Abstract
Inactivation of the retinoblastoma gene Rb leads to defects in cell proliferation, differentiation, or apoptosis, depending on specific cell or tissue types. To gain insights into the genes that can modulate the consequences of Rb inactivation, we carried out a genetic screen in Drosophila to identify mutations that affected apoptosis induced by inactivation of the Retinoblastoma-family protein (rbf) and identified a mutation that blocked apoptosis induced by rbf. We found this mutation to be a new allele of head involution defective (hid) and showed that hid expression is deregulated in rbf mutant cells in larval imaginal discs. We identified an enhancer that regulates hid expression in response to developmental cues as well as to radiation and demonstrated that this hid enhancer is directly repressed by RBF through an E2F binding site. These observations indicate that apoptosis of rbf mutant cells is mediated by an upregulation of hid. Finally, we showed that bantam, a miRNA that regulates hid translation, is expressed in the interommatidial cells in the larval eye discs and modulates the survival of rbf mutant cells.
Keywords: Rb family protein Rbf, hid, apoptosis, Drosophila, eye development, bantam
Introduction
The retinoblastoma protein pRb is a prototype tumor suppressor often mutated or inactivated in cancers (reviewed in (Classon and Harlow, 2002; Weinberg, 1995). pRb functions by binding to the E2F family of transcription factors and regulates a variety of normal cellular processes including cell proliferation, differentiation, as well as apoptosis. In mammals, eight E2F, and three DP family members have been identified (reviewed in (DeGregori and Johnson, 2006). These E2F and DP proteins form different complexes with extensive functional overlap within each family; therefore, an analysis of the in vivo role of Rb/E2F proteins in mammals is very complicated. In contrast, the Rb/E2F proteins in Drosophila are much simpler and yet are highly conserved. There are only one DP (dDP), two dE2F (dE2F1 and dE2F2), and two Rb family proteins (RBF and RBF2) in Drosophila (Du et al., 1996; Dynlacht et al., 1994; Ohtani and Nevins, 1994; Sawado et al., 1998; Stevaux et al., 2002). The two E2F proteins in Drosophila correspond to the two distinct classes of E2F proteins in mammals: dE2F1 mainly functions as a transcription activator (Du, 2000) similar to the activating E2Fs (E2F1-3), while dE2F2 mainly functions to mediate active repression similar to the repressive E2Fs (E2F4-5) in mammalian systems (Frolov et al., 2001). Similar to mammalian Rb, RBF can bind to both the activating E2F (dE2F1) as well as the repressive E2F (dE2F2), while RBF2 binds specifically to dE2F2, similar to the preferential binding of p107/p130 to the repressive E2F proteins in mammals (Stevaux et al., 2002). Therefore, Drosophila provides an attractive model system to study the Rb/E2F proteins.
Although Rb is widely expressed in most cell types, in vivo studies of Rb inactivation revealed tissue specific defects in cell proliferation, differentiation, and apoptosis (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992). Similarly, RBF is also broadly expressed in all the cells in the Drosophila developing eye but inactivation of rbf in different cells of the developing eye exhibited distinct effects (Du, 2000; Firth and Baker, 2005). For example, inactivation of rbf in the developing eye led to ectopic S phase posterior to the MF but not in the MF (Du, 2000; Firth and Baker, 2005). In contrast, cells near the MF but not in the posterior exhibited significantly increased apoptosis in the absence of RBF (Du, 2000; Moon et al., 2006). Therefore, different cells can respond differently to the inactivation of Rb (RBF) in both flies and in mammalian systems. Understanding the mechanism by which cells respond differently to inactivation of Rb will be critical to our understanding of how Rb functions as a tumor suppressor. In addition, since Rb is functionally inactivated in a majority of human cancers, approaches that specifically increase the apoptosis of Rb mutant cells will have therapeutic potential.
In an effort to identify genes that affected the consequence of Rb inactivation on cell proliferation, differentiation, and apoptosis, we carried out a genetic screen and identified a mutation that blocked apoptosis induced by rbf mutation. We found this mutation to be a new allele of hid. Additionally, we showed that hid expression is increased in rbf mutant cells. We identified an enhancer that mediates hid expression in response to developmental cues as well as to radiation and showed that this hid enhancer is negatively regulated by RBF through an E2F binding site. Finally, we showed that bantam, a regulator of hid translation, is expressed in the interommatidial cells in the eye discs and modulated the survival of rbf mutant cells.
Results
Identification of w138, a mutation that blocked apoptosis induced by rbf inactivation
Inactivation of rbf in the developing eye disc led to ectopic S phase in both the anterior half of the MF and posterior to the MF, and significantly increased apoptosis in and immediately anterior to the MF (Du, 2000; Firth and Baker, 2005; Moon et al., 2006). In the adult compound eye, rbf mutant clones occupied slightly smaller areas compared to the control wild-type clones (white ommatidia, Fig. 1A and B) and displayed mild roughness. To identify genes that modulate the effect of rbf inactivation, we carried out a genetic screen on the left-arm of the third chromosome (3L) using the eyFLP/FRT system (see Materials and Methods). We isolated a mutation, w138, which significantly expanded rbf mutant clone areas in adult eyes. Double mutant clones of rbf and the newly isolated recessive w138 mutation occupied a large majority of the adult eye tissues (Fig. 1C), whereas the clones of w138 mutation alone occupied only a little bit more areas than that of the control WT clones (Fig. 1E and B). These observations indicate that w138 and rbf exhibited a synergistic effect on mutant clone size in the adult eyes and suggest that w138 modulates the effect of rbf inactivation.
Figure 1. Modification of the rbf mutant phenotype by mutations of w138 and hid.

A) rbf mutant clones (A, white ommatidia) occupied slightly smaller areas in adult eyes as compared to the control wild-type clones (B, white ommatidia). (C and D) Mutation of w138 or hid in conjunction with rbf dramatically increased areas occupied by the mutant clones (C, rbf;w138, white ommatidia; D, rbf;hid, white ommatidia). (E and F) The morphology of w138, or hid single mutant clones was relatively normal with occasional extra number of interommatidial bristles. Mutant clones were generated using eyFLP/FRT method. Anterior of the eye is to the left and dorsal is up. (G-K) Mosaic clone analysis of activated Caspase 3 or pyknotic nuclei in third instar eye disc. Mutant clones were marked by absence of GFP shown in green. (G) rbf mutant clones spanning the MF displayed significantly elevated level of anti-activated Caspase 3 staining (Cas3 in magenta, white arrow). (H) rbf; w138 double mutant clones in the MF lack Cas3 staining. (I-K) High magnification images of DAPI staining to detect nuclear pyknosis in the MF region of the eye discs. (I) rbf mutant clones in the MF contained extensive pyknotic nuclei (bright magenta dots, white arrows). (J and K) Nuclear pyknosis was not observed in either rbf; w138 (J) or rbf; hid (K) double mutant clones. In all figures anterior of the eye disc is to the left and dorsal is up. Arrowhead points the MF groove.
