Abstract
The prion infection is a conversion of host encoded prion protein (PrP) from its cellular isoform PrPC into the pathological and infectious isoform PrPSc; the conversion process was investigated by in vitro studies using recombinant and cellular PrP and natural PrPSc. We present a brief summary of the results determined with our in vitro conversion system and the derived mechanistic models. We describe well characterized intermediates and precursor states during the conversion process, kinetic studies of spontaneous and seeded fibrillogenesis and the impact of the membrane environment.
Key words: prion protein conversion, seeding, fibril, dimer, precursor state, kinetics, membrane
Introduction
The molecular key event in prion diseases is the conformational change of the host-encoded prion protein, denoted as PrPC, into the disease-causing isoform, PrPSc.1 Because prions do not contain any genetic information in the form of nucleic acid2,3 the information for prions is enciphered in the structure of the pathological isoform. In contrast to other neurodegenerative diseases like Alzheimer disease, Huntington's disease etc., prion diseases are not only of spontaneous or hereditary etiology but are also transmissible. Prion replication occurs by converting PrPC to PrPSc; in the transmissible case the process is induced by invading PrPSc and the pool of PrPC is replenished by the cellular synthesis of PrPC. Several mechanistic models have been proposed for this conformational transition. Most experimental data support the model of seeded polymerization.4 The conformational change of PrPC into PrPSc results in a fundamental change of its biophysical properties. PrPC is membrane-bound, rich in α-helical secondary structure, soluble in mild detergents, sensitive against Proteinase K (PK) digestion and noninfectious, whereas PrPSc is β-sheet-rich, aggregated, after truncation of its N-terminal segment (aa 23–89/90) resistant to further PK digestion and it is the major or only component of the infectious particle.1
The eukaryotic PrPC is post-translationally modified; carrying two N-glycosylations and a glycosyl phosphatidyl inositol (GPI) anchor. The GPI anchor attaches PrP to the cell membrane.5 Like many other GPI-anchored proteins, PrPC is enriched in specific membrane-microdomains called rafts.6 With only minor variation, recombinant (rec) PrP of nearby all susceptible species exhibits a C-terminal globular domain, consisting of three α-helices and a small antiparallel β-sheet7–9 whereas the N-terminus is highly flexible without well-defined structure. The 1D-NMR spectrum of natural PrPC from bovine brain, carrying the two N-glycosylations but lacking the GPI-anchor showed no significant structural differences between anchorless PrPC and recPrP in solution.10 PrPC in vivo is anchored to the cell membrane. Therefore the effect of membrane binding on the structure of PrP is of functional interest.
Synthetic prions were formed from amyloidic recPrP, demonstrating that conversion of purified recPrP is sufficient for the generation of infectivity, albeit at very low titers.11,12 Prion infectivity was also generated by protein-misfolding cyclic amplification (PMCA)13 in extracts from noninfected animals.11,14 To investigate the molecular mechanism of prion formation, we pursued two different strategies. In one approach, the spontaneous conversion of PrPC into a PrPSc-like form was investigated, which is a model for the sporadic form of prion disease. In another approach, the PrPSc-dependent conversion of PrP, which simulates exogenous prion infection, was studied. The conformational transitions of PrP were induced by solvent conditions, particularly by lowering the concentration of SDS; in the absence of NaCl polymorphic aggregates were formed, and in the presence of NaCl amyloidic fibrils. We compared spontaneous and seeded fibrillization, analyzed the structure of intermediates during fibrillization and could describe the kinetics of fibril formation in terms of a theoretical description published earlier.15 Since in vivo PrPC is anchored to the membrane and is presented in this form for the contact with PrPSc, we analyzed the structure of PrPC in the membrane-anchored state and could describe this structure as particularly prone to the conversion into PrPSc.
