Abstract
A focused library of TX-67 (C10 hemi-succinate) analogs has been prepared, including C7 regioisomers, esters, amides, and one-carbon homologs. These were prepared to investigate whether the lack of TX-67 interaction with P-glycoprotein (Pgp) is due to the presence of the carboxylic acid moiety and whether this phenomenon was restricted to C10 analogs. Tubulin stabilization ability, cytotoxicity, and Pgp interactions were evaluated. All carboxylic acid analogs and several of the amides had no apparent interactions with Pgp at the concentrations used, whereas the ester variants displayed characteristics of Pgp substrates. Furthermore, it was demonstrated that hydrogen-bonding properties were significant with respect to Pgp interactions. Calculations of LogD and cross-sectional areas revealed that these analogues are predicted to partition into the membrane and can compete for Pgp binding sites. The anionic and amide introduction strategy may allow for delivery of paclitaxel into the CNS and may be a potential approach for the delivery of other, structurally complex and lipophilic non-CNS permeable drugs.
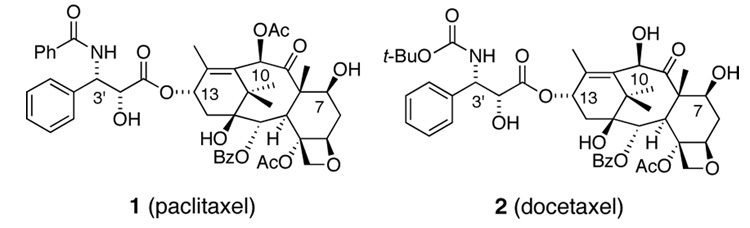
Paclitaxel (Taxol, 1, Figure 1), a structurally complex natural product derived from the bark of the Pacific Yew, is one of the most studied and active anti-cancer agents known.1–3 Although its clinical success is remarkable, the efficacy of the parent compound has limitations.4,5
Figure 1.
Paclitaxel and docetaxel
One such shortcoming is paclitaxel’s inability to cross the blood-brain barrier (BBB).6,7 Accordingly, paclitaxel is not an effective treatment for primary or metastatic brain cancer despite its potent anti-proliferative activity. In addition to paclitaxel’s well known anti-tumor properties, it has been shown to protect primary cortical neurons from beta amyloid (Aβ)-induced cell death as well as being non-toxic to primary cortical neurons.8 Indeed, a paclitaxel derivative that could permeate the CNS is highly desirable from both the standpoint of chemotherapy as well as an investigational therapy for neurofibrillary pathology.
A primary mechanism limiting the distribution of paclitaxel and other highly lipophilic substances into the brain is active efflux by the multidrug resistance gene product 1 (MDR1) P-glycoprotein (Pgp).9–11 We have recently described a series of recognition elements required for Pgp interactions based upon the analysis of over one hundred known Pgp substrates.12–14 This analysis revealed clusters of spatially distinct hydrogen bond acceptor units, which correlated, in their relative frequency, with the strength of Pgp interaction. We have demonstrated that deletion or modification of these “recognition elements” in paclitaxel reduces interaction with Pgp in bovine brain microvessel endothelial cells.15,16 These studies also bolstered our ascertation that the C10 region of paclitaxel is particularly important for Pgp affinity.
This same examination12–14 of known Pgp substrates also revealed that compounds that carry a negative charge at physiological pH, such as those that contain a carboxylic acid, sulphonate, or nitro group, with few exceptions, are not substrates for Pgp efflux.12–14 One caveat to this observation was that chemical structures that contain additional recognition elements, maintain their affinity for Pgp.