Since inactivation of rbf affects cell proliferation as well as apoptosis, we carried out assays to determine if mutation of w138 affected cell proliferation or apoptosis induced by rbf inactivation. Although BrdU incorporation assay did not reveal striking difference between rbf single and rbf;w138 double mutant clones, apoptosis in the MF observed in rbf mutant clones was completely blocked by mutation of w138. As shown in Figure 1G, rbf mutant clones (marked by the absence of GFP) near the MF exhibited a significant level of apoptosis as shown by increased activated Caspase-3 (Cas3) staining and increased TUNEL positive cells (Fig.1G and data not shown). In addition, the mutant clones near the MF accumulated condensed apoptotic pyknotic nuclei as shown by DAPI staining (white arrows, Fig. 1I). These observations are consistent with the previous reports that inactivation of rbf leads to increased apoptosis near the MF (Du, 2000; Moon et al., 2006). In contrast, no increased Cas3 staining, no TUNEL positive cells, and no pyknotic nuclei were observed in rbf;w138 double mutant clones near the MF (Fig. 1H, J, and data not shown), indicating that mutation of w138 blocked apoptosis of rbf mutant cells near the MF. Previous studies have shown that apoptotic cells in the developing discs induced compensatory proliferation by producing signaling proteins such as Wingless (Wg) (Fan and Bergmann, 2008; Huh et al., 2004; Perez-Garijo et al., 2004; Ryoo et al., 2004). Indeed, ectopic Wg expression was observed in rbf mutant cells near the MF and was blocked in rbf;w138 double mutant clones (data not shown), indicating w138 is also required for the compensatory proliferation pathway. In conclusion, our data indicate that w138 mutation blocks apoptosis induced by the inactivation of rbf.
w138 is a novel allele of hid
To identify the gene that corresponded to the w138 mutation, we genetically mapped the w138 mutation. w138 is a partially lethal mutation, the homozygote survivors exhibited a number of visible phenotypes, including non-transparent wings, degenerated humeral bristles, and additional interommatidial cells in the pupal retina (data not shown). Using the 3L chromosome deficiencies, w138 was mapped to the chromosomal interval 75C1-2 (between the left breakpoint of Df(3L)H99 and the right breakpoint of Df(3L)X25). This region contains at least 7 annotated genes including the Drosophila apoptosis activators hid and grim according to the FlyBase genome annotation (Grumbling and Strelets, 2006). hid and grim, as well as reaper (rpr) encode a family of related pro-apoptotic proteins that inactivate the function of Drosophila IAP proteins (Bergmann et al., 2003; Ryoo et al., 2002).
Interestingly, the non-transparent wing, additional interommatidial cells, and degenerated humeral bristles phenotypes of w138 is similar to those of the viable mutations of the components of the apoptosis pathway, such as mutations of hid, dronc and Dark (Abbott and Lengyel, 1991; Rodriguez et al., 1999; Xu et al., 2005). Complementation tests showed that the transheterozygotes of w138/hidPZ 05014, w138/Df(3L)H99, w138/Df(3L)X25, and hidPZ 05014 /Df(3L)H99 all exhibited reduced viability with survivors that displayed the pupal and adult phenotypes described above. In contrast, the deletion chromosome Df(3R)X38, which lacks rpr but still retains hid and grim, complemented w138 mutation. Taken together, these complementation tests indicate that w138 is likely an allele of hid.
To characterize the w138 mutation at molecular level, we performed sequencing analysis. A set of DNA fragments corresponding to the hid coding regions were amplified by PCR from w138 adult flies or from transheterozygotes of w138 and w300 (w300 is a different mutant isolated from the same screen). Two different size PCR fragments were obtained from w138/w300 genomic DNA while only a single size fragment, corresponding to the smaller PCR fragment was amplified from w138 homozygote genomic DNA (Fig. 2B). Sequencing analysis of these fragments revealed that the w138 mutation is associated with a 312bp deletion in the second exon of hid (Fig. 2A and C), which results in frame shift and premature stop. Therefore, w138 is a new allele of hid, which we refer to as hidw138. hidw138 can still encode an 176 amino-acid (a.a.) protein including the RHG domain and two MAPK phosphorylation sites (Ser-121 and Thr-148) (Bergmann et al., 1998; Chai et al., 2003). In contrast the wild-type hid encodes a 410a.a. protein with a C-terminus hydrophobic region that required for mitochondria localization which is absent from hidw138 (Fig. 2D)
Figure 2. hidw138 is new allele of hid.
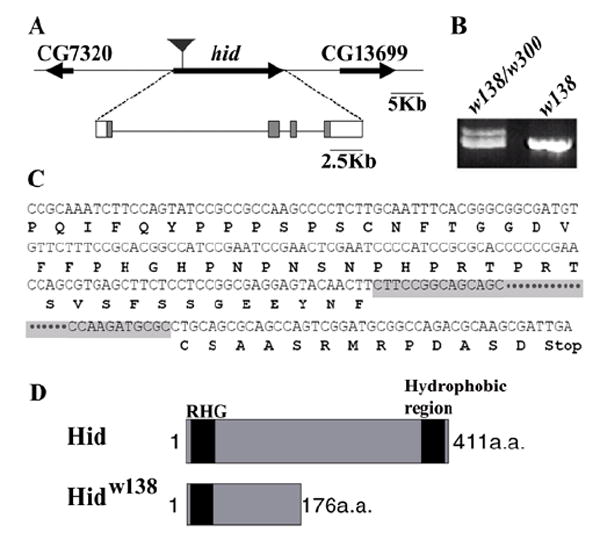
(A) Schematic representation of the hid locus. An approximate 60Kb region surrounding the hid locus is shown. A solid black arrow represents the hid transcript unit. The black inverted triangle represents the location of the P-element insertion in the hidPZ line. The hid ORF is shown by shaded boxes and the 5’ and 3’ UTR are shown by open boxes. (B) PCR product of hid exon2-4 from different genomic DNA. Two different size PCR fragments were obtained from w138/w300 transheterozygotes, a single fragment corresponding to the shorter one was obtained from w138 homozygote. (C) The sequence of w138 mutation. Amino acids sequence is shown beneath the nucleotide sequence. Deleted 312bp DNA sequence in the second exon is highlighted in gray. This deletion introduced a frame shift leading to the coding of 13-amino-acids followed by a stop codon. (D) Schematic representation of open reading frames of wild-type Hid and Hidw138.
To further compare the effect of hidw138 mutation with previously identified hid alleles on rbf-induced apoptosis, we used a strong loss-of-function allele, hid PZ 05014 (See Materials and Methods). The rbf; hid PZ double mutant clones displayed similar phenotypes as that of the rbf; hidw138 double mutants in adult compound eyes (Fig. 1D and C). Additionally, characterization of the apoptosis phenotype in the third instar eye disc revealed a block of nuclear pyknosis (by DAPI staining) (Fig. 1K) and an inhibition of TUNEL and activation of Caspase 3 in the MF. These phenotypes are identical to those of the rbf; hidw138 double mutant clones. These observations further support the idea that w138 is a loss of function allele of hid.