Basics of the Conformational Transitions of the Prion Protein
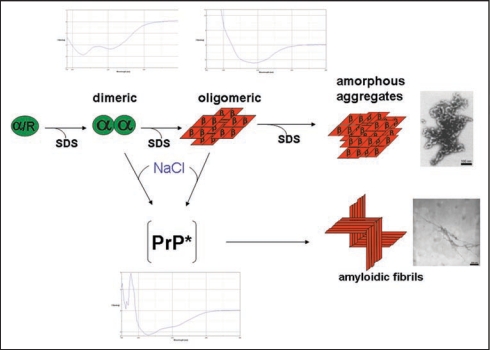
The mechanism of spontaneous prion protein conversion was studied systematically with recPrP. Our in vitro conversion system16 is based on the solubilization of PrP in low concentrations (0.2% w/v) of sodium dodecyl sulfate (SDS) under otherwise physiological conditions. Most extended and systematic studies were carried out with recPrP (90–231), i.e., without the N-terminus, corresponding to the amino acid sequence of hamster prions PrP 27–30;17 similar results were obtained with the natural SHa PrP 27–30.16 Varying the SDS concentration (from 0.2% SDS down to 0.0001%) at neutral pH and room temperature, different conformations could be established. The conformations were characterized with respect to secondary structure as determined by CD spectroscopy and to molecular mass, as determined by fluorescence correlation spectroscopy and analytical ultracentrifugation: α-helical monomers, soluble α-helical dimers, soluble but β-sheet rich oligomers of a minimal size of 12–14 PrP molecules, and insoluble amorphous aggregates of β-sheet rich structure were observed. A high activation barrier was found between the α-helical dimers and the β-sheet rich oligomers.17 In the upper part of Figure 1 the different structures are represented schematically together with their CD-spectra and the EM picture of the polymorphic aggregates. In 0.2% SDS PrP is present in an α-helical and particular denatured conformation denoted with α/R. For every state a well defined number of SDS molecules bound to PrP was determined as described in original literature.17
Figure 1.
Structures involved in the in vitro conversion of rec PrP(90–231) induced by lowering SDS concentration. The structures analysed within the in vitro conversion system are summarized. Exemplarily for the methods used to analyse these structures, beneath the different intermediates the CD-spectra are shown. Next to the aggregated states the electron micrograph of these structures are shown. The upper part represents the conversion without added NaCl, the lower part with added NaCl, respectively. Modified according to reference 18.
Structural Model of the PrP Dimer
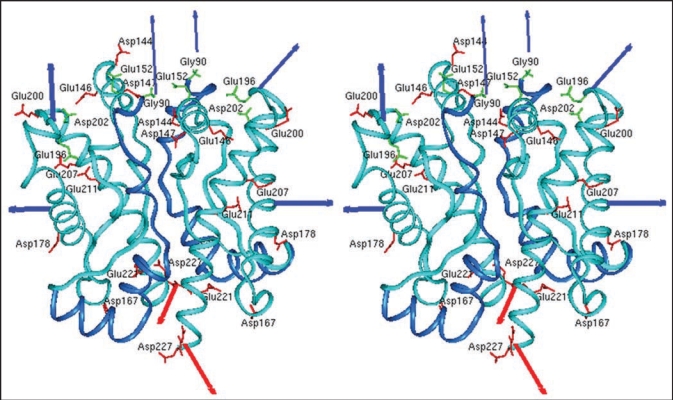
Our studies described above but also studies from other laboratories19,20 have indicated a critical role of PrP dimers in the conversion process. Consequently, we studied the structure of dimers in more derail and have chosen for those investigation conditions which stabilize dimers or drive the dimer-oligomer equilibrium completely to the side of dimers, respectively.21 Rec PrP(90–231) in 0.06% SDS 10 mM phosphoric buffer pH 6.8 without additional NaCl is 100% dimeric. As a structural approach we use covalent cross-links; the crosslinker EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide) forms inter- and intramolecular bonds between directly neighbored amino groups and carboxyl groups. The bonds were identified by tryptic digestion and subsequent mass spectrometric analysis. Intra- and intermolecular cross-links between N-terminal glycine and three acidic amino acid side chains in the globular part of PrP were identified showing that the N-terminal amino acids (90–124) are not as flexible as known from NMR analysis. When the cross-linked sites were used as structural constraint, molecular modelling calculations yielded a structural model for PrP dimer and its monomeric subunit, including the folding of amino acids 90–124 in addition to the known structure (Fig. 2). Molecular dynamics of the structure after release of the constraint indicated an intrinsic stability of the domain of amino acids 90–124.22 The structural model as shown in Figure 2 could be well in accordance with anchoring the dimer to the membrane via the glycolipid anchor. As demonstrated by arrows the two glycolipid anchors are directed to the lower side where the membrane would be present, whereas the glycosyl groups and the N-termini are directed into the water phase.
Figure 2.
