Abstract
Nucleic acid sequences of the prion gene (PRNP) were examined and genotypes compiled for 76 white-tailed deer from northern Illinois, which previously tested positive for chronic wasting disease (CWD), and 120 negative animals selected to control for geographic location and age. Nine nucleotide polymorphisms, seven silent and two coding, were found in the sampled population. All observed polymorphisms except two of very low frequency were observed in both negative and positive animals, although five polymorphic loci had significantly different distributions of alleles between infected and non-infected individuals. Nucleotide base changes 60C/T, 285A/C, 286G/A and 555C/T were observed with higher than expected frequencies in CWD negative animals suggesting disease resistance, while 153C/T was observed more than expected in positive animals, suggesting susceptibility. The two coding polymorphisms, 285A/C (Q95H) and 286G/A (G96S), have been described in white-tailed deer populations sampled in Colorado and Wisconsin. Frequency distributions of coding polymorphisms in Wisconsin and Illinois deer populations were different, an unexpected result considering the sampled areas are less than 150 km apart. The total number of polymorphisms per animal, silent or coding, was negatively correlated to disease status. The potential importance of silent polymorphisms (60C/T, 153C/T, 555C/T), either individually or cumulatively, in CWD disease status has not been previously reported.
Key words: CWD, PRNP, synonymous polymorphism, cumulative polymorphisms, haplotype
Introduction
Chronic wasting disease is the only prion disease established in free ranging animals and is found in white-tailed deer, mule deer, elk and moose.1,2 Factors related to CWD transmission are of particular interest because of its spread in the wild and ability to become established under natural population densities. Environmental transmission of CWD has been demonstrated in captive herd holding pens contaminated with carcass residue or fecal material,3,4 and may also occur by infectious prions present in blood and saliva of deer with CWD.5 The first reported occurrences in wild deer and elk were found in Colorado and Wyoming in 1985. CWD has since spread slowly, often displaying large geographical gaps.6
The first positive cases of CWD east of the Mississippi river were detected during 2002 in Wisconsin,7 and another outbreak was discovered in Illinois later that same year.8 It is unclear whether the two outbreaks are related or of independent origin despite their geographic proximity of less than 150 km. Substantial differences exist between the sites, ranging from a moderately rugged agriculture and deciduous forest mix with distributed deer habitat in the Wisconsin outbreak to flat, large-field agriculture and highly urban landscapes with narrowly connected small habitat fragments in the Illinois outbreak.
Prion protein sequence is important in many aspects of prion disease including etiology, pathology and transmission. A number of PrP polymorphisms that alter resistance to prion disease have been documented in many species,9 and several may alter CWD susceptibility in deer. For example Q95H and G96S polymorphisms have been reported in white-tailed deer from Colorado,10 and south central Wisconsin,11,12 although small sample sizes precluded a clear conclusion about genetic susceptibility. In mule deer the S225F polymorphism was weakly related to CWD susceptibility.13 A S138N polymorphism reported in deer,11,14 has subsequently been shown to reside within a processed pseudogene.10,15 White-tailed deer from the western United States may also have a G65E or A116G polymorphism,16 although these have not been related to CWD.
This research examined the relationship between prion polymorphisms and CWD disease status in free-ranging white-tailed deer from northern Illinois as indicated by allelic frequency distributions of positive and negative animals. Additionally, comparisons between PRNP polymorphisms in Illinois deer and those observed in other CWD outbreaks will provide a better assessment of the underlying mechanisms that link PRNP with CWD susceptibility and resistance.
Results
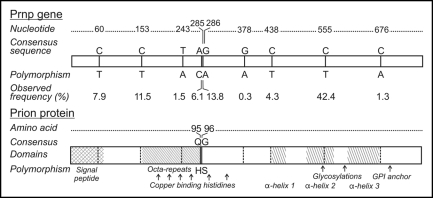
PRNP sequences were determined for 196 deer, 76 CWD positives and 120 negative controls. In the 196 samples, nine single nucleotide polymorphisms (SNP) were detected (Fig. 1), eight of which had been previously described (nucleotide 286G/A,14 nucleotides 60C/T, 153C/T, 438C/T, 555C/T,16 nucleotide 285A/C,11 nucleotides 243T/A, 438C/T,10 nucleotide 676C/A EMBL AY425673).
Figure 1.
Nucleotide and amino acid database consensus sequence and polymorphisms observed in Illinois white-tailed deer. Numbers indicate the nucleotide or deduced amino acid sequence from a consensus. Observed frequency (# of polymorphic alleles/total # of alleles* 100) of polymorphisms and domains within the prion protein are also indicated.
Blast and literature searching indicated the 378G/A SNP has not been previously reported, consistent with our observation in a single individual. Seven of these polymorphic loci were silent and translated to a synonymous amino acid sequence while two of the SNP translated to a change in amino acid sequence of the protein; nucleotide 285A/C to amino acid Q95H and 286G/A to G96S. All of the observed allele frequencies of polymorphic loci (Fig. 1) were less than 50% which indicated that the database derived consensus sequence was also the wild type genotype in Illinois white-tailed deer.
Polymorphisms and CWD status.
Five of the nine polymorphic alleles were significantly related to CWD status according to Chisquare tests (Table 1). Loci 60, 285, 286 and 555 had higher than expected frequencies of polymorphisms in CWD negative animals while the opposite was true at locus 153 where observed frequencies of polymorphic alleles were higher than expected in CWD positive animals. Logistic regression also indicated that polymorphisms at loci 60, 285 and 286 were protective against CWD (p < 0.05) as each had odds ratios less than one (Table 1). All loci were in Hardy-Weinburg equilibrium (HWE) except for locus 286, which was found to significantly deviate from the expected frequency (HWE p < 0.01).
Table 1.
Ratio of observed and expected allele frequencies for nucleotide polymorphisms
| Allele | Total frequency | Frequency in negative population | Frequency in positive population | Odds ratio | Wald 95% confidence internal |
| 60C | 92.1 | 88.8 | 97.4 | 0.21 | 0.07–0.62 |
| *60T | 7.9 | 11.2 | 2.6 | ||
| 153C | 88.5 | 92.5 | 82.2 | 2.66 | 1.41–5.03 |
| *153T | 11.5 | 7.5 | 17.8 | ||
| 243T | 98.5 | 98.3 | 98.6 | ns | |
| 243A | 1.5 | 1.7 | 1.4 | ||
| 285A | 93.9 | 91.7 | 98.0 | 0.21 | 0.06–0.72 |
| *285C | 6.1 | 8.3 | 2.0 | ||
| 286G | 86.2 | 79.6 | 96.7 | 0.13 | 0.05–0.34 |
| *286A | 13.8 | 20.4 | 3.3 | ||
| 378G | 99.7 | 99.6 | 100 | ns | |
| 378A | 0.3 | 0.4 | 0 | ||
| 438C | 95.7 | 95.0 | 96.7 | ns | |
| 438T | 4.3 | 5.0 | 3.3 | ||
| 555C | 57.6 | 53.8 | 63.8 | 0.66 | 0.43–1.00 |
| *555T | 42.4 | 46.2 | 36.2 | ||
| 676C | 98.7 | 97.9 | 100.0 | ns | |
| 676A | 1.3 | 2.1 | 0 |
Bold text indicates loci where nucleotide sequence resulted in a change of expressed protein; other polymorphisms were synonymous.