Ectopic hid expression in rbf mutants
The observation that mutation of hid blocked rbf-induced apoptosis prompted us to examine whether the levels of hid mRNA and protein differed in WT and rbf mutant eye discs. Using the anti-Hid antibody (Haining et al., 1999), a very low level of Hid protein was detected in WT eye discs (Fig. 3A). Similarly, only a background level of hid mRNA was detected in eye discs by in situ hybridization or by RT-PCR of total RNA isolated from eye-antennal discs (Fig. 3F and 3L). Coincide with this very low level of hid expression, very few apoptotic cells were observed as detected by Acridine Orange (AO) staining (Fig. 3I). In contrast to WT eye discs, transheterozygote of rbf15Δ/120 eye discs exhibited significant increase in apoptosis near the MF as reported earlier (Fig. 3J). The increased apoptosis was correlated with strong punctuated staining of Hid protein in the MF of the rbf15Δ/120 eye discs (Fig. 3B). A high level of Hid protein accumulation was also observed in the rbf mutant clones spanning the MF (Fig. 3D, Hid shown in magenta). Interestingly, while blocking apoptosis by using a mutation of dronc did not block Hid protein accumulation, Hid staining changed from a punctuate and perinuclear pattern in rbf mutant clones to a diffused cytoplasmic pattern in rbf;dronc double mutant clones (Fig. 3D, D’, E and E’). These observations are consistent with reports that showed Hid protein is localized in the mitochondria (Abdelwahid et al., 2007; Haining et al., 1999) and suggest that the possibility that Dronc is required for the mitochondria localization of Hid protein.
Figure 3. rbf mutant discs exhibit increased hid expression and elevated level of apoptosis in the MF.
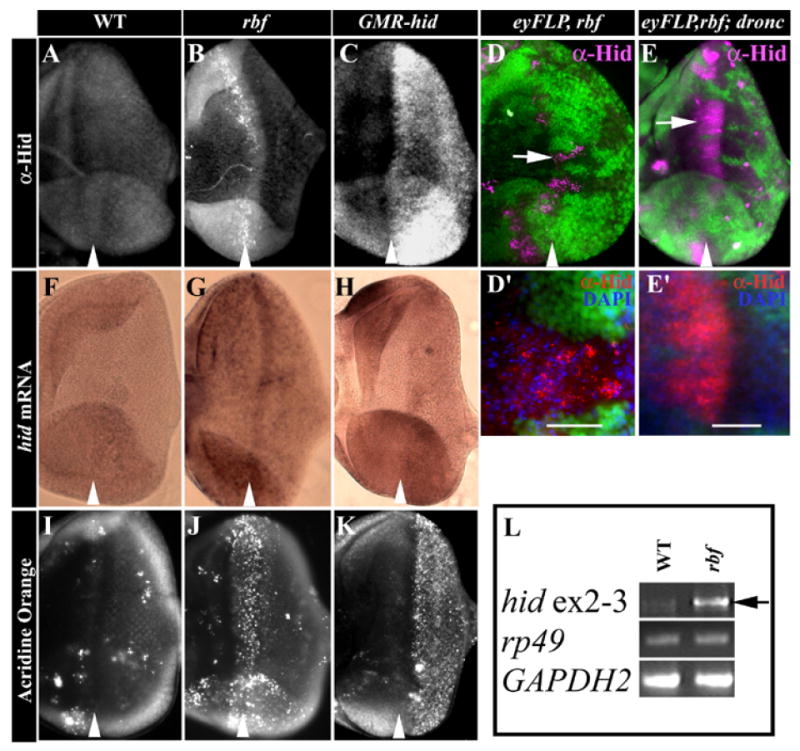
(A-E) Immunostaining of anti-Hid (CL1C3) antibody in third instar eye discs. (F-H) in situ hybridization to determine hid transcripts. (I-K) Acridine Orange staining to detect cell death. (A, F and I) WT eye disc, (B, G and J) rbf15Δ/120 eye discs, and (C, H and K) GMR-hid eye discs. While WT eye discs were mostly negative for anti-Hid labeling in eye discs (A), Hid protein was highly accumulated in the MF of rbf mutant discs (B), in the mutant clones of rbf (D) and rbf;dronc (E) that span the MF, and in the in the posterior in GMR-hid discs (C). Hid staining was shown in magenta in (D and E). (D’ and E’) High magnification image that show Hid was localized in punctate pattern and surrounded by pyknotic nuclei in rbf clones (D’) as compared to a diffused cytoplasmic localization in rbf;droncO1 double mutant clones in (E’). No significant hid mRNA is detected in WT eye discs (F) and only scattered AO labeled cells were observed in WT eye disc (I). In contrast, elevated hid mRNA was detected throughout the rbf mutant eye discs with the highest level observed in the MF (G), which correlated with significantly elevated AO staining in the MF of the rbf mutant discs (J). As expected, elevated hid expression (H) and AO staining (K) was observed in the posterior of GMR-hid discs. Arrowhead point the MF groove. In all figures anterior of the eye disc is to the left and dorsal is up. (L) Reverse transcription-PCR analysis demonstrated increased level of hid mRNA in rbf mutant discs (black arrow). The levels of ribosomal protein 49 (rp49) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH2) RNA were used as controls.
While increased apoptosis and Hid protein were mostly observed near the MF of rbf mutant discs, deregulated hid mRNA was observed throughout the entire eye disc; although the highest hid mRNA level was observed in the MF (Fig. 3G). The observed hid mRNA up-regulation in rbf mutant eye-antennal eye discs was further confirmed by RT-PCR analysis (Fig. 3L, arrow). In contrast, the level of rpr mRNA was not significantly different between WT and rbf mutant (data not shown). These observations are consistent with the previous finding that deficiencies that removed hid suppressed apoptosis in rbf mutant discs while a deficiency that removed both reaper and sickle did not (Moon et al., 2006). In conclusion, hid protein and mRNA is upregulated in rbf mutants and is required for apoptosis induced by the inactivation of rbf.
An E2F binding site in the hid 5’ regulatory sequences is occupied by dE2F proteins in vivo
To examine the possibility that hid is a direct transcriptional target of Rbf/E2F proteins, we analyzed the hid non-coding DNA sequence to identify conserved consensus E2F binding sites using the VISTA Browser (http://pipeline.lbl.gov/cgi-bin/gateway2) multiple alignments. The 14 Kb hid 5’ flanking sequences and 11 Kb intron 1 sequences (Fig. 2A) from D. melanogaster and D. pseudoobscura were analyzed and two consensus E2F binding sites in conserved sequence blocks were identified: one at -1415 bp (-5’TTTCGCGC3’-) in the hid 5’flanking sequence (hid 5’F) and one at +2183 bp (-5’TTTGGCGC3’-) in hid intron 1.