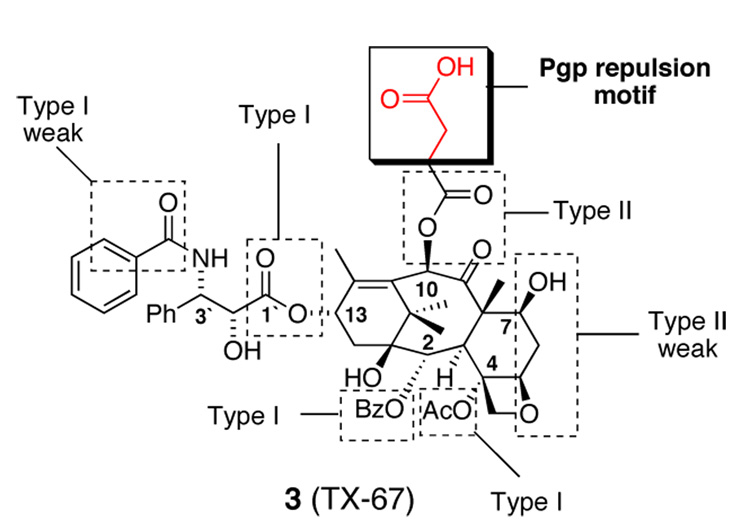
With this in mind we prepared a C10 modified taxane, TX-67 (3, Figure 2), which only differs from paclitaxel by the addition of an acetic acid unit to the terminus of the C10 acetyl ester.17 TX-67 (3) contains all of the recognition elements of paclitaxel, however, our studies suggest Pgp efflux mechanisms are substantially reduced or absent completely.17,18 Compound 3 demonstrates improved distribution across the BBB without co-administration of Pgp inhibitors both in vitro and in situ.17
Figure 2.
TX-67 recognition and repulsion elements.
Herein we describe the synthesis and biological evaluation of a focused library of TX-67 analogs that include C7 regioisomers, esters, amides, and one-carbon homologs. These were made to determine if the acid functionality was essential for Pgp evasion and if this phenomenon was restricted to only C10 analogs.
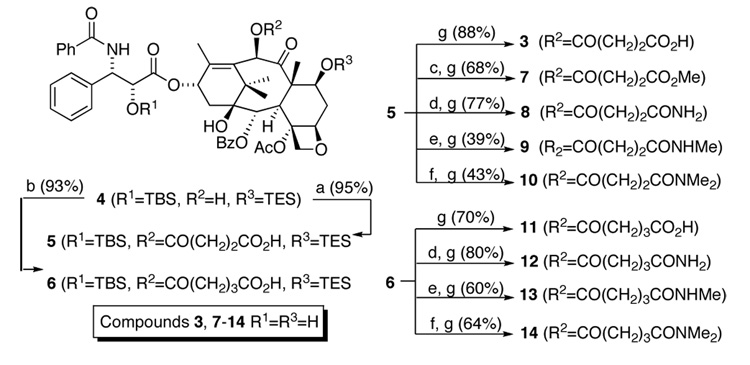
The C10 functional group analogs were prepared from common intermediate 4 (Scheme 1). TX-67 (3) was prepared as previously described17 while compounds 7–10 were accessed via acid 5. Acid 5 is cleanly and quantitatively converted to the methyl ester via treatment with diazomethane (Scheme 1), which, after treatment with HF-pyridine solution provides methyl ester 7 in good yield. The primary amide analog 8 is prepared via mixed anhydride formation with BOC2O followed by amide formation via ammonium bicarbonate (NH4CO3) under basic conditions. The N-methyl and N,N-dimethyl, amides (9 and 10 are generated via standard coupling procedures (Scheme 1). The described functional group transformations were each followed by fluoride anion assisted protecting group removal. Compounds 11–14 were prepared in the same manner from glutaric acid derivative 6.
Scheme 1.
Synthesis of C10-modified TX-67 analogs. Reagents and conditions: (a) succinic anhydride, DMAP, toluene 85 °C; (b) glutaric anhydride, DMAP, toluene 85 °C; (c) CH2N2, THF; (d) NH4HCO3, THF, BOC2O, pyridine; (e) NH2Me HCl, EDCI, NMM, CH2Cl2; (f) NHMe2 HCl, EDCI, NMM, CH2Cl2; (g) HF-pyridine, pyridine.