Indicates a significant association (p < 0.05) between allele and CWD status using chi-square expected frequency for presence/absence of each allele obtained from PROC FREQ. Odds ratios were obtained with PROC LOGISTIC using each allele as a class variable regressed against CWD as an outcome. Only odds ratios with significant (p < 0.05) parameter estimates are reported. Polymorphisms with non-significant odds ratios and 95% confidence limits overlapping 1 are denoted by ns.
The relationship of CWD with polymorphic genotype frequency (Table 2) was similar to that for polymorphic allele frequency. Individuals heterozygous for polymorphisms at 60, 285 and 286 had significantly reduced odds ratios. However, the low number of individuals homozygous for these polymorphisms prevented calculation of meaningful risk reduction for these genotypes. Polymorphism at locus 555 was more common (42.4% overall) and individuals with a homozygous 555T/T genotype (Table 2) had a reduced odds ratio of 0.29 for CWD. However, the odds ratios for the polymorphic 555T allele (Table 1), as well as for the heterozygous genotype 555C/T (Table 2), overlapped 1 and so were not considered significant.
Table 2.
Ratio of observed and expected genotype frequencies for nucleotide polymorphisms
| Locus | Genotype | n | Frequency in negatives | Frequency in positives | Odds ratio | Wald 95% confidence interval |
| *60** | CC | 166 | 78.4 | 94.7 | ||
| CT | 29 | 20.8 | 5.3 | 0.21 | 0.07–0.63 | |
| TT | 1 | 0.8 | 0 | nc | ||
| *153 | CC | 155 | 85.8 | 68.4 | ||
| CT | 37 | 13.3 | 27.6 | 2.60 | 1.25–5.40 | |
| TT | 4 | 0.8 | 4.0 | 5.94 | 0.60–58.52 | |
| 243 | TT | 191 | 97.5 | 97.4 | ||
| TA | 4 | 1.7 | 2.6 | ns | ||
| AA | 1 | 0.8 | 0 | ns | ||
| *285** | AA | 173 | 83.3 | 96.1 | ||
| AC | 22 | 15.8 | 3.9 | 0.22 | 0.06–0.76 | |
| CC | 1 | 0.8 | 0 | nc | nc | |
| *286 | GG | 153 | 68.3 | 93.4 | ||
| GA | 32 | 22.5 | 6.6 | 0.21 | 0.08–0.59 | |
| AA | 11 | 9.2 | 0 | nc | nc | |
| 378 | GG | 195 | 99.2 | 100 | ||
| GA | 1 | 0.8 | 0 | ns | ||
| AA | 0 | 0 | 0 | ns | ||
| 438 | CC | 179 | 90 | 93.4 | ||
| CT | 17 | 10 | 6.6 | ns | ||
| TT | 0 | 0 | 0 | ns | ||
| *555 | CC | 65 | 31.6 | 35.5 | ||
| CT | 96 | 44.2 | 56.6 | 1.14 | 0.60–2.16 | |
| TT | 35 | 24.2 | 7.9 | 0.29 | 0.11–0.80 | |
| 676 | CC | 192 | 96.7 | 100 | ||
| CA | 3 | 2.5 | 0 | ns | ||
| AA | 1 | 0.8 | 0 | ns |
Bold text indicates loci where nucleotide sequence resulted in a change of expressed protein; other polymorphisms were synonymous.
Indicates a significant association (p < 0.05) between each genotype and CWD status using chi-square expected frequency obtained from PROC FREQ. Odds ratios were obtained with PROC LOGISTIC using each SNP as a class variable regressed against CWD as an outcome. Only odds ratios with significant (p < 0.05) parameter estimates are reported. Polymorphisms with non-significant odds ratios and 95% confidence limits overlapping 1 are denoted by ns.
Indicates loci with significant linkage disequilibrium (p < 0.01). “nc” indicates no cases so odds ratios were not calculable.
The polymorphism at locus 153 was a risk factor for CWD (Table 1), with 153T alleles more than doubling the risk of being CWD positive (odds ratio of 2.66). Similarly, heterozygous genotypes resulted in a 2.6-fold increase in CWD risk (Table 2). Although only four deer were found to be polymorphic homozygous at 153 (153T/T), this genotype produced a near 6-fold increase in the odds of CWD. Confidence limits for 153T/T, however, included 1 and so were not considered significant.
Statistical power to detect differences between positive and negative animals was good for most alleles although the low frequencies observed for some polymorphic alleles limited power. For example, the polymorphic coding alleles had power of 0.70 and 0.99 with an α of 0.05 and a two-tailed test. In contrast, the frequency of individual polymorphic alleles at 243, 378, 438 and 676 resulted in a power of less than 0.50 limiting the potential to detect an actual difference between positive and negative animals in this study.
Linkage disequilibrium between loci was observed in both positive and negative populations with the strongest association occurring between loci 60 and 285 (χ2 = 70.5, p < 0.01). Fifteen deer were polymorphic at either locus 60 or locus 285 and nineteen deer had both polymorphisms. Including both correlated loci in a multiple logistic regression model resulted in adjusted odds ratios of 0.18 and 0.21 for heterozygous genotypes 60C/T or 285A/C respectively.
Haplotype frequencies were estimated from unphased genotypes and the nine most common haplotypes (frequencies greater than 1%) are reported in Table 3. These haplotypes were present in 97% of deer sampled. Thirty-three different haplotypes were reconstructed in the sampled population with three haplotypes being distinct to the CWD positive population and nine of the haplotypes restricted to negative animals. The permutation test performed on the haplotypes determined that CWD positive and negative animals had significantly different distributions of haplotype frequencies (p < 0.01), suggesting that genetic variation between CWD positive and negative populations exceeds genetic variation within the two groups. Unfortunately, low frequencies in the majority of haplotypes prevented us from using the phased data in further exploratory analyses.
Table 3.