To determine if these two consensus E2F binding sites are occupied by dE2F proteins in vivo, we used Chromatin Immunoprecipitation (ChIP) assay in eye-antennal disc. As shown in Fig. 4, ChIP with anti dE2F1 antibody led to significant enrichment of the DNA fragment containing the 5’ E2F binding site but not the DNA fragment containing the intron 1 E2F binding site (Fig. 4A). These observations indicate that the 5’ E2F binding site but not the intron 1 E2F binding site is occupied by dE2F1 in vivo. We also carried out ChIP analysis using the anti dE2F2 antibody. In comparison to dE2F1, the ChIP using anti dE2F2 antibody led to modest enrichment of the DNA fragment containing the 5’ E2F binding site and not the DNA fragment containing the intron 1 E2F binding site (Fig. 4A). As a positive control for dE2F2 ChIP experiments, we observed the 5’ flanking sequence of Arp53D (Dimova et al., 2003) was significantly enriched in anti dE2F2 ChIP experiments (Fig. 4A). These data indicate that in eye-antennal disc, the E2F binding site in hid 5’F but not the one in intron 1 is occupied by dE2F proteins in vivo. It appears that this E2F binding site is bound mostly by dE2F1 and to a lesser degree dE2F2 in vivo. This is similar to the observation reported previously in SL2 cells (Moon et al., 2005).
Figure 4. Both dE2F1 and dE2F2 bind to the consensus E2F binding site in the hid 5’ regulatory region.
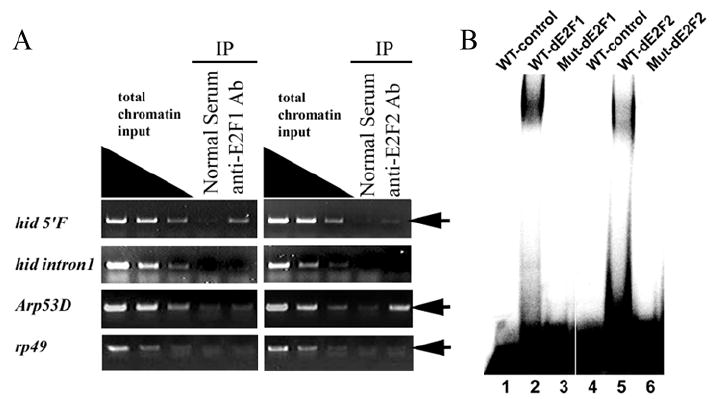
A) ChIP assay using antibodies against dE2F1 and dE2F2 proteins exhibited different degree of enrichment of the DNA fragment containing the E2F binding site at −1.4Kb in hid 5’F (first row, arrow). In contrast, ChIP with either the dE2F1 or the dE2F2 antibody failed to enrich the DNA fragment containing the introns 1 E2F site at +2.1Kb (second row). The E2F binding site in 5’ flanking sequence of Arp53D was used as a positive control for dE2F2 ChIP assay. Anti-dE2F2 highly enriched target DNA fragment (third row, arrow). rp49 was used as a negative control. No enrichment of DNA fragment was detected (fourth row, arrow). Nonspecific IgG (Normal Serum) was used as internal negative control. (B) DNA fragments containing either the WT or mutant E2F binding site of the hid 5’F locus were used as probes and recombinant dE2F1/dDP (lanes 1-3) or dE2F2/dDP (lanes 4-6) proteins were used in an EMSA assay as indicated. wild-type E2F binding site probe (WT, lanes 1, 2, 4, and 5), E2F binding site mutated probe (Mut, lanes 3 and 6).
To further demonstrate that dE2F proteins indeed bind specifically to the observed consensus hid 5’ E2F binding site, we carried out in vitro electrophoretic mobility shift assay (EMSA). As shown in Fig. 4B, both dE2F1/dDP and dE2F2/dDP heterodimer bound to the probe containing the WT E2F binding site from the hid 5’F (Fig. 4B; lanes 2 and 5) but not to a probe containing the mutated E2F site (Fig. 4B, lanes 3 and 6). These observations demonstrate that the E2F binding site in hid 5’F can interact with the two dE2F proteins specifically in vitro and is occupied by dE2F proteins in vivo in larval eye discs.
An enhancer element that regulates hid expression during development and in response to radiation
Since only the E2F binding site in hid 5’F is occupied by dE2F proteins in vivo, we further characterized the role of this E2F binding site on hid transcription. A 2.2Kb fragment containing the hid 5’ E2F binding site was cloned into the pH-stinger vector (Barolo et al., 2004) to generate WT transgenic lines (Fig. 5A, hid5’F-WT) and the GFP reporter expression was examined in transgenic animals during development.
Figure 5. hid 5’F drives reporter GFP expression in imaginal discs.

(A) Schematic representation of hid enhancer constructs: Black lines indicate non-coding DNA, open boxes indicate reporter GFP and lacZ, “X” represents mutated E2F site. (B and C) Very low level of GFP reporter expression are observed in 3rd instar eye and wing discs from WT constructs (hid5’F-WT). (D and E) Mutation of the consensus E2F binding site dramatically upregulated GFP reporter expression in eye and wing discs. (F-K) Eye and wing imaginal discs 4-5 hr after gamma-irradiation (4Gy). (F and G) A high level of GFP reporter expression from hid5’F-WT was observed in most cells in the eye disc (F) and wing disc (G). (H and I) Acridine Orange staining of irradiated discs show increased cell death in the eye disc (H) as well as the wing disc (I). (J and K) Irradiation induced cell death in the eye disc (J) and wing disc (K) was suppressed by removal of hid. (L-N) Increased expression of hid reporter in rbf mutant background. In situ hybridizations of GFP reporter expression in WT and rbf15Δ/120 eye discs are shown in (L) and (M), respectively. Increased β-gal reporter level was also observed in rbf mutant clones (N, magenta, arrow). The position of the MF groove was indicated by white arrows. Anterior of eye disc is to the left and dorsal is up. Anterior of the wing disc is to the left and ventral is up. (O) Sequence of the conserved hid 5’ E2F site in Drosophila species. E2F binding site is underlined. E2F mutant sequence of mutated E2F binding site is shown at the bottom. Mutated bases are shown in lowercase letters. (P-S’) The hid 5’F responds various biological cell death stimuli. GFP reporter expression from whole third instar larva from hid5’F-WT (P) and hid5’F-E2FMut (Q). (R) A 42hr APF pupal retina showed high level of hid 5’F-WT GFP reporter expression in ommatidia at the periphery of the eye disc and in some interommatidial cells. (S-S’) High magnification image of 42hr APF pupal retina showed upregulation of GPF reporter expression from hid5’F-WT enhancer in some interommatidial cells.
Apoptosis plays little role during larval wing and eye disc development (Milan et al., 1997) and there were very few apoptotic cells in WT eye and wing discs at this stage. Consistent with this, very low levels of GFP reporter expression from the hid5’F-WT observed in WT eye-antennal or wing discs (Fig. 5B, C). When these tissues were exposed to exogenous death stimuli such as ionizing radiation, high levels of apoptosis were induced in these discs (Fig. 5H, I) and there was a significantly higher level of GFP reporter expression from hid5’F-WT (Fig. 5F and G). Hid induction by ionizing radiation is important for radiation induced apoptosis since apoptosis induced by irradiation in eye and wing discs was significantly blocked in hidw138 mutant background (Fig. 5J and K). In addition, although the level of GFP reporter expression in larval stages was quite low, a significantly higher level of the GFP reporter expression from hid5’F-WT was observed in pupal stages in tissues with significant amount of developmentally regulated apoptosis such as in ommatidia at the periphery of the eye disc and in interommatidial cells during mid-pupation (Fig. 5R-S’). During retina development, ommatidia at the periphery of the eye undergo apoptosis in response to Wg signaling, which activates the expression of hid, rpr, and grim (Lin et al., 2004). In addition, extra interommatidial cells are removed by apoptosis during pupal stage and this developmental regulated apoptosis is completely dependent on the presence of Hid (Miller and Cagan, 1998; Yu et al., 2002). In conclusion, the 2.2Kb hid5’F-WT fragment contains regulatory elements that control hid transcription in response to developmental cues as well as to stress signals such as ionizing radiation.