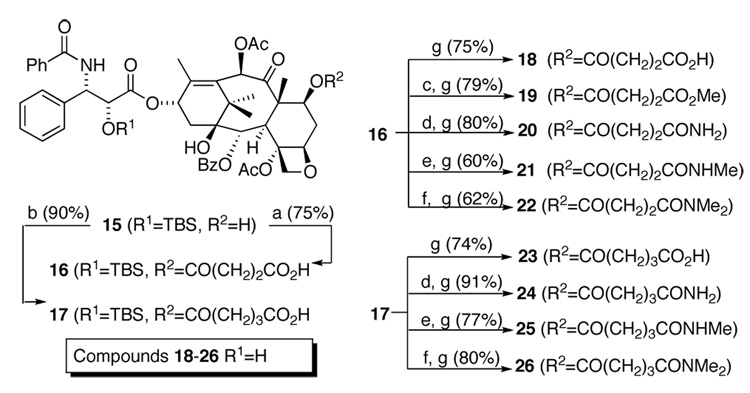
The desired C7 analogs were accessed in short synthetic sequences from common intermediate 15 (Scheme 2). The C7 succinnic acid analog 18 was prepared by acylating the C7 hydroxyl group with succinic anhydride, giving 16, followed by removing the 2’OTBS protecting group. The methyl ester analog 19 was generated from a coupling between acid 16 and MeOH followed by the same deprotection. The remaining compounds 21–26 were synthesized as described for the C10 analogs.
Scheme 2.
Synthesis of C7 TX-67 analogs.19 Reagents and conditions: (a) succinic anhydride, DMAP, toluene 85 °C; (b) glutaric anhydride, DMAP, toluene 85 °C; (c) MeOH, EDCI, DMAP; (d) NH4HCO3, THF, BOC2O, pyridine; (e) NH2Me HCl, EDCI, NMM, CH2Cl2; (f) NHMe2.HCl, EDCI, NMM, CH2Cl2; (g) HF-pyridine, pyridine.
All compounds were first evaluated for tubulin assembly ability and their effectiveness against the MCF7 breast cancer cell line and the drug resistant breast cancer cell line NCI/ADR-RES (Table 1).
Table 1.
ED50 ratios (compound/paclitaxel) for in vitro tubulin assembly and cytoxicity.
| Entry | Compound | Tubulin Assembly | MCF7 | NCI/ADR-RES |
|---|---|---|---|---|
| 1 | paclitaxel (1) | 1.0 | 1.0 | 1.0 |
| 2 | TX-67 (3 – C10 CO2H) | 1.7 | 13.3 | 1.3 |
| 3 | C10 CH2CO2H (11) | 1.8 | 27.9 | 5.8 |
| 4 | C7 CO2H (18) | 3.8 | >30.0 | 5.8 |
| 5 | C7 CH2CO2H (23) | 1.0 | 8.8 | 13.6 |
| 6 | C10 CO2Me (7) | 1.0 | 2.5 | 0.4 |
| 7 | C7 CO2Me (19) | 1.2 | 0.54 | 0.5 |
| 8 | C10 CONH2 (8) | 0.4 | 10 | 6.0 |
| 9 | C10 CH2CONH2 (12) | 1.8 | 10 | 5.3 |
| 10 | C7 CONH2 (20) | 0.6 | 3.2 | 0.6 |
| 11 | C7 CH2CONH2 (24) | 0.6 | 5.0 | 6.5 |
| 12 | C10 CONHMe (9) | 0.8 | 7.9 | 6.0 |
| 13 | C10 CH2CONHMe (13) | 1.8 | 2.3 | >6.3 |
| 14 | C7 CONHMe (21) | 0.8 | 2.2 | 1.0 |
| 15 | C7 CH2CONHMe (25) | 0.8 | 3.7 | 3.2 |
| 16 | C10 CONMe2 (10) | 1.6 | 2.1 | 2.7 |
| 17 | C10 CH2CONMe2 (14) | 2.2 | 5.6 | >6.3 |
| 18 | C7 CONMe2 (22) | 1.1 | 10.8 | 0.8 |
| 19 | C7 CH2CONMe2 (26) | 0.7 | 3.9 | 3.0 |
Paclitaxel has a mean ED50 of 3.2 nM ± 1.8 and 1.5 µM ± 1.3 in the MCF-7 and NCI/ADR-RES lines, respectively.
In the tubulin assembly assay, all analogs maintained similar activity to the parent compound (entries 2–19, Table 1). Against the MCF7 breast cancer cell line most analogs maintained effectiveness. The carboxylic acid analogs (entries 2–5), as a functional group class, showed the largest drop in activity (approx. 20-fold). The methyl ester variants (entries 6 and 7) maintained similar potency as compared to paclitaxel while the amides (entries 8–19) generally experienced an approximate 5-fold reduction in cytotoxicity. Most analogs were similar to paclitaxel against the NCI/ADR-RES cancer cell line. We had originally hypothesized an increase in effectiveness of these analogs against the Pgp over-expressing cancer cell line MCF7-ADR. However this was not the case as none of the analogues in Table 1 had superior potency compared to paclitaxel.