Reconstructed frequencies of common haplotypes for CWD positive animals and controls
| Haplotype definition | Frequency in (-) animals | Frequency in (+) animals |
| CCTAGGCCC | 33.0 | 39.8 |
| CCTAGGCTC | 28.1 | 31.6 |
| CCTAAGCTC | 12.2 | 2.6 |
| CTTAGGCCC | 10.3 | 16.3 |
| TCTCGGCCC | 4.6 | 0.0 |
| CCTAGGTCC | 3.9 | 2.8 |
| TCTAGGCCC | 1.4 | 1.8 |
| TCTAGGCCA | 1.3 | 0.0 |
| CCTCGGCCC | 1.1 | 1.5 |
Haplotypes were generated from unphased genotypes with the Bayesian (ELB) algorithim in Phase ver 2.1. The distribution of haplotype frequencies was significantly different between CWD positive and CWD negative animals (p < 0.01). Only haplotypes with frequencies greater than 1% are reported.
In the population sampled, 89% of the CWD positive animals had the consensus genotypes at both coding polymorphisms while 57% of the negative animals had the consensus genotypes at these loci (285A = Q95 and 286G = G96). No deer with two coding SNP, either homozygous for either polymorphic coding allele, or heterozygous for both coding alleles, were CWD positive although negative deer with these genotypes were observed (p < 0.01).
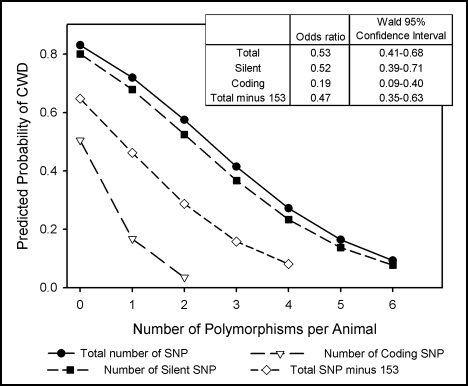
Because it appeared an additional copy of coding SNP reduced the percentage of CWD positive animals, we examined the relationship of multiple polymorphisms and disease status (Fig. 2). If deer were grouped by the cumulative number of polymorphic alleles per animal, a significant Wald Chi-Square was observed for the model (p < 0.01). Reverse coding or omission of locus 153 from models did not alter outcome, as both models were significant and produced similar odds ratio estimates (p < 0.01).
Figure 2.
Probability of CWD as predicted by the number of polymorphic alleles. Total polymorphisms, silent polymorphisms, coding polymorphisms and total polymorphisms omitting locus 153 were used as independent variables in separate logistic regression models with CWD as the outcome variable. Odds ratios were estimated using the number of heterozygous and homozygous SNP summed across all loci. Coding for locus 153 was reversed because wild-type alleles conferred susceptibility (2 = homozygous for the consensus genotype, 1 = heterozygous, 0 = homozygous for the polymorphism), which resulted in a higher predicted probability of CWD than in a model omitting locus 153.
To examine relative contributions of expressed and silent changes to CWD resistance, coding and silent polymorphisms were modeled separately and combined for multivariate analysis. Single variable models produced odds ratios of 0.19 for each coding polymorphic allele, and odds ratios of 0.52 for each silent polymorphic allele (Fig. 2). Multivariate modeling of silent and coding polymorphisms resulted in significant Wald Chi-Square (p < 0.05) tests for both variables and significant Log Likelihood ratio tests (p < 0.01). Including independent variables for both coding and silent polymorphisms also decreased AIC values compared to single variable models, indicating that multivariate analysis improved the overall fit of the regression model.
We found no relationships between CWD status and age or gender although the experimental design was not intended to test this question. Polymorphisms were not related to age or gender according to chi-square tests. Protease resistant PrP was detected by immunohistochemistry in retropharyngeal lymph nodes of all CWD positive deer and in obex from just over 75% of positive deer. No relationship was apparent between genotypes and presence or absence of protease resistant PrP in the obex.
The non-coding pseudogene was present in approximately 15% of the animals sampled. Sequencing of the pseudogene PCR products revealed that the observed polymorphisms were indeed present in the functional PRNP gene nucleotide sequences, and were not detected in the pseudogene. All pseudogene PCR products displayed a 138S/N substitution, as had been previously reported.10,15 Presence of the pseudogene was not significantly correlated to CWD test status, age or gender of the animal according to chi-square tests.
Geographic variation in polymorphisms.
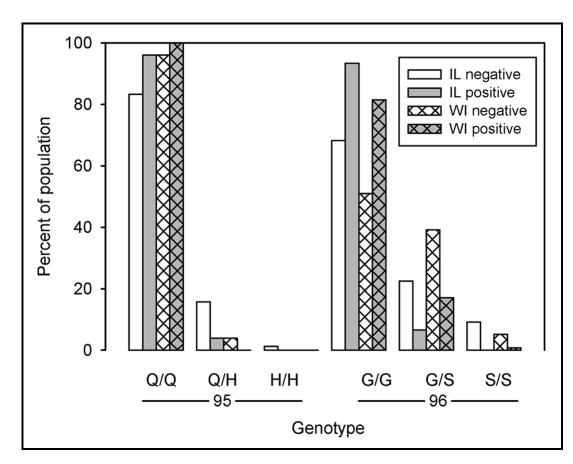
Polymorphisms with significant association to disease status were distributed across the study area and have been found in other geographical locations. Low frequency polymorphisms had a non-uniform distribution across the study area, as would be expected for a gene introduced by a recent migrant or mutation, but low numbers limited spatial analysis. The same coding polymorphisms observed in Illinois have been reported in Wisconsin deer 150 km to the northwest. Based on our observed genotype frequencies and published data,12 the proportion of deer with consensus genotypes were similar. Approximately 57% of the Illinois negative deer had a homozygous consensus sequence compared to 51% of Wisconsin negative deer, while 90% of the Illinois positive deer had a consensus sequence compared to 82% of the Wisconsin CWD positive deer. However, genotype frequencies of the two coding polymorphisms (Fig. 3) differed according to chi-square test (p < 0.05). Illinois had a higher frequency of the Q95H polymorphism than Wisconsin with 17% percent of negative deer heterozygous or homozygous for Q95H in Illinois while only 4% of Wisconsin negative deer had this polymorphism. Conversely, G96S was more common in the Wisconsin herd with 44% of negative and 17% of CWD positive deer heterozygous or homozygous for this polymorphism compared to 32% of negative and 7% of CWD positive deer from Illinois. Deer with either SNP had a lower frequency of CWD than those with the consensus in both states.
Figure 3.
