Abstract
The importance of cell separation in plant development cannot be overemphasized. The polygalacturonases (PGs) are the one of cell wall hydrolytic enzyme families that has been associated with various cell separation processes in plant development including seed germination, dehiscence, organ abscission, and fruit ripening. Both Arabidopsis and rice PG gene family have expanded in a lineage-specific fashion after the split more than 150 million years ago. Tandem duplications and large-scale duplications are the major contributors to the current PG family size in Arabidopsis. The spatial expression analysis of the 66 Arabidopsis PG family members have led us to conclude that different duplication mechanisms affect the expression divergence differently. This becomes more apparent when temporal examination of expression is conducted in five developmental stages of floral organ abscission in Arabidopsis. Nine distinct patterns of PGs are identified during floral organ abscission in Arabidopsis. Four PGs are specifically upregulated during abscission associated with cell separation process. Careful understanding of relationships among Arabidopsis PGs in a context of evolution together with expression analysis of these PGs will facilitate the functional study of PGs specifically in floral organ abscission in Arabidopsis.
Key Words: cell separation, polygalacturonases, gene duplication, abscission
Cell separation processes have been recognized for their involvement in plant development and more importantly, agricultural traits such as pollen dehiscence and the abscission of organs including leaves, floral organs, and fruits.1–6 Among many cell wall hydrolytic enzymes, the PGs (polygalaturonases) have been shown to be associated with a wide range of these plant developmental programs such as seed germination, organ abscission, pod and anther dehiscence, pollen grain maturation, fruit softening and decay, xylem cell formation, and pollen tube growth.4,7–9 Although the roles of the PG members in the various developmental programs have been hypothesized, many scientists still face challenges in developing a comprehensive understanding of the biological functions of the PG family, due to both size and possible redundancy.10,11
It has been previously reported that PGs from both tomato and Arabidopsis located in tandem clusters were derived from tandem duplications.10,12 In addition, it has been shown by several groups that the Arabidopsis genome contains large blocks of related regions derived from whole genome duplication events.13–15
We have recently conducted a comparative analysis of PGs from Arabidopsis and rice in attempts to address following questions: (1) the patterns and extent of expansion of PG gene family in Arabidopsis and rice, (2) the mechanisms that contribute to the expansion of this gene family, (3) the degree of both spatial and temporal expression divergence amongst Arabidopsis PGs, and (4) possible mechanisms of duplicates retention and the biological roles in floral organ abscission.16
In this study, we have shown that with the identification of the nodes that lead to each Arabidopsis-specific and rice-specific subfamily, there are at least 21 immediate ancestors before the split between these two organisms.16 Subsequent expansion events have been followed in a lineage specific mode in both organisms. We found that in Arabidopsis the PG family is the product of both tandem duplications and large-scale duplications, similar to other gene families such as the NBS-LRR17 and the RLK/Pelle gene family.18 As a matter of fact, more than one third of the Arabidopsis PGs (24 out of 66) were found in tandem clusters. In addition, using both AGI (Arabidopsis Genome initiative) and BHW (Blanc, Hokamp, Wolfe) blocks almost 90% (59 out of 66) of the PGs were included within large-scale duplication events.13,15,16
Interestingly, the correlation between the synonymous substitution rate (Ks) and the expression profile for the related PGs in the tandem clusters was not significant. However, the related PGs in the duplicated blocks from large-scale duplication events tend to have a similar expression pattern within the five major tissues (flowers, siliques, stems, rosette and cauline leaves, and roots). Thus, indicating tandem-duplicated PGs have higher levels of expression divergence compared with PGs found in the large-scale duplication blocks.
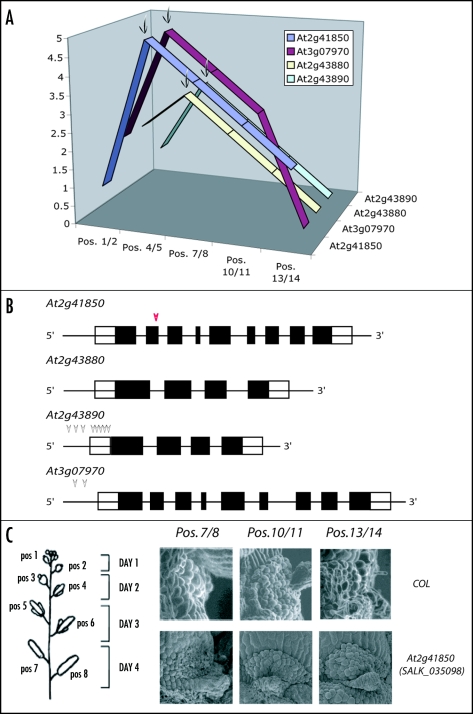
Further examination of PG expression during specific aspects of plant growth such as the five developmental stages of floral organ abscission has led us to conclude that expression divergence between all of the PGs that showed no difference at the tissue level most likely have differences. Thus, by looking more closely at specific tissue types and time of development, the divergence in expression patterns can be revealed. This is also supported by the hypothesis that duplication mechanisms may contribute to divergence of expression differently. In addition, by identifying nine distinct patterns of PG expression during the five developmental stages of floral organ abscission, we provided candidate PGs important for studies of abscission. Considering the proposed roles of PGs in pectin modification and/or breakdown, four of the PGs (At2g41850, At2g43880, At2g43890, and At3g07970) that showed upregulated expression during floral organ abscission associated with cell separation were identified as “best candidates” for understanding floral organ abscission (Fig. 1A).