Model of the prion protein dimer. Stereo presentation of the model for the PrP dimer. The structure of segment 92–124 (blue) is the result of Kaimann et al.,22 the structure of segment 125–228 (cyan) is taken from the NMR analysis.23 Red arrows represent the glycolipid anchor; thick blue arrows, glycosyl groups; and thin blue arrows, N-termini. Figure according to reference 22.
Mechanism of Fibril Formation
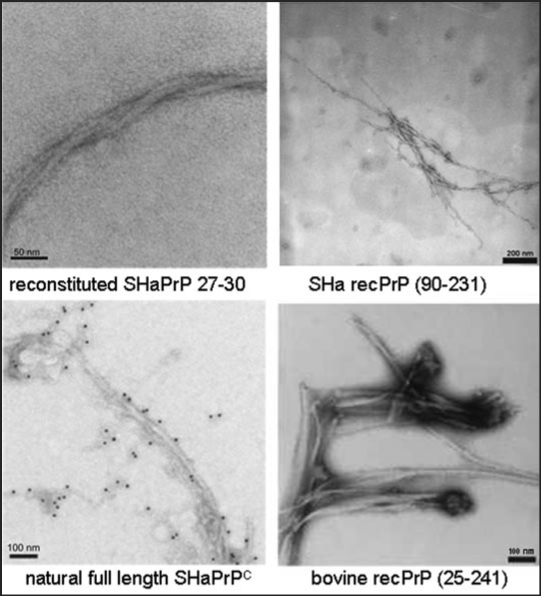
As shown in Figure 1, addition of 250 mM NaCl to the conditions of α-helical dimers or β-sheet oligomers shifts PrP into a still soluble state that allows, however, fibril assembly to proceed over several weeks. Fibril formation was assayed by electron microscopy (Fig. 3), and staining with gold particles linked to anti-PrP-antibodies (Fig. 3, left side, lower part) showed clearly that the fibrils were formed of PrP. Fibril formation was achieved with a large variety of PrP as SHa rec PrP 90–231, SHa rec PrP 23–231, with SDS solubilized SHa PrP 27–30 (prion rods), native SHaPrPC purified from brain tissue and full length bovine rec PrP (Fig. 3).24–26 When fibrils were formed from SHa, either solubilized PrP 27–30, PrPC or rec PrP and were bioassayed in transgenic mice overexpressing SHa PrP, infectivity could not be detected, whereas amyloid fibrils formed of rec mouse PrP(89–230) were infectious11 in transgenic mice and in the second passage in wildtype mice. Possible reasons for these deviating results are discussed in the original literature.24
Figure 3.
Fibrils formed of different PrP preparations by the in vitro conversion system in presence of NaCl. Electron micrographs of fibrils formed of different PrP preparations are shown. Recombinant (recPrP) as well as fully translational modified PrP (reconstituted SHaPrP 27–30, natural full length PrPC) with hamster (SHa) sequenz as well as full length recombinant bovine PrP (bovine rec PrP 25–241) were used. Figure rearranged according to reference 24 and 26.
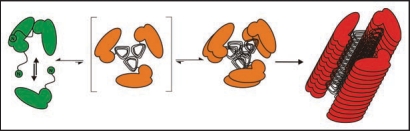
The structure of the intermediated state PrP* (Fig. 1) is decisive to switch aggregates formation from the polymorphic pathway into the fibrillar pathway. Therefore, we called this state fibril precursor state and characterized it by different methods; CD spectroscopy (cf. lower spectrum in Fig. 1) showed a mixture of α-helical and random coil secondary structure. We assume that the α-helical part is identical or close to that which is also present in the fibrillar structure. Studies with analytical ultracentrifugation demonstrated a monomer-dimer equilibrium, and the crosslinking studies as described above showed the same well defined intermolecular contact site as in the stable dimer, but in lower concentration as expected for the monomer-dimer equilibrium. In Figure 4 the mechanistic model for spontaneous fibril formation is depicted. The fibril structure is taken from a model in the literature27 is underplayed, which postulates a trimeric subunit. The trimeric subunit would be formed by β-sheets of the flexible parts of the monomers and dimers which exhibit then also intermolecular contacts. This trimer or even a stack of two trimers would present a steady-state and only a stack of several trimers would stabilize and drive the fibril formation.
Figure 4.
Mechanistic model of PrP fibrillization. A proposed mechanistic model depicts the preamyloid state in a monomer-dimer equilibrium, stationary state of trimer, stable nucleus of two trimers, and a growing fibril. Figure according to reference 25.