Distribution of coding polymorphisms Q95H and G96S. Bars represent the percent of sampled population of the indicated genotype within disease status. Distribution of coding polymorphisms for sampled populations of Illinois deer are represented by open bars. Distribution of polymorphisms for Wisconsin deer (hatched bars) were calculated from published results.12
Discussion
Polymorphisms that alter resistance to prion disease are present in many species.9 For example, in sheep PrP containing the V136 R154 Q171 polymorphic sequence confers scrapie susceptibility while a combination of A136 R154 R171 alleles confers scrapie resistance.27,28 Selection for the resistant alleles has been used in the European Union to reduce the prevalence of scrapie.29 Wildlife managers are similarly interested in the potential of genetic resistance in managing CWD, both for understanding or predicting spread of disease, and for the possibility of use in repopulation following herd reductions. Furthermore, information about polymorphisms and susceptibility in additional species could contribute to the general understanding of prion disease mechanisms.
This study identified allelic variants at nucleotide positions 60, 285, 286 and 555 (codons 20, 95, 96 and 185) of the white-tailed deer prion gene associated with resistance and at nucleotide 153 (codon 51) associated with susceptibility to CWD. The association with resistance to CWD of polymorphism 285A/C which codes for Q95H and 286G/A which codes for G96S, confirms previous reports of a genetic trend or association in enclosed herds,10 and in a free-ranging white-tailed deer population.12 Recently it was reported that S96 (286G/A) transgenic mice inoculated with CWD displayed high levels of resistance to multiple strains of the disease when compared to their G96 transgenic counterparts,30 which is consistent with the absence of CWD positive deer homozygous for serine at amino acid 96 in the present study area.
Although frequencies of individual polymorphisms ranged from 0.3–42.4% (Fig. 2), the majority (85%) of deer tested had at least one of the statistically significant nucleotide sequence changes, and half displayed more than one. Deer from Illinois exhibited low allelic diversity of polymorphisms when compared to deer from the western United States but the frequencies of the two observed coding alleles were similar between regions. Heaton et al.16 found seveb coding and 13 synonymous polymorphisms in white-tailed deer from Wyoming, with frequencies of G96S reported at 20%, close to the 13.8% observed in Illinois deer. O'Rourke, et al.10 reported three coding polymorphisms and a G96S frequency of 13% in Nebraska deer although Q95H was present in only 1% of that population, lower than the 6.1% observed in Illinois. Discrepancies between geographic locations in allelic distributions are also apparent in the frequencies of synonymous mutations 60C/T, 153C/T and 555C/T which Heaton et al.16 report as 3, 6 and 20%, all lower than the 7.9, 11.5 and 42.4% in the Illinois study. Observed frequencies of the two coding polymorphisms, Q95H and G96S, also differed between central Wisconsin and northern Illinois, which implies some degree of genetic isolation exists between these two locations though they are only separated by approximately 150 km.
Collectively these findings suggest that regional variations in PRNP might be a result of barriers to gene flow between regions or a function of geographic distance. Dissimilarities in genetic composition between the Illinois and Wisconsin herds could be attributed to barriers such as notable habitat variation between the two study sites,31 or differences in patterns of deer dispersal in agricultural environments compared to forested environments.19 Differences in the proportion of even fairly rare individual genotypes with a degree of genetic resistance might be important in CWD spread within discrete populations. By analogy with herd immunity,32 resistant individuals might reduce efficiency of transmission by direct animal contact disproportionately to their numbers.
The two coding polymorphisms at residues 95 and 96 are in the metal binding region of the protease resistance PrPsc core, consisting roughly of residues 90–230.33 Binding of copper or other metals in this domain may modulate conformational change, self-association and PrPsc propagation.34,35 The role of copper in propagation of prion disease is clearly complex as both copper chelation,36 and conversely, copper addition,37 have been reported to delay prion disease in vivo. Similarly either deletion,38,39 or insertion,40,41 of additional copper binding octapeptide repeats may increase prion disease susceptibility. In vitro, copper attenuates conversion of PrP into PrPsc fibrils,42 but also enhances protease resistance of PrP,43 as well as PrPsc fibrils.42,44 The N-terminal domain of PrP binds up to six Cu2+ ions. Copper binding occurs at histidine in four of the five octapeptide repeats as well as with the two histidine residues at amino acids 99 (H95 in mouse, H96 in human PrP) and 114 (H110 in mouse, H111 in human PrP). At higher copper concentrations, each Cu2+ ion is coordinated by a single histidine imidazole and an amide nitrogen from glycine.45,46 At lower copper concentrations, all six histidines are involved in binding two Cu2+ ions.34 The Q95H polymorphism may represents a gain in potential copper binding in vivo. If this is the case, copper binding would appear protective since Q95H animals were more likely to be CWD negative.
An alternative explanation for the relationship of the coding polymorphisms to disease status is the close proximity of Q95H and G96S to proteolytic cleavage sites at amino acid residues 82 and 78.47 In human Creutzfeldt-Jakob disease (CJD), polymorphisms in the amino acid sequence have been associated with alterations in location of these cleavage sites.48 In addition, polymorphisms quite distal to cleavage sites may alter conformation sufficiently to change proteolytic susceptibility. Prion protein from humans homozygous for M129 has a secondary cleavage site shifted towards the N-terminus while V129 results in an additional cleavage site shifted towards the C-terminus of the protein.48 Similar observations have been made in elk where an M132L polymorphism shifts the cleavage site towards the C-terminus.49
Synonymous or silent polymorphisms are non-coding and do not alter the amino acid sequence of translated protein. Furthermore, none of our observed synonymous polymorphisms have been previously related to CWD susceptibility. So it was initially surprising that one of the lower odds ratios for CWD corresponded to the 60C/T polymorphism which is not only silent but also codes a region cleaved during processing of PrP. The 60C/T polymorphism is correlated with the 285A/C coding polymorphism and linkage is one of several ways that synonymous polymorphisms may be associated with altered phenotype. First, a synonymous sequence may be linked to an expressed sequence elsewhere in the same protein or in a different protein. Such a case occurs in atypical, asymptomatic scrapie, where two synonymous polymorphisms in PRNP have an apparent linkage with the ARH allele, which is under-represented in atypical scrapie. This linkage results in the synonymous polymorphisms also being under-represented in atypical scrapie despite not coding for a protein difference.50 Linkage that may occur between closely related proteins is illustrated by a synonymous polymorphism in the prion related doppel gene that is associated with BSE in Fleckvieh cattle.51 Similarly, a synonymous polymorphism within acetyl-coenzyme A carboxylase, the rate limiting enzyme in lipogenesis, that is positively associated with both milk fat and lactose content,52 indicates potential linkage across multiple proteins.
