Abstract
Plant pathogenic microbes deliver effector proteins inside host cells to modulate plant defense circuitry and enable parasitic colonization. As genome sequences from plant pathogens become available, genome-wide evolutionary analyses will shed light on how pathogen effector genes evolved and adapted to the cellular environment of their host plants. In the August 2007 issue of Plant Cell, we described adaptive evolution (positive selection) in the cytoplasmic RXLR effectors of three recently sequenced oomycete plant pathogens. Here, we summarize our findings and describe additional data that further validate our approach.
Key words: plant-microbe interactions, effectors, gene families, positive selection
A diverse number of plant pathogens, including bacteria, oomycetes, fungi and nematodes, deliver effector proteins inside host cells to modulate plant defense circuitry and enable parasitic colonization.1–8 Because these so-called cytoplasmic effectors function inside plant cells and produce phenotypes that extend to plant cells and tissues, their genes are expected to be the direct target of the evolutionary forces that drive the antagonistic interplay between pathogen and host.9,10 In a study published in the August 2007 issue of Plant Cell, we and our collaborators examined the extent to which positive selection (adaptive evolution) has shaped the evolution of the cytoplasmic effectors of three recently sequenced oomycete plant pathogens Phytophthora sojae, Phytophthora ramorum, and Hyaloperonospora parasitica (Genome Sequencing Center at Washington University).11
Oomycete RXLR Effectors are Modular Proteins
Four oomycete Avr proteins have been described in the past three years and were found to contain a secretory signal peptide followed by a conserved domain featuring the motif RXLR, flanked by a high frequency of acidic (D/E) residues.1,3,12 The RXLR motif defines a domain that functions in delivery of effector proteins into host cells.13 It is similar in sequence and position and is functionally interchangeable with the plasmodial host translocation (HT)/Pexel motif that functions in delivery of parasite proteins into the cytoplasm of red blood cells of mammalian hosts.14 Also, the RXLR motif is not required for the effector activities of P. infestans AVR3a when this protein is directly expressed inside plant cells consistent with a role in targeting rather than effector activity.15 Altogether these findings led to the view that oomycete RXLR effectors are modular proteins with two major functional domains.3 While the N-terminal domain encompassing the signal peptide and RXLR leader functions in secretion and targeting, the remaining C-terminal region carries the effector activity and operates inside plant cells.
Ab Initio Identification of RXLR Effectors: Rationale
In the initial part of our study, we aimed to develop a method for ab initio identification of RXLR effector genes in the sequenced genomes. Our approach was to first determine the defining features of validated oomycete RXLR effectors in order to develop a robust set of data mining criteria. We therefore, developed an unbiased list of 43 oomycete RXLR proteins consisting of validated effectors and their closest homologs. Also, to objectively address the extent to which the tetrapeptide RXLR sequence is overrepresented and positionally constrained in Phytophthora, we examined the distribution of the RXLR sequence in the proteomes of these species compared to 46 other eukaryotes. These analyses indicated that the RXLR sequence is significantly overrepresented and positionally constrained in the secretomes of Phytophthora relative to other eukaryotes and formed the basis of the ab initio algorithm.
Ab Initio Identification of RXLR Effectors: Further Validation
Since the publication of our study, two new avirulence genes, PsAvr1a and PsAvr3a, were reported from Phytophthora sojae by Mark Gijzen laboratory, London, Ontario, Canada (GenBank accessions ABQ81647 and ABO47652). Interestingly, PsAvr1a and PsAvr3a fulfill our criteria for RXLR effectors and were identified by our ab initio algorithm (Supplemental Table S2 of the Win et al. paper). In Table 1, we list the features of PsAvr1a and PsAvr3a, and their 34 homologous genes. The mean values for protein size, position of RXLR, and position of EER sequence obtained with this new set of validated RXLR effectors are remarkably similar to those we reported earlier.
Table 1.