Figure 1.
Floral organ abscission specific PGs and their T-DNA insertion lines. (A) Expression of four PGs that showed specific upregulation during floral organ abscission is demonstrated. X-axis represents the developmental stages of floral organ abscission. Pos. 1/2 and Pos. 4/5 represent pre-abscission, Pos. 7/8 represents during abscission, and Pos. 10/11 and Pos. 13/14 represent post-abscission. Y-axis represents the relative level of expression of these four PGs and the names of each gene are shown in the z-axis. Four arrows indicate the positions that are associated with cell separation and upregulation of the PGs during abscission. (B) Gene structures of abscission specific PGs and the positions of the T-DNA insertions in each mutant. Each arrow indicates the position of the T-DNA insertion for the mutant. At2g41850 (SALK_035098, red arrow head) is further examined for petal abscission zones using SEM as shown in (C). (C) Schematic diagram of flower positions in wild type in Arabidopsis and SEM of petal abscission zones from SALK_035098 and wild type. Flower position 1 denotes the flower position where white petals protrude right after anthesis and subsequent positions correspond to older stages of flowers. The days after anthesis are demonstrated with flower positions in the right. Note that at flower position 7/8 floral organs are abscised. When comparing during-and post-abscission (Pos. 7/8, Pos. 10/11 and Pos. 13/14) between the mutant and wild type, difference of a delay in floral organ abscission is not observed.
In order to better understand the functions of these PGs in floral organ abscission, we also isolated homozygous T-DNA insertion lines (http://signal.salk.edu/cgi-bin/tdnaexpress) for three of these PGs (At2g41850, At2g43890, and At2g07970) associated with cell separation during abscission. We examined these mutants for a delay in floral organ abscission (Fig. 1B and C); however, nearly all of the single insertion lines examined did not display any notable phenotype regarding floral organ abscission even though in the T-DNA disruption line of At2g41850 showed slight delay positions around 7/8 (Fig. 1 and 1C). One possible explanation is that even though each of these PGs is specifically upregulated right before or during the cell separation process of abscission, there is still redundancy in their functions. One of the approaches to resolve this problem may be the generation of multiple mutants. For example, At2g41850 and At3g07970 are derived from the same ancestor. Thus, examination of double mutants between these two diverged PGs may provide a clue as to the abscission function. In addition, At2g43880, At2g43890 and At2g41850 are also derived from the large-scale duplication events while they share different immediate ancestors. Making double mutants between the two PGs At2g43890 and At2g41850 may provide a clue as to PG function during development. Ultimately, generating triple mutants among At2g41850, At2g43890, and At3g07970 may also provide insights into the possible functions of these PGs in floral organ abscission. Studying functions of a large gene family has been always a challenge due to the size and the redundancy of the family, but these recent findings will facilitate this task.
From our study, we addressed the questions as to how PG gene family has expanded and/or duplicated in the evolution, how the duplicates were retained with the possible biological functions, and how functional studies of PGs in floral organ abscission can be conducted in the future.
Abbreviations
- PG
polygalacturonase
- AGI
Arabidopsis Genome Initiative
- BHW
Blanc, Hokamp, Wolfe
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3541
References
- 1.Rose JKC, Bennett AB. Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends in Plant Sci. 1999;4:176–183. doi: 10.1016/s1360-1385(99)01405-3. [DOI] [PubMed] [Google Scholar]
- 2.Cosgrove DJ. Expansive growth of plant cell walls. Plant Physiol Biochem. 2000;38:109–124. doi: 10.1016/s0981-9428(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JA, Elliott KA, Gonzalez-Carranza ZH. Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol. 2002;53:131–158. doi: 10.1146/annurev.arplant.53.092701.180236. [DOI] [PubMed] [Google Scholar]
- 4.Hadfield KA, Bennett AB. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998;117:337–343. doi: 10.1104/pp.117.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts JA, Whitelaw CA, Gonzalez-Carranza ZH, McManus MT. Cell separation processes in plants: Models, mechanisms, and manipulation. Ann Bot. 2000;86:223–235. [Google Scholar]
- 6.Patterson SE. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie AB. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999;121:419–428. doi: 10.1104/pp.121.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape. Plant Mol Biol. 2001;46:469–479. doi: 10.1023/a:1010619002833. [DOI] [PubMed] [Google Scholar]
- 9.Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y. Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA. 2002;99:15794–15799. doi: 10.1073/pnas.232590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torki M, Mandaron P, Mache R, Falconet D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene. 2000;242:427–436. doi: 10.1016/s0378-1119(99)00497-7. [DOI] [PubMed] [Google Scholar]
- 11.Markovic O, Janecek S. Pectin degrading glycoside hydrolases of family 28: Sequence-structural features, specificities and evolution. Protein Eng. 2001;14:615–631. doi: 10.1093/protein/14.9.615. [DOI] [PubMed] [Google Scholar]
- 12.Hong SB, Tucker ML. Genomic organization of six tomato polygalacturonases and 5′ upstream sequence identity with tap1 and win2 genes. Mol Gen Genet. 1998;258:479–487. doi: 10.1007/s004380050758. [DOI] [PubMed] [Google Scholar]
- 13.Arabidopsis Genome Initiative, author. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 14.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 15.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Shiu SH, Thoma S, Li WH, Patterson SE. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006 doi: 10.1186/gb-2006-7-9-r87. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]