Alternatively to linkage effects, synonymous polymorphisms may cause phenotypic variation by less well understood mechanisms such as by altering RNA stability, tRNA binding or translational protein folding. There is considerable evidence for conformational variants of PrPsc that transmit distinct pathologies or prion disease strains into animals with identical protein sequence,9 so it is conceivable a translation folding event could alter disease susceptibility. There is also some evidence that RNA mediates enhanced PrP folding during PrPsc conversion,53 and so polymorphic mRNA might also alter translational folding and subsequently, potential for PrPsc formation.
This scenario has some support in a recent observation that multiple synonymous polymorphisms have an additive effect on protein function, similar to our observation in Figure 2. The MDR1 gene encodes a membrane transport protein important in multiple drug resistance which contains a single synonymous polymorphism, 3435C/T, that sometimes has an effect on drug transport,54 as a result of increased mRNA turnover. However, with the addition of two or three further synonymous polymorphisms in MDR1, drug transport is consistently reduced at equivalent mRNA and protein levels in a variety of cell lines.55 Conformation dependent antibody inhibition indicated reduced function was a result of altered protein folding which would have become progressively different as the number of synonymous polymorphisms increased. The inverse relationship between number of SNP, whether silent or coding, and CWD status could be the result of a similar process.
Although we examined only polymorphisms within PRNP, recent reports of multiple insertion-deletion polymorphisms located in the promoter region of bovine PRNP that are significantly associated with BSE,56,57 demonstrate an additional mechanism for synonomous polymorphisms to impact disease. Changes in the promoter region of PRNP apparently alter binding of transcription factors which in turn affect PrP expression.58,59
In addition to genetics, both environmental and physiological factors such as deer densities, habitat composition, and age must be considered when interpreting disease occurrence and pathology. In Wisconsin for example, it appears that older deer have a higher prevalence of CWD than younger animals.7 Differential binding capacities of various soil particles to PrPsc,60 suggests that environmental factors, such as soil composition, could influence CWD infection and transmission. These epigenetic factors may have contributed to the inability of our data to confirm a relationship between genotypes and histochemical locations of resistant PrP in lymph node or obex, which can be indicative of disease progression.10
In general, it appears likely that most white-tailed deer residing in the northern Illinois study area have some degree of genetic susceptibility to CWD despite differences in genotypic frequencies between positive and negative animals. Except for SNP with low frequency (2.5% or less), CWD was observed in deer heterozygous for all observed polymorphisms. The association between the level of allelic variation within the study area and CWD status has interesting implications for CWD management and understanding of CWD transmission in free ranging animals. If further research shows that genetic diversity within the PRNP gene does reduce disease incidence or transmission, it would emphasize the importance for CWD management teams to understand and manage ecological barriers to gene flow in deer while conducting disease surveillance efforts.
Methods
Deer sampling.
A CWD surveillance program for free-ranging wild white-tailed deer, consisting of public hunting in the traditional late fall season and sharp shooting by field biologists in the early spring, was conducted by the Illinois Department of Natural Resources (IDNR) between Winter 2002 and Spring 2006. Hunter harvested deer were located to the nearest section, based on hunter description of locations using detailed topographical maps at deer check stations. A section is a 1.6-by-1.6 square km parcel of known location. Deer population control efforts by field biologists provided detailed spatial information about individual deer including Global Positioning System (GPS) coordinates and time of harvest. Demographic data, including sex and age estimated by dentition were collected from all deer sampled.
Samples of obex and retropharyngeal lymph nodes were tested using USDA approved immunohistochemical procedures to detect protease resistant PrP at the Illinois Department of Agriculture Diagnostic Laboratories in Galesburg or Centralia and most positive samples were confirmed at the National Veterinary Services Laboratory. When possible, frozen tissue samples were archived for both CWD positive and negative deer.
Samples from all CWD positive deer with frozen tissue available were used for DNA extraction, PCR and PRNP sequencing. Given the nature of sample collection during surveillance efforts, we could not do a perfect paired-case control design though for each CWD positive case, negative controls were selected on the basis of age, sex and geographic location. We controlled for age estimated by dentition because of reported age dependency for CWD prevalence in Wisconsin.7 The prevalence of CWD has also been reported to be gender dependent,17 so whenever possible, both female and male negative controls of the same age were selected for each CWD positive deer. Furthermore, with the geographic spread of new CWD cases over time and the fragmented habitat in the study area, we felt it was important to also control for geographic variation in the samples. Deer harvest locations were known to the nearest section (1.6 × 1.6 km) and controls were selected on this basis. To check for geographic control we used ArcGIS and calculated that 66% (50 of 76) of positives were from the same section as their matched negative control, 18% (14 of 76) from an adjacent section, and 16% (12 of 76) were 3.2–8.2 km from the controls. These longer distances are not problematic when compared to deer movements in habitat similar to ours (i.e., intensive row-crop agriculture). For example, Brinkman et al.18 found the mean distance between summer and winter home range was 10.1 km for female deer in a landscape dominated by row-crops and over 85% of all deer moved. Furthermore, winter home range sizes average 5.2 km2 (∼2 sections). In habitats similar to our study area, Nixon et al.19 evaluated white-tailed deer dispersal in Illinois agricultural environments, their findings indicate average distance moved between 30 and 44 km for males and 37 to 41 km for females in East central, West central and Northern Illinois. Thus, 89% of our controls likely fell within the same home range as the positive and the other 15% were within seasonal movement ranges. Whenever possible control animals were selected from the same harvest year as the CWD case.
The study area included four counties in northern Illinois with less than 1/3 of the area containing suitable deer habitat as described by Roseberry and Woolf.20 The mean age of deer in the study was estimated to be 1.8 years based on dentition and consisted of 72 males (30 CWD positive) and 124 females (46 CWD positive). All of the deer tested were free-ranging but all came from counties with a human population exceeding two/hectare, which is three times greater than the state average population density excluding the city of Chicago. Both CWD positive and control animals were sampled from deer habitat within metropolitan and rural areas.
PCR.
Genomic DNA was isolated from skeletal muscle using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) in accordance with manufacturer's instructions. Forward and reverse PCR primers previously published,11 were used to amplify the coding region for the mature prion protein. The forward primer, CWD-13 (5′-TTTTGCAGATAAGTCATCATGGTGAAA-3′) overlaps the junction of Intron 2 and Exon 3 of the prion gene. The reverse primer CWD-LA (5′-AGAAGATAATGAAAACAGGAAGGTTGC-3′) was located 850 bp downstream to the forward primer according to the PRNP numbering in GenBank (Accession #AF156185). To ensure that the observed polymorphic alleles were contained within the functional gene, an additional primer set was utilized to screen for the presence of the non-coding PrP pseudogene. The pseudogene primer set used forward primer 369 (5′-CAACCAAGTCGAAGCATCT-3′) and reverse primer 224 (5′-AGAAGATAATGAAAACAGGAAG-3′).10 If the pseudogene was detected in an animal, both the pseudogene and functional gene PCR reactions were sequenced to determine the true origin of polymorphisms.