New validated RXLR effectors. The new validated effectors are based on two Phytophthora sojae avirulence proteins, PsAvr1a and PsAvr3a, reported by the laboratory of Mark Gijzen, London, Ontario, Canada and their homologs (E value <10−4)
| Description | Accessiona | Species | Evidence | Lengthb | Signal Peptide Lengthb | SignalP v2.0 HMM score | SignalP v2.0 NN score | RXLR Positionc | EER positionc |
| ABQ81647, | |||||||||
| Avirulence effector protein PsAvr3a | Ps_scaffold_80_R245 | P. sojae | Avr effector | 111 | 20 | 0.998 | 0.910 | 43 | |
| ABo47652, | |||||||||
| Avirulence effector protein PsAvr1a | Ps_scaffold_1058_F4 | P. sojae | Avr effector | 121 | 25 | 0.999 | 0.813 | 54 | 64 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_103_F268 | P. ramorum | homolog | 121 | 21 | 0.994 | 0.850 | 54 | |
| Unknown protein similar to PsAvr1a | Pr_scaffold_13_F1570 | P. ramorum | homolog | 129 | 25 | 0.998 | 0.880 | 48 | 68 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_17_F1241 | P. ramorum | homolog | 112 | 21 | 0.997 | 0.815 | 51 | 59 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_207_F26 | P. ramorum | homolog | 138 | 23 | 0.997 | 0.898 | 56 | 69 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_251_R3 | P. ramorum | homolog | 139 | 21 | 1 | 0.894 | 54 | 73 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_26_R566 | P. ramorum | homolog | 141 | 21 | 1 | 0.898 | 54 | 75 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_26_R615 | P. ramorum | homolog | 152 | 21 | 1 | 0.884 | 58 | 86 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_34_F586 | P. ramorum | homolog | 293 | 21 | 1 | 0.938 | 52 | 67 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_50_R933 | P. ramorum | homolog | 140 | 23 | 1 | 0.822 | 52 | |
| Unknown protein similar to PsAvr1a | Pr_scaffold_52_F517 | P. ramorum | homolog | 151 | 22 | 0.994 | 0.811 | 52 | 70 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_64_F233 | P. ramorum | homolog | 293 | 21 | 1 | 0.947 | 52 | 67 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_64_F343 | P. ramorum | homolog | 294 | 21 | 1 | 0.932 | 52 | 67 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_65_R231 | P. ramorum | homolog | 162 | 21 | 1 | 0.953 | 58 | 83 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_75_F477 | P. ramorum | homolog | 136 | 23 | 0.999 | 0.870 | 53 | 69 |
| Unknown protein similar to PsAvr1a | Pr_scaffold_91_R166 | P. ramorum | homolog | 154 | 21 | 0.999 | 0.892 | 57 | 81 |
| Unknown protein similar to PsAvr1a | Ps_scaffold_118_R508 | P. sojae | homolog | 98 | 21 | 0.998 | 0.869 | 54 | 71 |
| Unknown protein similar to PsAvr1a | Ps_scaffold_122_R489 | P. sojae | homolog | 125 | 25 | 0.999 | 0.858 | 48 | 68 |
| Unknown protein similar to PsAvr1a | Ps_scaffold_27_R1297 | P. sojae | homolog | 305 | 21 | 0.996 | 0.951 | 51 | |
| Unknown protein similar to PsAvr1a | Ps_scaffold_3_R4103 | P. sojae | homolog | 130 | 21 | 0.994 | 0.863 | 54 | 70 |
| Unknown protein similar to PsAvr1a | Ps_scaffold_36_F644 | P. sojae | homolog | 137 | 23 | 1 | 0.856 | 53 | 74 |
| Unknown protein similar to PsAvr1a | Ps_scaffold_68_F347 | P. sojae | homolog | 162 | 21 | 1 | 0.898 | 50 | 61 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_1497_R5 | P. ramorum | homolog | 126 | 19 | 0.997 | 0.934 | 41 | 56 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_33_F760 | P. ramorum | homolog | 126 | 19 | 0.998 | 0.942 | 41 | 56 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_33_F786 | P. ramorum | homolog | 125 | 19 | 1 | 0.932 | 41 | 56 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_33_R44 | P. ramorum | homolog | 128 | 19 | 0.998 | 0.942 | 41 | 56 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_34_R60 | P. ramorum | homolog | 127 | 19 | 0.997 | 0.943 | 41 | 56 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_6_R2337 | P. ramorum | homolog | 203 | 20 | 1 | 0.941 | 43 | 61 |
| Unknown protein similar to PsAvr3a | Pr_scaffold_6_R2603 | P. ramorum | homolog | 204 | 20 | 1 | 0.935 | 43 | 61 |
| Unknown protein similar to PsAvr3a | Ps_scaffold_106_F265 | P. sojae | homolog | 131 | 20 | 0.999 | 0.930 | 45 | |
| Unknown protein similar to PsAvr3a | Ps_scaffold_106_R557 | P. sojae | homolog | 131 | 20 | 0.999 | 0.930 | 45 | |
| Unknown protein similar to PsAvr3a | Ps_scaffold_24_F382 | P. sojae | homolog | 137 | 20 | 1 | 0.954 | 44 | |
| Unknown protein similar to PsAvr3a | Ps_scaffold_31_F1779 | P. sojae | homolog | 167 | 20 | 1 | 0.934 | 43 | 56 |
| Unknown protein similar to PsAvr3a | Ps_scaffold_31_R1171 | P. sojae | homolog | 120 | 20 | 1 | 0.950 | 40 | 58 |
| Unknown protein similar to PsAvr3a | Ps_scaffold_87_F189 | P. sojae | homolog | 145 | 22 | 1 | 0.879 | 43 | 75 |
| Means | 155.94 | 21.1 | 0.99 | 0.90 | 48.9 | 66.6 | |||
| Means | 158.3 | 20.7 | 0.99 | 0.86 | 45.0 | 62.1 | |||
| reported by | |||||||||
| Win et al | |||||||||
| (2007) |
The two P. sojae avirulence proteins were reported after we applied the gene mining pipeline described in Win et al (2007) and therefore validate the approach. This list of 36 genes complements the 43 validated effectors described in Table 1 of Win et al. (2007).