The E2F binding site in the hid 5’ regulatory region negatively regulates hid expression
To determine if the E2F binding site in hid 5’F regulates hid transcription, the same base pair substitutions that disrupted the binding of dE2F2 (Fig. 4B) was introduced into the 2.2 Kb hid5’F-WT fragment to generate hid 5’F with mutated E2F binding sequence (Fig.5A, hid5’F-E2FMut). In contrast to hid5’F-WT, transgenic flies carrying hid5’F-E2FMut displayed significantly increased GFP expression in larval eye and wing discs (Fig. 5D and E). In addition, elevated GFP level was also observed in many other larval cells in hid5’F-E2FMut animal, indicating a relative broad requirement of this E2F binding site to repress hid expression at this stage (Fig. 5P,Q). To further characterize if RBF repressed the hid5’F-WT enhancer activity, we examined the level of GFP reporter expression in rbf mutant background. In situ hybridization results showed an increased hid5’F-WT reporter expression in rbf mutant discs, with the highest level observed near the MF (Fig. 5L,M). Similarly, increased β-gal reporter was also observed rbf mutant clones (Fig. 5N, β-gal in magenta). These results, in conjunction with the observed effect of the E2F binding site mutation, indicate that the hid5’F-WT enhancer is repressed by the RBF/dE2F proteins in the larval stage.
bantam microRNA attenuates rbf induced cell death in posterior larval eye disc
Although deregulated hid expression in the absence of rbf was most obvious around the MF in the eye disc, a lower level of deregulated hid expression was also observed in other parts of the eye disc (Fig. 3G). These observations suggest that additional post-transcriptional mechanisms also contribute to the observed induction of apoptosis near the MF in the absence of rbf. In support of this idea, EGFR signaling, which was shown to negatively regulate Hid transcription as well as Hid activity (Bergmann et al., 1998; Kurada and White, 1998), was found to regulate the sensitivity of rbf mutant cells to apoptosis (Moon et al., 2006). Since Hid is also regulated at the translational level by bantam (Brennecke et al., 2003; Hipfner et al., 2002), we examined the possibility that bantam may also regulate the sensitivity of rbf mutant cells to apoptosis in vivo.
bantam is expressed in a spatio-temporally restricted manner throughout development. We first generated transgenic lines to monitor developmentally regulated bantam expression. The important regulatory element for bantam was mapped between the restriction enzyme sites BamHI and SpeI (Brennecke et al., 2003) (Fig. 6A). Since all bantam enhancer trap lines are clustered within a small region upstream of the bantam hairpin sequence (Brennecke et al., 2003; Wilson et al., 2008), we generated bantam reporter lines (ban 5’F-lacZ) using the BamHI-EcoRV bantam 5’ flanking fragment and compare the pattern of β-gal expression with that of the bantam-sensor. The bantam-sensor has been used previously as a reporter for bantam miRNA activity (Brennecke et al., 2003). Cells with low level of bantam-sensor would indicate high bantam miRNA activity. Consistent with this, the expression of β-gal reporter from the ban 5’F-lacZ transgene was complementary to the previously generated bantam-sensor (Brennecke et al., 2003). For example, in posterior eye disc, β-gal from the ban5’F-lacZ was highly expressed in interommatidial cells but not in the ommatidia clusters in contrast to the expression of bantam-sensor, which is very high in the ommatidia clusters and low in the interommatidial cells (Fig. 6B-B”). The complementary expression pattern between ban 5’F-lacZ reporter and bantam-sensor also extends to the optic lobes, wing and leg discs (data not shown). In situ hybridization of ban5’F-lacZ also confirmed the expression of the lacZ reporter in the MF and the posterior interommatidial cells (Fig. 6C). These data indicate that the 2.35Kb ban 5’F fragment was sufficient to drive bantam expression in the MF and in the interommatidial cells.
Figure 6. Spatial expression of bantam miRNA in developing eye disc and its modulation of rbf-induced apoptosis.
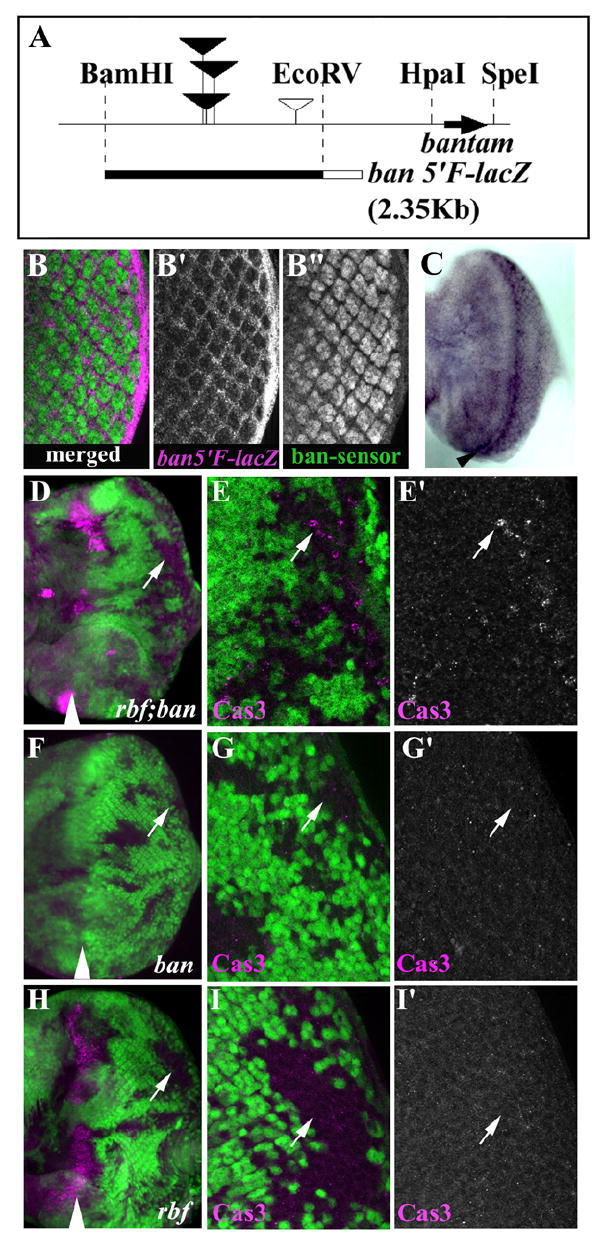
(A) Schematic representation of bantam flanking sequence. Bantam hairpin coding region is shown by a black arrow. Filled inverted triangles indicate P-element insertion sites that disrupted bantam function. An open inverted triangle indicates P-element insertion site associated non-bantam mutation. The 2.35Kb BamHI and EcoRV fragment was used to generate ban-5’F-lacZ shown at the bottom (filled black bar). The open box represents the LacZ reporter. β-gal expression from ban5’F-lacZ (magenta) and GFP expression from ban-sensor-GFP (green) are complementary (B-B”). High bantam expression is seen in interommatidial cells (β-gal, B’) while high bantam sensor is seen in photoreceptor cells (GFP, B”). Merged image is shown in (B). (C) In situ hybridization with DIG labeled lacZ RNA probe to ban5’F-lacZ eye disc. High level of expression in the MF and posterior interommatidial cells were seen. The MF groove indicates black arrowhead. (D-I’) bantam miRNA attenuates rbf induced cell death. Low (D) and high (E, E’) magnification images of rbf; ban double mutant cells showed increased Cas3 staining (magenta) in posterior mutant clones. Low (F) and high magnification (G, G’) images of bantam mutant clones showed background level of Cas3 staining. Low (H) and high magnification (I, I’) images of rbf mutant clones showed very low Cas3 labeling in the posterior, while clones spanning the MF showed extensive Cas3 labeling.