Seeded Fibril Formation
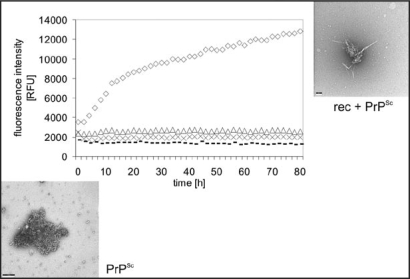
As a model for the infection process fibril formation was also studied when induced by seeds of PrPSc. As seeds we used natural PrPSc purified from brain homogenate by PTA precipitation under avoidance of any proteinase K digestion;28 rec PrP was applied as substrate. The fibrillization conditions were taken exactly from the spontaneous fibrillization. Fibrillization of PrP without seeds takes up to several weeks, as described above, whereas seed-enhanced fibrillization was achieved within hours to days (Fig. 5). A lag phase is followed by exponential growth and saturation. We monitored and quantitatively described the kinetics of seeded fibril formation, including dependence of the reaction on substrate and seed concentrations, using the fluorescence increase of thioflavin T. In Figure 5 it also shown nicely that fibrils are growing out of an amorphous PrPSc aggregate.25
Figure 5.
Seeded fibril formation. RecPrP (80 ng/µl) seeded with purified PrPSc (diamonds) forms amyloid fibrils readily compared with controls: recPrP + uninfected 1.6 × 10−4 brain equivalents per µl (x); recPrP alone (triangles); purified PrPSc alone (dashes). Fibril formation was monitored by using the ThT fluorescence assay. Electron micrographs: PrPSc aggregates after NaPTA precipitation, recPrP + PrPSc after seeding assay. Figure rearranged according to reference 25.
Kinetics of Prion Amplification
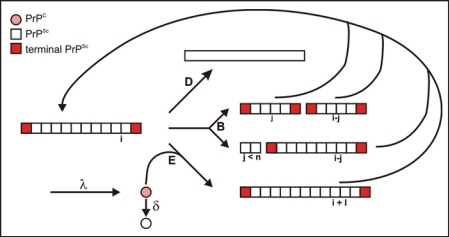
Seeded fibrillization serves as a model for prion amplification and vice versa the kinetics of prion amplification was analyzed in the literature15 as a seed dependent fibrillization process. In Figure 6, the authors used parameters for seed-dependent growth (reaction E), degradation (reaction D), and/or breakage of fibrils (reaction B), synthesis (λ) and degradation of monomeric PrP (δ). Obviously, the calculations apply to homogeneous solution conditions only, i.e., growth and breakage occur simultaneously as established in our experiments, whereas in PMCA the conditions are varied in a cyclic manner. The shape of our experimental curves is in agreement with the scheme depicted in Figure 6. The lag phase followed by a steep increase is interpreted as an exponential growth in which both growth and generation of new seeds contribute to the curve; saturation is due to the limited monomer supply and competing reactions. The incubation time was defined as the duration required to create a well defined number of infectious particles;15 it corresponds to the in vitro lag phase, which is in our experiments a fixed low level of fluorescence intensity. For more details of the corresponding experimental and calculatory parameters see Stöhr et al.25 The calculations15 imply that the lag-phase tl depends on the concentration of seeds (CPrPSc) and the concentration of monomeric PrP (CPrP), respectively, as: C1, C2, C3 and C4 are constants that are not further evaluated here. The experimental data could well be interpreted in terms of the formulas given above and confirmed the fibrillization scheme in Figure 6 in respect to the three features: exponential growth, dependence on CPrPSc, and dependence on CPrP. The limited set and accuracy of the data did not allow us to evaluate parameters like growth rate, degradation rate, etc. It is not possible to extrapolate the in vitro lag phase to the typical incubation time in hamster that is 103 times longer for the equivalent ID50. If one considers, however, lower effective PrPC levels (10x) and a clearance factor of PrPSc of 100,29 in hamster, one would expect similarly long incubation times. Also the saturation levels could not be interpreted quantitatively since degradation and denaturation of PrP, degradation of fibrils and formation of polymorphous aggregates compete with fibril formation.
Figure 6.
Reaction scheme of seeded fibrillization. For parameter, see text. Figure according to reference. 25.