GenBank accession number is provided where available. Otherwise, accession numbers correspond to sequences listed in Table S2 of Win et al (2007).
Length in amino acids.
Position counting from N-terminus.
Patterns of Positive Selection are Consistent with the Modular Structure of RXLR Effectors
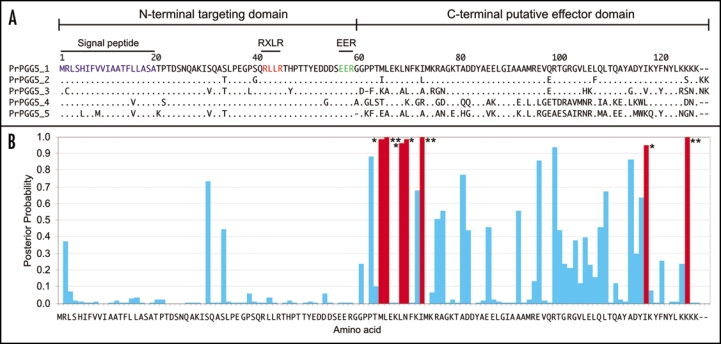
The genome-wide catalogs of RXLR effector genes from the three oomycete species revealed complex and diverse sets of RXLR effector genes that have undergone relatively rapid birth and death evolution. We obtained robust evidence of positive selection in more than two thirds of the examined paralog families of RXLR effectors. Positive selection has acted on paralogous RXLR gene families targeting for the most part the C-terminal region. These findings are consistent with the view that RXLR effectors are modular proteins with the N-terminus involved in secretion and host translocation and the C-terminal domain dedicated to modulating host defenses inside plant cells. In Figure 1, we illustrate the remarkably biased distribution of the positively selected sites towards the C-terminal region for PrPGG5, one of the paralogous gene groups of P. ramorum.
Figure 1.
An example of a paralogous gene group (PGG) with evidence of positive selection focused mainly on the C-terminal effector domain. (A) Multiple sequence alignment of the five Phytophthora ramorum proteins that form PrPGG5. Identical amino acids are indicated by dots. (B) Posterior probabilities estimated by Bayes Empirical Bayes analysis for the model M8 in PAML software package were plotted for each amino acid site in PrPGG5. Positively selected sites are indicated by “*”. *p > 95% and **p > 99%.
Conclusion
In summary, we reported and validated a method for ab initio mining of RXLR effectors in oomycete genome sequences. We applied this method to develop genome-wide catalogs of RXLR effectors and demonstrate that adaptive evolution has shaped the structure of these genes. Future studies will determine the extent to which the positively selected genes and residues identified in our study are functionally important.
Footnotes
Previously published online as a Plant Signaling & behavior E-publication: www.landesbioscience.com/journals/psb/article/5182
References
- 1.Birch PR, Rehmany AP, Pritchard L, Kamoun S, Beynon JL. Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 2006;14:8–11. doi: 10.1016/j.tim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 4.O'Connell RJ, Panstruga R. Tete a tete inside a plant cell: Establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 2006;171:699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- 5.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 6.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 7.Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA. 2006;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang G, Dong R, Allen R, Davis EL, Baum TJ, Hussey RS. A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol Plant Microbe Interact. 2006;19:463–470. doi: 10.1094/MPMI-19-0463. [DOI] [PubMed] [Google Scholar]
- 9.Dawkins R, Krebs JR. Arms Races between and within species. Proc Royal Soc London Series B. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 10.Dawkins R. The Extended Phenotype: The Long reach of the Gene. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- 11.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 12.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PR, Beynon JL. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007 doi: 10.1038/nature06203. In press. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharjee S, Hiller NL, Liolios K, Win J, Kanneganti TD, Young C, Kamoun S, Haldar K. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bos JI, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PR, Kamoun S. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]