A 40 µl reaction was assembled for amplification of the functional gene and pseudogene consisting of 24.2 µl of deionized water, 0.8 µl of 10 mM dNTP, 8 µl of TaqMaster, 4 µl of 10X TaqBuffer, 0.4 µl of Eppendorf Taq (5 Units/µl), 0.8 µl of 10 µM forward primer, 0.8 µl of 10 µM reverse primer and 1 µl (≈20 µg /µl) of genomic DNA. Thermal cycler conditions for pseudogene and functional gene PCR are as previously described.10,11
PRNP sequence evaluation.
PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega) and then sequenced at the University of Illinois Keck Center for Functional and Comparative Genomics using a BigDye Terminator Sequencing kit and an ABI 3730XL DNA Sequencer (Applied Biosystems). To check sequence accuracy and repeatability, replicate samples of DNA were submitted and also sent to two off-site sequencing centers. The Keck Center yielded the highest quality reactions with 99.8% sequence homology between duplicates.
CodonCode Aligner software (CodonCode Corp.,) was used to evaluate chromatograms produced from the ABI sequencing reactions. Only nucleotide sequences possessing Phred quality scores of 20 or higher were imported into Biology Workbench for further analysis. The CLUSTALW and FASTA alignment programs (Biology Workbench, SDSC) were used to align each sequence with the consensus to identify Single Nucleotide Polymorphisms (SNP).
Observed polymorphisms were compared to a databank derived consensus sequence. All previously published white-tailed deer prion nucleotide sequences from the GenBank and EMBL-Bank databases were used to compile a consensus PRNP sequence using MultAlin software,21 with a protein translation generated from the nucleotide consensus sequence using Six Frame software (Biology WorkBench, SDSC). A Blast search indicated the consensus nucleotide sequence was not equivalent to any individual published sequence. The similar AF156185.1 (EMBLCDS:AAF80284) had T at the 438C/T SNP while AY286008.1(EMBLCDS:AAP37447) had T at the 555C/T SNP. The protein translated sequence of the consensus sequence was equivalent to both of these entries.
Statistical analysis.
Logistic regression and chi-squared tests were performed with SAS for Windows, version 9.1 (SAS Institute Inc.,). Chi-square tests were performed for each polymorphic allele and genotype to check for differences between observed and expected frequencies in both CWD negative and positive animals using an α level of 0.05. When needed, the Mantel-Haenzel Chi-square Test was employed to correct for low frequency genotypes. Deviations from Hardy-Weinburg equilibrium were tested using the program GenAlEx version 6.22
Additionally, logistic regression was used to determine the relationship between polymorphisms and disease status. Each allele was classified as 1 (polymorphic) or 0 (consensus) and regressed against the probability of being CWD positive. Genotypes were also compared to consensus genotypes in an additive model as discussed by North et al.23 In this method genotypes were classified as 0 = homozygous for the consensus genotype, 1 = heterozygous, 2 = homozygous for the polymorphism. For all logistic regression models, odds ratios less than 1 were equated with probability of disease below 50% and were therefore considered protective against CWD. Odds ratios greater than 1 were equated with probability of disease greater than 50% and were therefore considered risk factors for CWD.24
To determine the cumulative effects of polymorphisms, the total number of polymorphic alleles per animal was summed across all loci and regressed against CWD status. Observed allele frequencies were higher than expected for locus 153, so coding for this variable was reversed for cumulative regression models (2 = homozygous for the consensus genotype, 1 = heterozygous, 0 = homozygous for the polymorphism). The total number of polymorphic alleles (0, 1, 2, 3, 4, 5 or 6) was examined in one model, the number of coding polymorphic alleles (0, 1 or 2) was examined in a second model, silent polymorphic alleles (0, 1, 2, 3, 4, 5 or 6) were evaluated in a third model, and coding and silent polymorphic alleles were tested simultaneously in a fourth model. To be sure that the susceptibility conferred by locus 153 did not have excessive influence, models were also run with this polymorphism omitted.
To determine our ability to detect significant differences in polymorphisms between positive and negative animals, power analyses were performed for each polymorphic allele and cumulative logistic regression predictor variables using SamplePower for Windows, version 2.0 (SPSS, Inc.,).
Haplotypes were generated from unphased genotypes using Phase, version 2.1.25,26 This program was also used to test for linkage disequilibrim between genotypes, and random distribution of haplotype frequencies between cases and controls. We also used logistic regression and chi-square tests to determine the relationship between disease status and sex, age and gender. Finally, allele frequencies of the Q95H and G96S coding polymorphisms were compared between the Illinois and Wisconsin outbreaks. Wisconsin allele frequencies were calculated from published data.12
Acknowledgements
The Illinois Department of Natural Resources was responsible for the collection of all samples and this project could not have been completed without the numerous Wildlife Biologists who generously donated both their time and labor to this study. The Illinois Department of Agriculture Animal Disease Laboratory in Galesburg also provided invaluable assistance. Funding for this project was provided by the Illinois Department of Natural Resources, the United States Geological Survey, the Federal Aid in Wildlife Restoration Project W-146-R, the Jonathan Baldwin Turner Scholarship Fund and the Environmental Council SURE grant program.