It was not the scope of this work but it should be noted, that seeded fibrillization can be applied as a sensitive diagnostic method cf also.30
Towards In Vivo Conditions
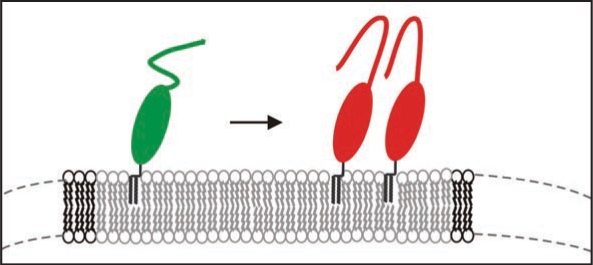
However, natural PrPC is post-translationally modified and carries two N-glycosylations and the natural GPI-anchor. We have isolated such PrPC from transfected CHO-cells and studied the thermodynamics of insertion of PrPC into raft-like domains of the membrane and analyzed secondary structural changes when bound to the membrane via its GPI anchor. With surface plasmon resonance it was determined that PrPC binds with an equilibrium constant of above 109 M−1 and can form close coverage in form of a monolayer on the membrane. In cooperation with the group of K. Gerwert we established a similar set-up, i.e., membrane anchored PrPC for structural studies and carried out FTIR measurements in the attenuated total reflection mode.31 We observed that native PrPC after membrane-anchoring assumed the same structure as in solution but above a threshold concentration of PrPC on the membrane a folding of the prion protein with intermolecular β-sheets was induced (Fig. 7). Such transition has never been observed in solution and is membrane specific. Earlier studies on PrP-membrane interaction could not observe such transition, most probably, because the quasi-native setup of raft like lipid membrane and PrPC anchored via the GPI anchor was not used. The membrane anchoring is thought to be a crucial event in development of prion diseases.
Figure 7.
Model of the structural transition of PrPC on the membrane above a certain threshold of PrPC concentration.
Synthetic and Natural Prions
Synthetic mammalian prions consist solely of recombinant PrP which was expressed in E. coli and had never been in contact with any animal.11 Consequently, prion infectivity was generated in vitro, but it was low if compared on an ID50/PrP basis with natural prions. Much higher infectivity could be generated with the PMCA method, when natural prions were amplified in a cellular extract as source for the substrate PrPC. At present it is not known which cellular factors are crucial for the effective prion amplification. Even in the absence of prion seeds but with the potential cellular factors present and PrPC as substrate infectivity could be generated by PMCA spontaneously. As described above membrane anchoring of PrPC might predispose the PrPC molecules for β-sheet rich intermolecular contacts, possibly as the first step for the conversion. Furthermore it is known that prions contain a polyglucose scaffold32 which might favour infectivity. In summary, prions can be generated in vitro from isolated prion protein without any other factors. One has to assume, however, that in vivo infectivity is raised substantially by cellular factors including membrane and polymetric sugar contacts.
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7060
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kellings K, Meyer N, Mirenda C, Prusiner SB, Riesner D. Further analysis of nucleic acids in purified scrapie prion preparations by improved return refocusing gel electrophoresis. J Gen Virol. 1992;73:1025–1029. doi: 10.1099/0022-1317-73-4-1025. [DOI] [PubMed] [Google Scholar]
- 3.Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. Search for a prion-specific nucleic acid. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Come JH, Fraser PE, Lansbury PT., Jr A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 6.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1997;272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 7.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 9.López Garcia F, Zahn R, Riek R, Wüthrich K. NMR structure of the bovine prion protein. Proc Natl Acad Sci USA. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornemann S, Schorn C, Wüthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legname G, Baskakov IV, Nguyen H-OB, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 12.Legname G, Nguyen H-OB, Baskakov IV, Cohen FE, DeArmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci USA. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 14.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masel J, Jansen VA. Prion kinetics. Biophys J. 2004;87:728. doi: 10.1529/biophysj.104.039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Post K, Pitschke M, Schäfer O, Wille H, Appel TR, Kirsch D, Mehlhorn I, Serban H, Prusiner SB, Riesner D. Rapid acquisition of beta-sheet structure in the prion protein prior to multimer formation. Biol Chem. 1998;379:1307–1317. doi: 10.1515/bchm.1998.379.11.1307. [DOI] [PubMed] [Google Scholar]