To determine whether bantam regulates rbf dependent cell death in third instar eye disc, we compared the level of apoptosis in rbf single and rbf; ban double mutant clones in the posterior of larval eye discs. Previous studies showed that although bantam is a negative regulator of hid, removal of bantam alone was insufficient to induce cell death in developing wing disc (Brennecke et al., 2003; Hipfner et al., 2002). Similarly, we did not observe activated Caspase 3 in bantam single mutant clones (Fig. 6F-G’). Increased activated Caspase 3 staining in rbf single mutant clones was mostly observed in the MF, with very low level in the posterior (Fig. 6H-I”). In contrast, significant increase in the level of activated Caspase 3 staining was observed in rbf; ban double mutant clones in posterior (Fig. 6D-E”, white arrows). Moreover the fluorescent signal of activated Caspase 3 staining in the MF was also much stronger in rbf; ban double mutant clones compare to the rbf mutant clones, suggesting that even more cells were under going apoptosis in the MF of rbf; ban double mutant clones (Fig. 6D, H). These results indicate that bantam antagonizes rbf-dependent apoptosis and that the spatial expression of bantam provides another level of control for the resistance or sensitivity of rbf mutant cells to apoptosis in the developing eye.
Discussion
hid is a key cell death regulator in Drosophila, which is upregulated in response to developmental cues as well as to various stress stimuli. In this manuscript, we show that hid is also upregulated and responsible for apoptosis in response to rbf inactivation. We have identified an enhancer element of hid, hid5’F, which regulates hid expression in response to developmental cues as well as to ionizing radiation. We demonstrate that RBF directly represses the expression of hid through a conserved E2F binding site in this enhancer. Finally we show that removal of bantam synergistically increases the apoptosis of rbf mutant cells without significantly affecting the apoptosis of WT cells.
Regulators of Hid activity modulates a cell’s sensitivity to apoptosis in response to rbf inactivation
Although RBF is ubiquitously expressed in developing eye discs, only cells near the MF display significant increased apoptosis in response to rbf inactivation, suggesting that RBF plays an important role in preventing apoptosis and that the sensitivity of rbf mutant cells to apoptosis is under an additional level of control. We identify hid as a key apoptosis target that is repressed by RBF and required for the apoptosis of rbf mutant cells. Interestingly, while deregulated hid expression can be observed throughout the rbf mutant eye discs, the highest hid expression was observed near the MF. These results point the contribution of transcriptional mechanisms to the increased sensitivity of MF cells to apoptosis in the absence of rbf and further characterization of the hid 5’ F enhancer will be necessary to elucidate the precise transcriptional mechanisms.
In addition to transcriptional regulation, Hid is also regulated by miRNA bantam at translational level and by survival signals such as EGFR signaling at the post-translational level. If hid is a key target of rbf in apoptosis regulation, it is expected that these additional regulators of Hid will modulate the sensitivity of rbf mutant cells to apoptosis. Indeed, removal of bantam together with rbf induced additional apoptosis in the posterior eye disc cells, indicating that bantam played an important role in preventing the apoptosis of rbf mutant cells in the posterior eye disc. The observed effect of bantam is consistent with its expression pattern, which is in the interommatidial cells in the posterior of the developing eye discs. Interestingly, removal of bantam alone was not sufficient to induce apoptosis in eye disc, similar to the observation in wing discs reported previously (Brennecke et al., 2003). These observations are consistent with the very low level of hid detected in WT eye or wing discs and suggest that bantam is not required during normal larval wing or eye development, but provides another level of fine tuning in regulating the sensitivity of disc cells to apoptosis in response to stress or developmental signals.
In addition to bantam, EGFR signaling, which regulates Hid expression as well as Hid protein activity (Bergmann et al., 1998; Kurada and White, 1998), was shown to be an important survival signal in the posterior eye disc and regulates the apoptosis of rbf mutant cells (Baker and Yu, 2001; Moon et al., 2006). EGFR signaling is required for the stepwise recruitment of all the cell types after R8 photoreceptor specification (Freeman, 1996). In contrast to bantam, EGFR signaling is required to prevent apoptosis in WT eye discs. Since EGFR activation depends on short range signals from the developing ommatidia clusters, interommatidial cells will likely receive lower level of EGFR signaling and bantam expression in these cells may provide an additional level of apoptosis control independent of EGFR signaling. Taken together, we suggest that the activity of Hid, which is regulated by multiple mechanisms at the transcriptional, translational, and post-translational levels, defines a cell’s sensitivity to apoptosis upon inactivation of rbf in vivo.
Hid, Reaper, and Grim can also induce apoptosis in vertebrate model systems, and their apoptosis activity is subject to regulation by inhibitors of mammalian cell death (Claveria et al., 1998; Evans et al., 1997; Haining et al., 1999; McCarthy and Dixit, 1998). These observations suggest that the proapoptotic function of these genes is conserved. Interestingly, Smac/Diablo, a mammalian functional homolog of Hid, was shown to be a direct target of E2F1 in mammalian cells (Xie et al., 2006) and activation of Ras signaling protected the Rb family null fibroblasts from apoptosis (Young and Longmore, 2004). Therefore, there are striking similarities about the apoptosis regulation of Rb mutant cells between flies and mammalian cells and it will be interesting to determine if the basic principles we learned here in flies can be applied to mammalian cells.
Hid enhancer element
Drosophila contains at least four proapoptotic proteins, Rpr, Hid, Grim and Skl, which are located on the third chromosome 75C region and responsible for most of the apoptosis regulation in flies (Chen et al., 1996; Christich et al., 2002; Grether et al., 1995; Srinivasula et al., 2002; White et al., 1994; White et al., 1996). There are large non-coding sequences between each individual proapoptotic gene, which may provide dynamic regulation of the expression of these genes in response to developmental cues and environmental stresses. The sequences that control rpr transcription have been reported previously (Brodsky et al., 2000). A 4 kb genomic fragment containing the p53 responsive element was found to be sufficient to induce rpr expression upon ionizing radiation through the direct binding of Drosophila p53 homologue (Brodsky et al., 2000). In addition, rpr transcription is also regulated by Hox protein Deformed during sculpting of the larval head (Lohmann et al., 2002) and by Ecdysone receptor complex during salivary gland histolysis (Jiang et al., 2000). However, precise transcriptional regulation of other proapoptotic genes, including hid, remains to be elucidated.
