Abstract
Nanopores hold great promise as single-molecule analytical devices and biophysical model systems because the ionic current blockades they produce contain information about the identity, concentration, structure, and dynamics of target molecules. The porin MspA of Mycobacterium smegmatis has remarkable stability against environmental stresses and can be rationally modified based on its crystal structure. Further, MspA has a short and narrow channel constriction that is promising for DNA sequencing because it may enable improved characterization of short segments of a ssDNA molecule that is threaded through the pore. By eliminating the negative charge in the channel constriction, we designed and constructed an MspA mutant capable of electronically detecting and characterizing single molecules of ssDNA as they are electrophoretically driven through the pore. A second mutant with additional exchanges of negatively-charged residues for positively-charged residues in the vestibule region exhibited a factor of ≈20 higher interaction rates, required only half as much voltage to observe interaction, and allowed ssDNA to reside in the vestibule ≈100 times longer than the first mutant. Our results introduce MspA as a nanopore for nucleic acid analysis and highlight its potential as an engineerable platform for single-molecule detection and characterization applications.
Keywords: DNA sequencing, protein engineering, bio-nanotechnology
Nanopore analysis is based on the principle that an ionic current signal flowing through a nanometer-scale pore is blocked when the pore is occupied by a target molecule. Nanopores hold great promise as single-molecule analytical devices and biophysical model systems because the characteristics of these ionic current blockades contain information about the identity, concentration, structure, and dynamics of the target molecules. This technique has been implemented primarily with the biological protein pore α-hemolysin (αHL) (1–3) and inorganic nanopores (4–6). Protein pores are particularly attractive for nanopore analysis because X-ray crystallography reveals their structure with angstrom resolution, self-assembly facilitates atomic reproducibility from one pore to the next, pore size and composition are well matched to a variety of biologically important target molecules, and protein engineering offers an effective tool to tailor their analytical capabilities.
The strengths of nanopore analysis with protein pores are well illustrated by the growing body of work using αHL to detect and characterize ssDNA and RNA (1, 7–19) and by the recent demonstration of dADP detection with an engineered version of the porin OmpF (20). The possibility of using base-specific modulation of the ionic current for rapid, inexpensive DNA sequencing has been a central motivating factor for these investigations. Although recent progress in recognizing and differentiating the bases and regulating DNA passage through the pore supports the feasibility of this approach (18, 19, 21–24), realization of de novo nanopore DNA sequencing remains elusive. The introduction of new protein pores capable of single-molecule detection and characterization of nucleic acids is of high scientific and technological relevance because it may provide the improved biophysical insight and analytical capabilities needed to realize this goal.
We identified Mycobacterium smegmatis porin A (MspA) as an excellent candidate nanopore for nucleic acid analysis. MspA is a channel-forming protein that constitutes the major diffusion pathway for hydrophilic solutes in M. smegmatis (25, 26). Of particular relevance for nanopore DNA sequencing is the geometry of the MspA channel (27). It has a ≈1-nm-long, ≈1-nm-wide constriction that is flanked by regions of significantly larger diameter (Fig. 1A). This geometry is distinct from the αHL channel, which has a ≈4-nm-long, ≈1- to 2-nm-wide transmembrane region directly adjacent to its ≈1-nm-diameter constriction. The MspA geometry may provide improved spatial resolution because it is plausible that the characteristics of the ionic current blockades are determined primarily by the polynucleotide segment threaded through the narrow region of the pore (10). In addition to its promising geometry, MspA is very robust and retains channel-forming activity after exposure to any pH from 0 to 14 and after extraction at 100 °C for 30 min and further incubation at 80 °C in the presence of 2% SDS for 15 min (28). MspA is also ideal for engineering via site-directed mutagenesis, retaining high levels of expression and channel-forming activity for a wide variety of amino acid replacements (29). These characteristics suggest that MspA is an excellent nanopore for analytical applications. We constructed two MspA mutants and demonstrated their capabilities for single-molecule detection of ssDNA.
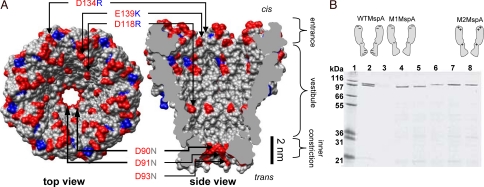
Fig. 1.