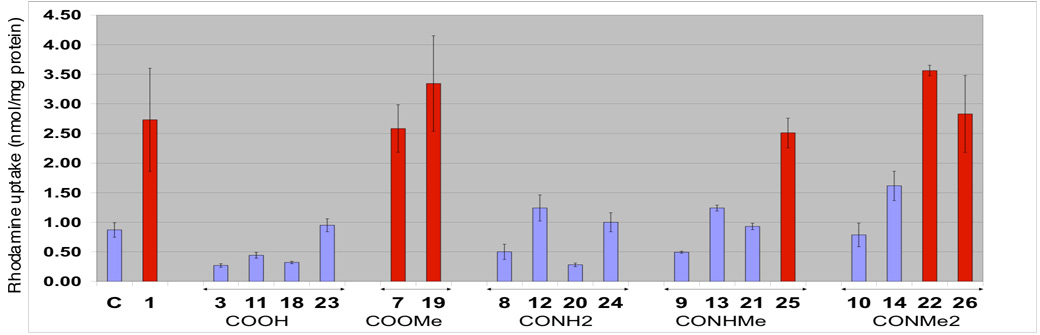
Our next screen was the rhodamine 123 uptake assay (Figure 3).20,21 This assay is a preliminary screen to evaluate a compound’s interaction with Pgp in bovine brain microvessel endothelial cells (BMECs). In this assay, rhodamine 123 is used as a surrogate Pgp substrate. The effect of the test compound on rhodamine 123 is determined by monitoring intracellular fluorescence. If the test compound has higher affinity to Pgp than rhodamine 123, then uptake of the latter will increase relative to the negative control.
Figure 3.
Rhodamine uptake results for compounds 1 (paclitaxel), C10 and C7 acids (3, 11, 18 and 23), C10 and C7 esters (7 and 19), C10 and C7 primary amides (8, 12, 20 and 24), C10 and C7 secondary amides (9, 13, 21, and 25), and C10 and C7 tertiary amides (10, 14, 22, and 26) in BMEC’s. Paclitaxel and the derivatives were present at a concentration of 10 µM. The concentration of rhodamine was 5µM. C = control. Compounds that significantly increase rhodamine 123 uptake are show as red bars. Compounds causing decreased or limited rhodamine 123 uptake are shown in blue.
All analogs containing a carboxylic acid functionality (3, 11, 18 and 23) showed no apparent interaction with Pgp. When the carboxylic acid is capped with a methyl group (methyl esters 7 and 19), a type I Pgp recognition element, in both the C7 and C10 series, a marked increase in rhodamine accumulation is observed. This is in agreement with our previous results suggesting that the carboxylic acid functionality is advantageous for Pgp evasion.17
Many members of the amide series (8–10, 12–14, 20, 21 and 24) did not significantly increase rhodamine accumulation. The weak interaction of secondary and primary amides with Pgp had already been suggested by us in an analysis of Pgp substrates and non-substrates. 12 As the secondary amides are converted from hydrogen bond donors (8, 9, 12, 13, 20, 21 and 25) to tertiary amides, hydrogen bond acceptors (10, 14, 22 and 26), increased rhodamine uptake is noted indicating that the molecules are interacting more strongly with Pgp. This is in accord with our hypothesis that H-bond donors will not interact significantly with Pgp and that H-bond acceptors will serve as substrates.
It is also of note that the succinic amides 8, 20, 9, 21 and 10 showed less rhodamine uptake than the corresponding one carbon homologs, the glutaric amides 12, 24, 13, 25, and 14, indicating that the length of the tether plays a role in binding to Pgp.
Our data further suggest that groups added to decrease Pgp interactions are generally more effective at the C10 position on the paclitaxel structure than on the C7 position. For example, the 10-linked methylamides 9 and 13 showed less rhodamine uptake than the O7-linked methylamides 21 and 25. This implies that the C10 region on these analogs has a very intimate relationship with Pgp
Our results suggest that Pgp evasion may not only be achieved by the introduction of a carboxylic acid moiety but also by the introduction of a primary or secondary amide into Pgp substrates.