In this study, we showed that a 2.2 Kb sequence immediately upstream of hid 5’UTR was sufficient to drive reporter expression in response to developmental cues and to environmental stress signals. For example, during pupal retina development, excess interommatidial cells are removed by apoptosis. Hid is required for this developmental regulated apoptosis (Yu et al., 2002) and mutation of hid led to increased interommatidial cells in the pupal retina. We found that hid 5’F enhancer conferred reporter expression specifically in the interommatidial cells of the pupal retinal. Furthermore, while only a background level of hid reporter expression was observed in WT larval eye discs, a developmental stage with little apoptosis, hid reporter expression was strongly induced in larval eye discs by ionizing radiation, which strongly induces apoptosis in discs that is dependent on the Hid activity. Therefore, the hid 5’F enhancer contains regulatory elements that control hid expression both in response to developmental cues as well as to ionizing radiation. Consistent with a previous report that showed Hid is negatively regulated by an E2F site in SL2 cells (Moon et al., 2005), we showed that hid 5’F enhancer is repressed by dE2F/RBF complexes through a conserved E2F binding site during development. These observations, however, are inconsistent with the observations that apoptosis observed in rbf mutants are suppressed when activation function of dE2F1 was removed (Moon et al., 2006). It is likely that there are additional targets regulated by dE2F1 that are also required for apoptosis in the absence of rbf. In addition, it should be pointed out that the hid 5’F enhancer is unlikely to contain all the regulatory elements controlling the expression of hid. For example, it was shown that an irradiation-responsive enhancer region (IRER) regulates both hid and rpr expression in response to ionizing radiation during embryogenesis (Zhang et al., 2008). Therefore multiple regulatory elements can control radiation induced hid expression possibly during different stages of development.
Mitochondria localization of Hid protein
Hid protein was found to be localized in the mitochondria in apoptotic cells depending on the presence of its C-terminal domain (Haining et al., 1999). Although initial studies suggested that the C-terminal hydrophobic region of Hid was not required for apoptosis induction when high level of Hid was expressed in HeLa cells, a recent study showed that the mitochondria localization played an important role for apoptosis in SL2 cells (Abdelwahid et al., 2007). The observation that hidw138, which lacks the C-terminal hydrophobic region, exhibited similar phenotype as another strong loss of function allele suggested the importance of Hid mitochondria localization in vivo. As reported earlier that Hid is localized predominantly in the mitochondria in apoptotic cells (Abdelwahid et al., 2007; Haining et al., 1999), we showed that Hid staining displayed a punctuate and perinuclear pattern in rbf mutant clones in near the MF (Fig. 3D, D’). Interestingly, blocking the apoptosis of rbf mutant cells by dronc mutation led to a lack of mitochondria localization of Hid. As shown in Fig. 3E, E’, Hid staining revealed a diffuse and cytoplasmic distribution in rbf;dronc double mutant cells. As reported earlier, cotransfection of BclXL blocked Hid mitochondria localization while cotransfection of p35, DIAP1, or XIAP, or treatment with cell-permeant cysteine proteases inhibitor peptide BOC-d-fmk did not (Haining et al., 1999). Since all these treatments inhibited Hid induced apoptosis (Haining et al., 1999), these observations suggest that Dronc is required for the localization of Hid protein to the mitochondria membrane.
Materials and methods
Drosophila genetics
Ethyl methanesulfonate (EMS)-induced rbf modifier screening was performed on the left arm of third chromosome (3L) in combination with the eyFLP/FRT-mediated mosaic clone technique (Newsome et al., 2000; Xu and Rubin, 1993). 3-day-old w; p{ry+, neoFRT80B} ry506 (BL#1988) males flies were fed overnight with 5mM EMS. These male flies were mated with rbf15aΔ,w, eyFLP; p{w+, rbf-G3} p{w+, Ubi-GFP} p{ry+, neoFRT80B} virgin females and cultured at 25°C. The p{w+, rbf-G3} is a rbf genomic rescue transgene integrated in an unknown site on the 3L-chromosome. F1 male flies that contain larger w- clones were subsequently crossed with w; TM3,Ser/TM6b,Tb balancer virgin females to establish balanced stocks. Balanced males were crossed with rbf15aΔ,w, eyFLP; p{w+, rbf-G3} p{w+, Ubi-GFP} p{ry+, neoFRT80B} or w, eyFLP; p{w+, Ubi-GFP} p{ry+, neoFRT80B} virgin females for retest and rbf dependence test, respectively. The w138 (=hidw138) and droncO1 were isolated from this screening. To map the w138 mutation, following deficiencies were used: 3L chromosome deficiencies from the Bloomington third-chromosome deficiency kit, Df(3L)H99, Df(3L)XR38, Df(3L)X25(White et al., 1994) and Df(3L)W4 (BL#2607). To generate w; hid PZ 05014 p{ry+, neoFRT80B}/TM3 act-GFP, Ser line, hidPZ 05014 (W05014-FlyBase, BL#11642) (Grether et al., 1995) was recombined with wild-type chromosome to segregate out unknown lethal background mutation(s) and subsequently recombined with w; p{ry+, neoFRT80B}. The following fly stocks were used in this study; rbf120-3-3 and rbf15aΔ, which was derived from imprecise excision of rbf15a hop3 (Du and Dyson, 1999), EP(3)3622, bantam-sensor (Brennecke et al., 2003). Fly stocks obtained from the Bloomington Stock Center are shown with Bloomington Stock (BL) number.
Molecular genetics
RT-PCR
Total RNA was prepared from 100 pairs of third instar eye-antenna discs that were dissected in 1xPBS. Transheterozygote of rbf120-3-3/rbf15aΔ female larva were used as rbf mutants and w1118 females as the control. Total RNA was prepared in with TRIZOL Reagent (GIBCO BRL) and RT-PCR (Enhanced Avian RT-PCR kit, SIGMA) reactions were done under the conditions supplied by the manufacturer. Oligo(dT)23 reverse primer and hid or GAPDH2 specific primers used for reverse transcription reaction.
Transgenic fly
A 2.2 Kb genomic fragment of hid 5’flanking sequence and a 2.35 Kb genomic fragment of bantam 5’ non-coding DNA sequence were amplified by PCR from w1118 genomic DNA. Nucleotide substitution of the consensus E2F binding site (from 5’-TTTCGCGC-3’ to 5’-TggCtaGC-3’) was made as shown previously in (Yamaguchi et al., 1995). Wild-type or mutated hid 5’F was subcloned into pH-Stinger or pH-Pelican vector (Barolo et al., 2004). Multiple transgenic lines were established and examined for reporter gene expression.