MspA structure and expression of MspA mutants. (A) Structure and charge distribution of wild-type MspA. Aspartate and glutamate residues are colored red, and argenine and lysine residues are colored blue. At our experimental pH 8, we expect the acidic (red) residues to be predominantly negatively charged and the basic (blue) residues to be positively charged. Locations and identities of mutations are indicated by arrows and labels. (B) Expression of MspA mutant porins. Raw extract (13 μL) was added to each lane. Gel was stained with Coomassie blue. Lane 1, protein mass marker. Lane 2, WTMspA. Lane 3, no MspA. Lane 4, mutant D90N/D91N/D93N (M1MspA). Lane 5, mutant D90N/D91N/D93N/D118R. Lane 6, mutant D90N/D91N/D93N/D118R/E139R. Lane 7, mutant D90N/D91N/D93N/D118R/E139K. Lane 8, mutant D90N/D91N/D93N/D118R/E139K/D134R (M2MspA). Mutants in lanes 5–7 were constructed, extracted, and assayed to ensure that expression and channel-forming activity were retained for each successive amino acid replacement. Diagrams above the gel show schematically the approximate location and polarity of the amino acids that we mutated in this investigation.
Results and Discussion
We first investigated wild-type MspA (WTMspA). Consistent with previous results (30), the purified protein demonstrated high channel-forming activity with a most frequent single-channel conductance of 4.9 nS in 1.0 M KCl at ≈20 °C (Fig. S1). In all our experiments the pores were oriented such that the “entrance” (Fig. 1A) was exposed to the cis compartment of our apparatus. The cis compartment was held at ground and positive voltage was applied to the trans compartment (Fig. S2a). Above ≈60 mV, WTMspA demonstrated frequent, spontaneous blockades of the ionic current in the absence of ssDNA (Fig. S2). Some spontaneous blockades were transient, and others required reversal of the voltage to reestablish the unblocked current level. Despite this behavior, there remained intervals of steady, unobstructed signal lasting tens of seconds for voltages up to ≈100 mV (Fig. S2). The addition of ≈2–8 μM dC50 ssDNA to the cis compartment did not lead to a noticeable enhancement or alteration of these blockade characteristics. Above ≈100 mV the spontaneous blockades were so frequent that ssDNA detection experiments were impractical.
One explanation for the apparent absence of ssDNA interactions with WTMspA is the high density of negative charge in the pore (Fig. 1A). Electrostatic interaction with the negatively-charged channel interior likely inhibits the entry of DNA into the pore. To address this issue we replaced the aspartate residues in the constriction zone with asparagines (Fig. 1A). The resulting MspA mutant D90N/D91N/D93N (M1MspA) was expressed and purified from the M. smegmatis strain ML16 that lacks most endogenous porins (31). The expression levels of M1MspA (Fig. 1B) and its channel-forming activity were similar to WTMspA, whereas the conductance was reduced by a factor of 2–3 (Fig. S1). Further, the frequency of spontaneous blockades was dramatically reduced in M1MspA, making it possible to conduct DNA detection experiments at voltages up to and above 180 mV (Fig. 2).
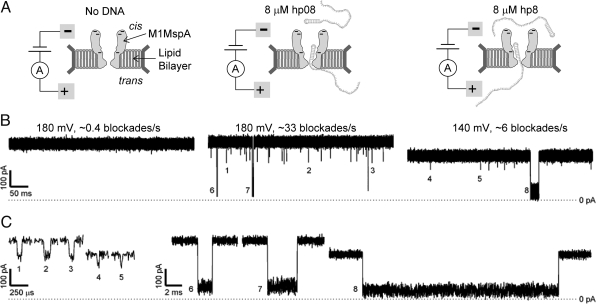
Fig. 2.
Detection of ssDNA hairpin constructs with M1MspA. (A) Schematic diagram of experiments. (B) Representative ionic current signal observed for M1MspA in the absence of DNA and the presence of 8 μM hp08 hairpin DNA at 180 and 140 mV. (C) Numbered blockades from traces in B shown at expanded time scales.
We used ssDNA hairpin constructs to investigate the interaction of DNA with M1MspA. Each construct had a 50-nt poly-dA overhang on the 3′ end, a dsDNA duplex region of variable length (8, 10, and 12 bp for constructs hp08, hp10, and hp12, respectively), and a 6-nt loop (Fig. 3). At 180 mV, the addition of ≈8 μM hp08 ssDNA to the cis compartment caused the rate of transient ionic current blockades to increase from 0.1–0.6 blockades per second to 20–50 blockades per second (Fig. 2). Blockade rates were proportional to DNA concentration and were strongly voltage-dependent, decreasing ≈3-fold for a 20-mV decrease in the applied voltage. Blockades long enough to be well-resolved were either partial blockades where the ionic current was reduced to between 80% and 50% of the unblocked level or deep blockades where the ionic current was reduced to less than 50% of the unblocked level (Fig. 2C). Blockades exhibiting both partial and deep subsegments were very rare. Partial blockades lasted tens to hundreds of microseconds and their dwell times increased with increasing voltage (Figs. 2C and S3). Deep blockades lasted hundreds of microseconds to hundreds of milliseconds and their dwell times decreased with increasing voltage (Fig. 3 and Fig. S3). We observed these trends in experiments with all three hairpins.