Abbreviations
- BSE
bovine spongiform encephalopathy
- CWD
chronic wasting disease
- GPS
global positioning system
- HWE
hardy-weinburg equilibrium
- IDNR
Illinois department of natural resources
- PRNP
prion gene
- PrP
encoded prion protein
- PrPsc
abnormal disease causing conformation of the prion protein
- SNP
single nucleotide polymorphism
- CJD
Creutzfeldt-Jakob disease
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/6321
References
- 1.Belay ED, Maddox RA, Williams ES, Miller MW, Gambetti P, Schonberger LB. Chronic wasting disease and potential transmission to humans. Emerging Infectious Diseases. 2004;10:977–984. doi: 10.3201/eid1006.031082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeten LA, Powers BE, Jewell JE, Spraker TR, Miller MW. A natural case of chronic wasting disease in a free-ranging moose (alces alces shirasi) J Wildl Dis. 2007;43:309–314. doi: 10.7589/0090-3558-43.2.309. [DOI] [PubMed] [Google Scholar]
- 3.Miller MW, Williams ES. Prion disease: Horizontal prion transmission in mule deer. Nature. 2003;425:35–36. doi: 10.1038/425035a. [DOI] [PubMed] [Google Scholar]
- 4.Williams ES, Young S. Spongiform encephalopathy of rocky mountain elk. J Wildl Dis. 1982;18:465–471. doi: 10.7589/0090-3558-18.4.465. [DOI] [PubMed] [Google Scholar]
- 5.Mathiason CK, Powers JG, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mason GL, Hays SA, Hayes-Klug J, Seelig DM, Wild MA, Wolfe LL, Spraker TR, Miller MW, Sigurdson CJ, Telling GC, Hoover EA. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006;314:133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 6.Sigurdson CJ, Aguzzi A. Chronic wasting disease. BBA-Molecular Basis of Disease. 2007;1772:610–618. doi: 10.1016/j.bbadis.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joly DO, Ribic CA, Langenberg JA, Beheler K, Batha CA, Dhuey BJ, Rolley RE, Bartelt G, Van Deelen TR, Samuel MD. Chronic wasting disease in free-ranging wisconsin whitetailed deer. Emer Infect Dis. 2003;9:599–601. doi: 10.3201/eid0905.020721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illinois Department of Natural Resources, author. WD information. 2007;2007:1. [Google Scholar]
- 9.Novakofski J, Brewer MS, Mateus-Pinilla N, Killefer J, McCusker RH. Prion biology relevant to bovine spongiform encephalopathy. J Anim Sci. 2005;83:1455–1476. doi: 10.2527/2005.8361455x. [DOI] [PubMed] [Google Scholar]
- 10.O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004;85:1339–1346. doi: 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- 11.Johnson C, Johnson J, Clayton M, McKenzie D, Aiken J. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of wisconsin. J Wildl Dis. 2003;39:576–581. doi: 10.7589/0090-3558-39.3.576. [DOI] [PubMed] [Google Scholar]
- 12.Johnson C, Johnson J, Vanderloo JP, Keane D, Aiken JM, McKenzie D. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006;87:2109–2114. doi: 10.1099/vir.0.81615-0. [DOI] [PubMed] [Google Scholar]
- 13.Jewell JE, Conner MM, Wolfe LL, Miller MW, Williams ES. Low frequency of PrP genotype 225SF among free-ranging mule deer (odocoileus hemionus) with chronic wasting disease. J Gen Virol. 2005;86:2127–2134. doi: 10.1099/vir.0.81077-0. [DOI] [PubMed] [Google Scholar]
- 14.Raymond G, Bossers A, Raymond L, O'Rourke K, McHolland L, Bryant P, III, Miller M, Williams E, Smits M, Caughey B. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19:4425–4430. doi: 10.1093/emboj/19.17.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brayton KA, O'Rourke KI, Lyda AK, Miller MW, Knowles DP. A processed pseudogene contributes to apparent mule deer prion gene heterogeneity. Gene. 2004;326:167–173. doi: 10.1016/j.gene.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Heaton MP, Leymaster KA, Freking BA, Hawk DA, Smith TP, Keele JW, Snelling WM, Fox JM, Chitko-McKown CG, Laegreid WW. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle and deer. Mamm Genome. 2003;14:765–777. doi: 10.1007/s00335-003-2283-y. [DOI] [PubMed] [Google Scholar]
- 17.Miller MW, Conner MM. Epidemiology of chronic wasting disease in free-ranging mule deer: Spatial, temporal and demographic influences on observed prevalence patterns. J Wildl Dis. 2005;41:275–290. doi: 10.7589/0090-3558-41.2.275. [DOI] [PubMed] [Google Scholar]
- 18.Brinkman TJ, Deperno CS, Jenks JA, Haroldson BS, Osborn RG. Movement of female white-tailed deer: Effects of climate and intensive row-crop agriculture. J Wildl Manage. 2005;69:1099–1111. [Google Scholar]
- 19.Nixon CM, Mankin PC, Etter DR, Hansen LP, Brewer PA, Chelsvig JE, Esker TL, Sullivan JB. White-tailed deer dispersal behavior in an agricultural environment. The American Midland Naturalist. 2007;157:212–220. [Google Scholar]
- 20.Roseberry JL, Woolf A. Habitat-population density relationships for white-tailed deer in illinois. Wildl Soc Bull. 1998;26:252–258. [Google Scholar]
- 21.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peakall R, Smouse PE. Genalex 6: Genetic analysis in excel. population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North BV, Curtis D, Sham PC. Application of logistic regression to case-control association studies involving two causative loci. Hum Hered. 2005;59:79–87. doi: 10.1159/000085222. [DOI] [PubMed] [Google Scholar]
- 24.Allison PD. Logistic Regression Using the SAS® System: Theory and Application. SAS Institute Inc; 1999. [Google Scholar]
- 25.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med. 2004;4:385–396. doi: 10.2174/1566524043360672. [DOI] [PubMed] [Google Scholar]
- 28.Roden JA, Nieuwhof GJ, Bishop SC, Jones DA, Haresign W, Gubbins S. Breeding programmes for TSE resistance in british sheep I. assessing the impact on prion protein (PrP) genotype frequencies. Prev Vet Med. 2006;73:1–16. doi: 10.1016/j.prevetmed.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Gubbins S, Roden JA. Breeding programmes for TSE resistance in british sheep II. assessing the impact on the prevalence and incidence of scrapie. Prev Vet Med. 2006;73:17–31. doi: 10.1016/j.prevetmed.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Meade-White K, Race B, Trifilo M, Bossers A, Favara C, Lacasse R, Miller M, Williams E, Oldstone M, Race R, Chesebro B. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol. 2007;81:4533–4539. doi: 10.1128/JVI.02762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewison AJ. Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: An individual-based approach. Mol Ecol. 2004;13:2841–2850. doi: 10.1111/j.1365-294X.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- 32.John TJ, Samuel R. Herd immunity and herd effect: New insights and definitions. Eur J Epidemiol. 2000;16:601–606. doi: 10.1023/a:1007626510002. [DOI] [PubMed] [Google Scholar]