Immunohistochemistry
The third instar imaginal discs were dissected and fixed as previously described (Tanaka-Matakatsu et al., 2007). Primary antibodies were used at following dilutions: mouse anti-Hid CL1C3, 1:100 (Haining et al., 1999), rabbit anti-Cleaved Caspase-3 (Asp175), 1:500 (Cell Signaling TECHNOLOGY), mouse anti-ß galactosidase JIE7, 1:500 (DSHB) and mouse anti-Wg, 1:10 (DSHB). Dye-conjugated secondary antibodies were from Jackson Immunoresearch, Inc. and were used at 1:500. DNA was counterstained with DAPI 1:1000 (stock 50 ug/ml).
TUNEL analysis was modified for use in eye discs (Lisi et al., 2000) and anti-DIG-Rhodamine used for signal detection. Acridine Orange staining was done by following the Protocol 12.7 in Drosophila Protocols (Wolff, 2000). Eye disc in situ hybridization was carried out similar to previously described (Du, 2000). All images were taken on Zeiss Axioscop or Zeiss ApoTome with AxioCam CCD cameras. Adult fly eye images were taken using a Leica dissection microscope. Male adult flies were used for data collection.
Electrophoresis mobility shift assay (EMSA)
The sequence of the probes are: Wild-type; 5’-ACTTTGCGCGCGAAAACGCTTGAA-3’, E2F mut; 5’-ACTTTGCGCtaGccAACGCTTGAA-3’. Forward and reverse oligonucleotides were annealed, end-labeled, and purified. Binding reactions were as previously described (Tanaka-Matakatsu and Du, 2008).
Chromatin Immunoprecipitation (ChIP)
The third instar larva were dissected, fixed on ice for 15min. Thirty pairs of eye discs were pooled in 250ul of ChIP lysis buffer and were processed essentially as described (Austin et al., 1999). Primer sequences are: hid 5’F Forward 5’-ttctctgtcattcccaactttgcgcgcg-3’, hid 5’F Reverse 5’- cgtaactggcattactttagatccat-3’, hid intron1 Forward 5’-agtaagacattcgactcgagtttggcg-3’, hid intron1 Reverse 5’-gtgctatggctttaagaaagaaacgccg-3’, Arp53D Forward 5’-gatttttggcctctgtatgtcatggcg-3’, Apr53D Reverse 5’-aggagaggttgctgaacgaatcatggtc-3’, rp49 Forward 5’-ccacgaattccca tcacaaacagaagcc-3’, rp49 Reverse 5’-aatatatcgcttggcatcgccatgaacg-3’.
Acknowledgments
We would like to thank Drs. H. Steller and S. Cohen, the Bloomington Stock Center, and the Developmental Studies Hybridoma bank at the University of Iowa for generously supplying fly stocks and reagents. We also thank Dr. J. Searle for reading this manuscript. M. T-M thanks H. Matakatsu for discussion. This work was supported by a grant from the National Institute of Health (GM 074197) and by a grant from the America Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott MK, Lengyel JA. Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics. 1991;129:783–9. doi: 10.1093/genetics/129.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–49. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–40. 442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–41. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Yang AY, Srivastava M. Regulators of IAP function: coming to grips with the grim reaper. Curr Opin Cell Biol. 2003;15:717–24. doi: 10.1016/j.ceb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–13. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–8. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. Grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–82. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Christich A, Kauppila S, Chen P, Sogame N, Ho SI, Abrams JM. The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim, and hid. Curr Biol. 2002;12:137–40. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–30. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–7. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Claveria C, Albar JP, Serrano A, Buesa JM, Barbero JL, Martinez AC, Torres M. Drosophila grim induces apoptosis in mammalian cells. Embo J. 1998;17:7199–208. doi: 10.1093/emboj/17.24.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–20. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development. 2000;127:367–79. doi: 10.1242/dev.127.2.367. [DOI] [PubMed] [Google Scholar]
- Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. Embo J. 1999;18:916–25. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10:1206–18. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Brook A, Dembski MS, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EK, Kuwana T, Strum SL, Smith JJ, Newmeyer DD, Kornbluth S. Reaper-induced apoptosis in a vertebrate system. Embo J. 1997;16:7372–81. doi: 10.1093/emboj/16.24.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–51. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–60. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Genes Dev. 2001;15:2146–60. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Grumbling G, Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–8. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:4936–41. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–37. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–6. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol Cell. 2000;5:445–55. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–29. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–94. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 2004;131:2409–18. doi: 10.1242/dev.01104. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–78. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann I, McGinnis N, Bodmer M, McGinnis W. The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell. 2002;110:457–66. doi: 10.1016/s0092-8674(02)00871-1. [DOI] [PubMed] [Google Scholar]
- McCarthy JV, Dixit VM. Apoptosis induced by Drosophila reaper and grim in a human system. Attenuation by inhibitor of apoptosis proteins (cIAPs) J Biol Chem. 1998;273:24009–15. doi: 10.1074/jbc.273.37.24009. [DOI] [PubMed] [Google Scholar]
- Milan M, Campuzano S, Garcia-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc Natl Acad Sci U S A. 1997;94:5691–6. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–35. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26:7601–15. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell. 2005;9:463–75. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–60. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Nevins JR. Functional properties of a Drosophila homolog of the E2F1 gene. Mol Cell Biol. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–8. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–9. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–8. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Sawado T, Yamaguchi M, Nishimoto Y, Ohno K, Sakaguchi K, Matsukage A. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem Biophys Res Commun. 1998;251:409–415. doi: 10.1006/bbrc.1998.9407. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Datta P, Kobayashi M, Wu JW, Fujioka M, Hegde R, Zhang Z, Mukattash R, Fernandes-Alnemri T, Shi Y, Jaynes JB, Alnemri ES. Sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr Biol. 2002;12:125–30. doi: 10.1016/s0960-9822(01)00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. Embo J. 2002;21:4927–37. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Thomas BJ, Du W. Mutation of the Apc1 homologue shattered disrupts normal eye development by disrupting G1 cell cycle arrest and progression through mitosis. Dev Biol. 2007;309:222–35. doi: 10.1016/j.ydbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–83. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–7. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–93. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T. Histological Techniques for the Drosophila Eye Part I: Larva and Pupa. In: Sullivan W, Ashburner M, Hawley SR, editors. Drosophila Protocols. Cold Spring Harbor laboratory Press; New York: 2000. pp. 201–227. [Google Scholar]
- Xie W, Jiang P, Miao L, Zhao Y, Zhimin Z, Qing L, Zhu WG, Wu M. Novel link between E2F1 and Smac/DIABLO: proapoptotic Smac/DIABLO is transcriptionally upregulated by E2F1. Nucleic Acids Res. 2006;34:2046–55. doi: 10.1093/nar/gkl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–34. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Hayashi Y, Matsukage A. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J Biol Chem. 1995;270:25159–65. doi: 10.1074/jbc.270.42.25159. [DOI] [PubMed] [Google Scholar]
- Young AP, Longmore GD. Ras protects Rb family null fibroblasts from cell death: a role for AP-1. J Biol Chem. 2004;279:10931–8. doi: 10.1074/jbc.M311814200. [DOI] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–78. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, Yao B, Wu SS, Zhou L. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev Cell. 2008;14:481–93. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