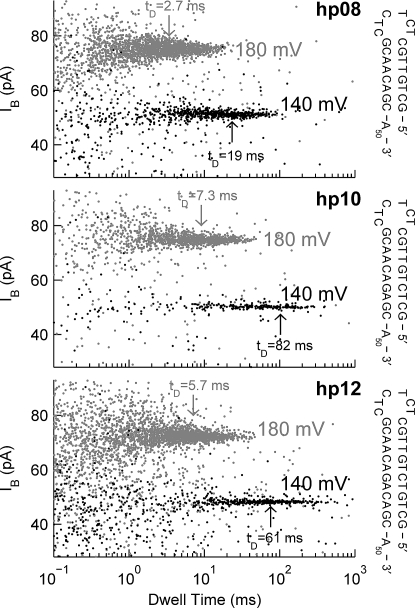
Fig. 3.
Characteristics of deep blockades from hairpin constructs in M1MspA. The coordinates of each point give the duration and average current of 1 deep blockade. Black and gray data were acquired at 140 and 180 mV, respectively. The mode of the log10 of the deep blockade dwell times, tD, is indicated for each dataset. Diagrams at right show the sequence of each hairpin construct.
In analogy to similar signals observed with αHL (16), we interpret the partial blockades as DNA entry into the M1MspA vestibule without threading of the single-stranded segment through the channel constriction. For this mechanism one expects only a moderate reduction of the ionic current, and the increase in dwell time with voltage (Fig. S3) most likely results from an increasing electrostatic barrier against escape of a DNA molecule from the vestibule back into the cis compartment (16). This explanation for the dwell time increase can be understood within a kinetic framework where decay of the polymer from the vestibule occurs via the two first-order processes of escape against the applied voltage gradient and threading of one end through the constriction. The lifetime is then the inverse of the sum of the rate constants for these processes. This lifetime will increase with voltage if (i) the escape rate constant decreases with voltage and (ii) its decrease dominates any changes in the threading rate constant.
For the deep blockades, the clear decrease in dwell times with increasing voltage is inconsistent with any process involving escape of the DNA back into the cis compartment. Both the degree of ionic current reduction and the voltage dependence of the dwell times are consistent with a process where the single-stranded polydA segment is driven through the ≈1-nm-diameter constriction until the ≈2.2-nm-diameter DNA duplex reaches the constriction and arrests translocation (Fig. 2A). The hairpin construct remains in this threaded configuration until either unzipping of the DNA duplex (9, 11, 12) or a conformational rearrangement of the M1MspA constriction zone allows translocation to be completed. We favor the unzipping mechanism of translocation completion because passage of a dsDNA helix would require the constriction to approximately double in diameter, disrupting the hydrogen bonds of the β-barrel flanking the constriction (27) and potentially exposing the hydrophobic regions of the protein and bilayer interior to water.
The hairpin deep blockades in M1MspA had very broad dwell time distributions that were not well described by simple exponentials or sums of exponentials (Fig. S4). We used the mode of the logarithm of the deep blockade dwell times, tD, corresponding in Fig. 3 to the dwell time with the highest density of blockades, to parameterize the distributions (Fig. S4). For all voltages, hp08 had the shortest tD. Below 160 mV, hp10 and hp12 had similar tD. However, above 160 mV hp10 had consistently longer tD than hp12. These observations are somewhat different than those from αHL, where hairpin blockade dwell time distributions were modeled with single exponentials and hairpins with larger standard free energies of formation consistently produced longer deep blockades (9, 12). Assuming the deep blockades are produced by translocation with duplex dissociation as the rate-limiting step, then this process is 10–100 times slower in M1MspA than in αHL (12). We were surprised by the observation that hp10 blockades persisted longer than hp12 blockades. We require further investigation before speculating on the origin of this observation and the differences between the hairpin data from MspA and αHL. All of the data displayed in Figs. 2 and 3 and Fig. S3 and S4, and all of the numbers given in this article, were derived from data taken on the same long-lived M1MspA pore. In six repeated experiments with hp10 at 180 mV, we observed an average unblocked current level of 340 ± 7 pA and an average tD of 9 ± 1 ms (mean ± SEM).
