [Google Scholar]
- 52.Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- 53.Pourtier-Manzanedo A, Vercamer C, Van Belle E, Mattot V, Mouquet F, Vandenbunder B. Expression of an Ets-1 dominant-negative mutant perturbs normal and tumor angiogenesis in a mouse ear model. Oncogene. 2003;22:1795–1806. doi: 10.1038/sj.onc.1206215. [DOI] [PubMed] [Google Scholar]
- 54.Richter K, Good PJ, Dawid IB. A developmentally regulated, nervous system-specific gene in Xenopus encodes a putative RNA-binding protein. New Biol. 1990;2:556–565. [PubMed] [Google Scholar]
- 55.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: A view from the ETS superfamily. Bioessays. 2005;27:285–298. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 56.Kawachi K, Masuyama N, Nishida E. Essential role of the transcription factor Ets-2 in Xenopus early development. J Biol Chem. 2003;278:5473–5477. doi: 10.1074/jbc.M211054200. [DOI] [PubMed] [Google Scholar]
- 57.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 58.Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rio C, Rieff HI, Qi P, Khurana TS, Corfas G. Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. [DOI] [PubMed] [Google Scholar]
- 60.Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- 61.Schmid RS, McGrath B, Berechid BE, Boyles B, Marchionni M, Sestan N, Anton ES. Neuregulin 1-erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex. Proc Natl Acad Sci USA. 2003;100:4251–4256. doi: 10.1073/pnas.0630496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patten BA, Peyrin JM, Weinmaster G, Corfas G. Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation. J Neurosci. 2003;23:6132–6140. doi: 10.1523/JNEUROSCI.23-14-06132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 64.Si J, Luo Z, Mei L. Induction of acetylcholine receptor gene expression by ARIA requires activation of mitogen-activated protein kinase. J Biol Chem. 1996;271:19752–19759. doi: 10.1074/jbc.271.33.19752. [DOI] [PubMed] [Google Scholar]
- 65.Tansey MG, Chu GC, Merlie JP. ARIA/HRG regulates AChR epsilon subunit gene expression at the neuromuscular synapse via activation of phosphatidylinositol 3-kinase and Ras/MAPK pathway. J Cell Biol. 1996;134:465–476. doi: 10.1083/jcb.134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Remy P, Baltzinger M. The Ets-transcription factor family in embryonic development: Lessons from the amphibian and bird. Oncogene. 2000;19:6417–6431. doi: 10.1038/sj.onc.1204044. [DOI] [PubMed] [Google Scholar]
- 67.Sharrocks AD. The ETS-domain transcription factor family. Nature Reviews. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 68.Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 69.Seidel JJ, Graves BJ. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes and Development. 2002;16:127–137. doi: 10.1101/gad.950902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneikert J, Lutz Y, Wasylyk B. Two independent activation domains in c-Ets-1 and c-Ets-2 located in non-conserved sequences of the ets gene family. Oncogene. 1992;7:249–256. [PubMed] [Google Scholar]
- 71.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]