- 33.Safar J, Wang W, Padgett MP, Ceroni M, Piccardo P, Zopf D, Gajdusek DC, Gibbs CJ. Molecular mass, biochemical composition and physicochemical behavior of the infectious form of the scrapie precursor protein monomer. Proceedings of the National Academy of Sciences. 1990;87:6373–6377. doi: 10.1073/pnas.87.16.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells MA, Jackson GS, Jones S, Hosszu LL, Craven CJ, Clarke AR, Collinge J, Waltho JP. A reassessment of copper(II) binding in the full-length prion protein. Biochem J. 2006;399:435–444. doi: 10.1042/BJ20060458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millhauser GL. Copper and the prion protein: Methods, structures, function and disease. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigurdsson EM, Brown DR, Alim MA, Scholtzova H, Carp R, Meeker HC, Prelli F, Frangione B, Wisniewski T. Copper chelation delays the onset of prion disease. J Biol Chem. 2003;278:46199–46202. doi: 10.1074/jbc.C300303200. [DOI] [PubMed] [Google Scholar]
- 37.Hijazi N, Shaked Y, Rosenmann H, Ben-Hur T, Gabizon R. Copper binding to PrPC may inhibit prion disease propagation. Brain Res. 2003;993:192–200. doi: 10.1016/j.brainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Beck JA, Mead S, Campbell TA, Dickinson A, Wientjens D, Croes EA, Van Duijn CM, Collinge J. Two-octapeptide repeat deletion of prion protein associated with rapidly progressive dementia. Neurology. 2001;57:354–356. doi: 10.1212/wnl.57.2.354. [DOI] [PubMed] [Google Scholar]
- 39.Capellari S, Parchi P, Wolff BD, Campbell J, Atkinson R, Posey DM, Petersen RB, Gambetti P. Creutzfeldt-jakob disease associated with a deletion of two repeats in the prion protein gene. Neurology. 2002;59:1628–1630. doi: 10.1212/01.wnl.0000035533.86833.28. [DOI] [PubMed] [Google Scholar]
- 40.Owen F, Poulter M, Lofthouse R, Collinge J, Crow TJ, Risby D, Baker HF, Ridley RM, Hsiao K, Prusiner SB. Insertion in prion protein gene in familial creutzfeldt-jakob disease. Lancet. 1989;1:51–52. doi: 10.1016/s0140-6736(89)91713-3. [DOI] [PubMed] [Google Scholar]
- 41.Castilla J, Gutierrez-Adan A, Brun A, Pintado B, Parra B, Ramirez MA, Salguero FJ, Diaz San Segundo F, Rabano A, Cano MJ, Torres JM. Different behavior toward bovine spongiform encephalopathy infection of bovine prion protein transgenic mice with one extra repeat octapeptide insert mutation. J Neurosci. 2004;24:2156–2164. doi: 10.1523/JNEUROSCI.3811-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bocharova OV, Breydo L, Salnikov VV, Baskakov IV. Copper(II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry. 2005;44:6776–6787. doi: 10.1021/bi050251q. [DOI] [PubMed] [Google Scholar]
- 43.Kuczius T, Buschmann A, WenLan Z, Karch H, Becker K, Peters G, Groschup MH. Cellular prion protein acquires resistance to proteolytic degradation following copper ion binding. Biol Chem. 2004;385:739–747. doi: 10.1515/BC.2004.090. [DOI] [PubMed] [Google Scholar]
- 44.Nishina K, Jenks S, Supattapone S. Ionic strength and transition metals control PrPSc protease resistance and conversion-inducing activity. J Biol Chem. 2004;279:40788–40794. doi: 10.1074/jbc.M406548200. [DOI] [PubMed] [Google Scholar]
- 45.Jackson GS, Murray I, Hosszu LLP, Gibbs N, Waltho JP, Clarke AR, Collinge J. Location and properties of metal-binding sites on the human prion protein. Proceedings of the National Academy of Sciences. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, Peisach J, Millhauser GL. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Z, O'Rourke KI, Dong Z, Jenny AL, Langenberg JA, Belay ED, Schonberger LB, Petersen RB, Zou W, Kong Q, Gambetti P, Chen SG. Chronic wasting disease of elk and deer and creutzfeldt-jakob disease: comparative analysis of the scrapie prion protein. J Biol Chem. 2006;281:4199–4206. doi: 10.1074/jbc.M509052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schultz-Schaeffer WJ, Kretzschmar HA, Head MW, Ironside JW, Gambetti P, Chen SG. Genetic influence on the structural variations of the abnormal prion protein. Proceedings of the National Academy of Sciences. 2000;97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rourke KI, Spraker TR, Zhuang D, Greenlee JJ, Gidlewski TE, Hamir AN. Elk with a long incubation prion disease phenotype have a unique prpd profile. Neuroreport. 2007;18:1935–1938. doi: 10.1097/WNR.0b013e3282f1ca2f. [DOI] [PubMed] [Google Scholar]
- 50.Saunders GC, Cawthraw S, Mountjoy SJ, Hope J, Windl O. PrP genotypes of atypical scrapie cases in great britain. J Gen Virol. 2006;87:3141–3149. doi: 10.1099/vir.0.81779-0. [DOI] [PubMed] [Google Scholar]
- 51.Balbus N, Humeny A, Kashkevich K, Henz I, Fischer C, Becker CM, Schiebel K. DNA polymorphisms of the prion doppel gene region in four different german cattle breeds and cows tested positive for bovine spongiform encephalopathy. Mamm Genome. 2005;16:884–892. doi: 10.1007/s00335-005-0052-9. [DOI] [PubMed] [Google Scholar]
- 52.Badaoui B, Serradilla JM, Tomas A, Urrutia B, Ares JL, Carrizosa J, Sanchez A, Jordana J, Amills M. Goat acetyl-coenzyme A carboxylase alpha: Molecular characterization, polymorphism and association with milk traits. J Dairy Sci. 2007;90:1039–1043. doi: 10.3168/jds.S0022-0302(07)71590-4. [DOI] [PubMed] [Google Scholar]
- 53.Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 54.Wang D, Sadee W. Searching for polymorphisms that affect gene expression and mRNA processing: Example ABCB1 (MDR1) AAPS J. 2006;8:515–520. doi: 10.1208/aapsj080361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 56.Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T. Analysis of sequence variability of the bovine prion protein gene (PRNP) in german cattle breeds. Neurogenetics. 2004;5:19–25. doi: 10.1007/s10048-003-0171-y. [DOI] [PubMed] [Google Scholar]
- 57.Haase B, Doherr MG, Seuberlich T, Drogemuller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T. PRNP promoter polymorphisms are associated with BSE susceptibility in swiss and german cattle. BMC Genet. 2007;8:15. doi: 10.1186/1471-2156-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sander P, Hamann H, Drogemuller C, Kashkevich K, Schiebel K, Leeb T. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 2005;280:37408–37814. doi: 10.1074/jbc.M506361200. [DOI] [PubMed] [Google Scholar]
- 59.Juling K, Schwarzenbacher H, Williams JL, Fries R. A major genetic component of BSE susceptibility. BMC Biol. 2006;4:33. doi: 10.1186/1741-7007-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson CJ, Phillips KE, Schramm PT, McKenzie D, Aiken JM, Pedersen JA. Prions adhere to soil minerals and remain infectious. PLoS Pathog. 2006;2:32. doi: 10.1371/journal.ppat.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]