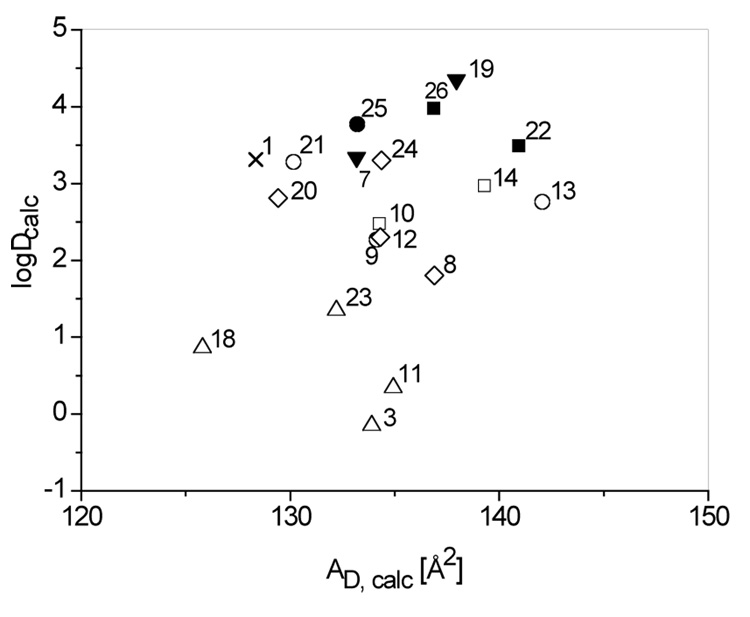
To compare the different analogs with respect to their ability to partition into the membrane and bind to Pgp we modeled the membrane-binding conformations and calculated the respective cross-sectional areas, AD. In addition we calculated the LogD values at pH 7.4 using an ionization constant pKa 4.2 for the carboxylic acid group (Figure 4).11 The former varied between LogD 0 to 4 and the latter between AD 125 to 143 Å2. The analogs of paclitaxel are thus clearly more hydrophobic and larger than rhodamine 123 with a logD −3.13 and an AD 69 Å2 and can therefore be predicted to compete for Pgp binding sites provided they carry the relevant recognition elements.
Figure 4.
Calculations of cross-sectional areas, AD and LogD values at pH 7.4 for paclitaxel and analogues. X = Paclitaxel, ∆ = CO2H, ▼ = CO2Me, ◊ = CONH2, ○ = CONHMe, ● = CONHMe, □ = CONMe2, ■ CONMe2 (filled symbols = Pgp substrates).
In summary, we have prepared a focused library of paclitaxel analogs in short synthetic sequences from the parent molecule. Biological evaluation in tubulin stabilization and cytotoxicity assays demonstrated that the designed analogs maintained the desired properties of paclitaxel. The rhodamine assay indicated that the introduction of esters at C10 and C7 increased rhodamine 123 uptake, whereas the placement of a carboxylic acid functionality on either the C7 or C10 position resulted in decreased uptake. Since carboxylic acid transporters are known to exist in the BBB, it should also be considered that the carboxylic acid functionality may be a substrate for an influx pump, in a similar fashion that paclitaxel acts as a substrate for cellular efflux. It is possible that this influx transport system is shuttling TX-67 into the cell, past the Pgp efflux system, allowing for the observed increase in BBB permeation. Furthermore, we have illustrated that positioning an acid or amide at C10 of paclitaxel is generally more effective than C7 attachment and that succinic acid and succinic amides lead to less rhodamine 123 uptake than the corresponding glutaric acid and amide analogs. We have also discovered that the hydrogen bonding character of the amide analogs plays a significant role and that the primary amides were generally superior compared to the secondary and tertiary amides analogs in evading interaction with Pgp.