- 17.Leffers K-W, Schell J, Jansen K, Lucassen R, Kaimann T, Nagel-Steger L, Tatzelt J, Riesner D. The structural transition of the prion protein into its pathogenic conformation is induced by unmasking hydrophobic sites. J Mol Biol. 2004;344:839–853. doi: 10.1016/j.jmb.2004.09.071. [DOI] [PubMed] [Google Scholar]
- 18.Riesner D, Birkmann E, Dumpitak C, Elfrink K, Kellings K, Leffers K-W, Nagel-Steger L, Stöhr J. von PrP-Monomeren zu PrP-Fibrillen und Infektiositaet. Nova Acta Leopoldina. 2006:61–77. (Ger). [Google Scholar]
- 19.Meyer RK, Lustig A, Oesch B, Fatzer R, Zurbriggen A, Vandevelde M. A monomer-dimer equilibrium of a cellular prion protein (PrPC) not observed with recombinant PrP. J Biol Chem. 2000;275:38081–38087. doi: 10.1074/jbc.M007114200. [DOI] [PubMed] [Google Scholar]
- 20.Rambold AS, Müller V, Ron U, Ben-Tal N, Winklhofer KF, Tatzelt J. Stress-protective signalling of prion protein is corrupted by scrapie prions. EMBO J. 2008;27:1974–1984. doi: 10.1038/emboj.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen K, Schäfer O, Birkmann E, Post K, Serban H, Prusiner SB, Riesner D. Structural intermediates in the putative pathway from the cellular prion protein to the pathogenic form. Biol Chem. 2001;382:683–691. doi: 10.1515/BC.2001.081. [DOI] [PubMed] [Google Scholar]
- 22.Kaimann T, Metzger S, Kuhlmann K, Brandt B, Birkmann E, Höltje H-D, Riesner D. Molecular model of an alpha-helical prion protein dimer and its monomeric subunits as derived from chemical cross-linking and molecular modeling calculations. J Mol Biol. 2008;376:582–596. doi: 10.1016/j.jmb.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Farr-Jones S, Ulyanov NB, Llinas M, Marqusee S, Groth D, Cohen FE, Prusiner SB, James TL. Solution structure of Syrian hamster prion protein rPrP(90–231) Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 24.Leffers K-W, Wille H, Stöhr J, Junger E, Prusiner SB, Riesner D. Assembly of natural and recombinant prion protein into fibrils. Biol Chem. 2005;386:569–580. doi: 10.1515/BC.2005.067. [DOI] [PubMed] [Google Scholar]
- 25.Stöhr J, Weinmann N, Wille H, Kaimann T, Nagel-Steger L, Birkmann E, Panza G, Prusiner SB, Eigen M, Riesner D. Mechanisms of prion protein assembly into amyloid. Proc Natl Acad Sci USA. 2008;105:2409–2414. doi: 10.1073/pnas.0712036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panza G, Stöhr J, Dumpitak C, Papathanassiou D, Weiss J, Riesner D, Willbold D, Birkmann E. Spontaneous and BSE-prion-seeded amyloid formation of full length recombinant bovine prion protein. Biochem Biophys Res Commun. 2008;373:493–497. doi: 10.1016/j.bbrc.2008.06.059. [DOI] [PubMed] [Google Scholar]
- 27.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birkmann E, Schäfer O, Weinmann N, Dumpitak C, Beekes M, Jackman R, Thorne L, Riesner D. Detection of prion particles in samples of BSE and scrapie by fluorescence correlation spectroscopy without proteinase K digestion. Biol Chem. 2006;387:95–102. doi: 10.1515/BC.2006.013. [DOI] [PubMed] [Google Scholar]
- 29.Safar JG, DeArmond SJ, Kociuba K, Deering C, Didorenko S, Bouzamondo-Bernstein E, Prusiner SB, Tremblay P. Prion clearance in bigenic mice. J Gen Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 30.Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, Prusiner SB. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci USA. 2007;104:20914–20919. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elfrink K, Ollesch J, Stöhr J, Willbold D, Riesner D, Gerwert K. Structural changes of membrane-anchored native PrPC. Proc Natl Acad Sci USA. 2008;105:10815–10819. doi: 10.1073/pnas.0804721105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appel TR, Dumpitak C, Matthiesen U, Riesner D. Prion rods contain an inert polysaccharide scaffold. Biol Chem. 1999;380:1295–1306. doi: 10.1515/BC.1999.165. [DOI] [PubMed] [Google Scholar]