The anionic introduction strategy and the introduction of a primary amide into the molecule may allow for delivery of paclitaxel into the CNS. This strategy could also hold promise for the delivery of other, highly complex, non-CNS permeable drugs, provided they are highly lipophilic.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (CA82801). B.J.T. would like to acknowledge the Department of Defense Breast Cancer Research Program for a predoctoral fellowship (DAMD17-02-1-0435). Paclitaxel was generously donated by Tapestry Pharmaceuticals, Boulder, CO. We thank Jacquelyn Huff for her excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Jordan MA. Curr. Med. Chem.: Anti-Cancer Agents. 2002;2:1. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 2.Suffness M, Wall ME. Taxol: Science and Applications. 1995;3 [Google Scholar]
- 3.Georg GI, Chen TT, Ojima I, Vyas DM, editors. Taxane Anticancer Agents: Basic Science and Current Status. Washington, DC: American Chemical Society; 1995. [Google Scholar]
- 4.Sparreboom A, van Asperen J, Mayer U, Schinkel AH, Smit JW, Meijer DK, Borst P, Nooijen WJ, Beijnen JH, van Tellingen O. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2031. doi: 10.1073/pnas.94.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouty-Boye D, Kolonias D, Wu CJ, Savaraj N, Lampidis TJ. Cancer. Res. 1995;55:1633. [PubMed] [Google Scholar]
- 6.Schinkel AH. Adv. Drug Del. Rev. 1999;36:179. doi: 10.1016/s0169-409x(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 7.Kemper EM, van Zandbergen AE, Cleypool C, Mos HA, Boogerd W, Beijnen JH, van Tellingen O. Clin. Cancer Res. 2003;9:2849. [PubMed] [Google Scholar]
- 8.Michaelis ML, Ranciat N, Chen Y, Bechtel M, Ragan R, Hepperle M, Liu Y, Georg G. J. Neurochem. 1998;70:1623. doi: 10.1046/j.1471-4159.1998.70041623.x. [DOI] [PubMed] [Google Scholar]
- 9.Fischer H, Gottschlich R, Seelig A. J. Membr. Biol. 1998;165:201. doi: 10.1007/s002329900434. [DOI] [PubMed] [Google Scholar]
- 10.Hitchcock SA, Pennington LD. J. Med. Chem. 2006;49:7559. doi: 10.1021/jm060642i. [DOI] [PubMed] [Google Scholar]
- 11.Gerebtzoff G, Seelig A. J. Chem. Inf. Model. 2006;46:2638. doi: 10.1021/ci0600814. [DOI] [PubMed] [Google Scholar]
- 12.Seelig A. Eur. J. Biochem. 1998;251:252. doi: 10.1046/j.1432-1327.1998.2510252.x. [DOI] [PubMed] [Google Scholar]
- 13.Seelig A, Blatter XL, Wohnsland F. Int. J. Clin. Pharmacol. Ther. 2000;38:111. doi: 10.5414/cpp38111. [DOI] [PubMed] [Google Scholar]
- 14.Gatlik-Landwojtowicz E, Aanismaa P, Seelig A. Biochemistry. 2006;45:3020. doi: 10.1021/bi051380+. [DOI] [PubMed] [Google Scholar]
- 15.Ge H, Vasandani V, Huff JK, Audus KL, Himes RH, Seelig A, Georg GI. Bioorg. Med. Chem. Lett. 2006;16:433. doi: 10.1016/j.bmcl.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 16.Spletstoser JT, Turunen BJ, Desino K, Rice A, Datta A, Dutta D, Huff JK, Himes RH, Audus KL, Seelig A, Georg GI. Bioorg. Med. Chem. Lett. 2006;16:495. doi: 10.1016/j.bmcl.2005.10.063. [DOI] [PubMed] [Google Scholar]
- 17.Rice A, Liu Y, Michaelis ML, Himes RH, Georg GI, Audus KL. J. Med. Chem. 2005;48:832. doi: 10.1021/jm040114b. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Faibushevich A, Turunen BJ, Yoon SO, Georg G, Michaelis ML, Dobrowsky RT. J. Neurochem. 2003;84:347. doi: 10.1046/j.1471-4159.2003.01526.x. [DOI] [PubMed] [Google Scholar]
- 19.All compounds prepared in this study showed spectroscopic properties in agreement with their structures.
- 20.Rose JM, Peckham SL, Scism JL, Audus KL. Neurochem. Res. 1998;23:203. doi: 10.1023/a:1022485026198. [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Dai H, Shaik N, Elmquist WF. Adv. Drug. Deliv. Rev. 2003;55:83. doi: 10.1016/s0169-409x(02)00172-2. [DOI] [PubMed] [Google Scholar]