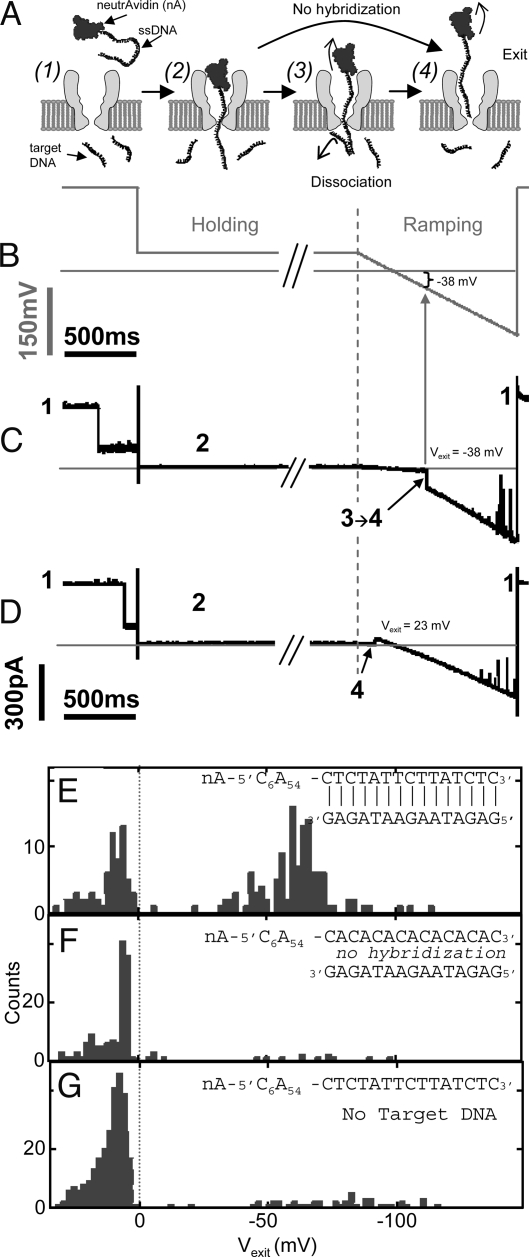
To obtain direct proof that DNA translocates through MspA, we used the transbilayer detection technique illustrated in Fig. 4 and pioneered by Nakane et al. (14). A ssDNA probe molecule with a bulky anchor complex at one end is electrophoretically driven into the nanopore. The free ssDNA end threads through the pore into the trans compartment until the anchor halts translocation. If the trans compartment contains short ssDNA target molecules that are complementary to the end of the ssDNA probe, then the probe and target can hybridize. If hybridization occurs, the probe is locked in a threaded configuration until the application of a sufficiently negative voltage causes the probe to dissociate from the target and exit into the cis compartment. If hybridization does not occur for stochastic reasons or because the probe end is not complementary to the target, or if there are no target molecules in the trans compartment, then a negative voltage is not needed for the probe to exit back into the cis compartment. The appearance of blockades that are only cleared by sufficiently negative voltage is evidence that the ssDNA probe has threaded through the nanopore to the trans compartment and hybridized to the target DNA.
Fig. 4.
Transbilayer probe experiments. (A) Animation of molecular configurations. (1) An unblocked pore. (2) A threaded ssDNA with nA arresting translocation of the nA–ssDNA complex. (3) Target DNA hybridized with nA–ssDNA disassociating at negative voltage. (4) The nA–ssDNA complex exiting from the pore at a voltage depending on the hybridization of the target DNA. (B) Time series of the applied voltage. A current blockade triggers a change from the 180-mV capture voltage to a holding voltage of 40 mV after delay of ≈200 ms. The holding voltage is maintained for 5 s to allow hybridization, and is then ramped negatively. (C and D) Current time series demonstrating nA–ssDNA exit at negative and positive voltages, respectively. Large current spikes occur because of instantaneous voltage changes and spontaneous pore closure at large negative voltage. (E–G) Exit voltage (Vexit) histograms. (E) Experiment where the probe is complementary to the target ssDNA molecules. (F) The same pore as in E, but with a probe that is not complementary to the target DNA. (G) A separate control using the same probe as in E, but without target DNA present in the trans compartment. A significant number of negative Vexit events are observed only in E, where the probe is complementary to the target. The infrequent occurance of negative Vexit events in F and G rule out the possibility that a majority of negative Vexit in E is caused by nonspecific probe–target association or by binding of the probe to the pore.
We used probe molecules comprised of 75-nt-long ssDNA molecules that were attached to a neutravidin (nA) anchor on their biotinylated 5′ end and had a heterogeneous 15-nt-long complementary sequence on their 3′ end. We observed indefinite deep current blockades when these probes were driven into the pore from the cis compartment with 180 mV. For the transbilayer experiments, we captured probe molecules with 180 mV. After a brief delay to ensure that the ssDNA was threaded as far as possible through the M1MspA pore, the voltage was reduced to 40 mV and held at that level for 5 s to allow one of the 15-nt-long target ssDNA to anneal to the probe's complementary end. The voltage was then ramped down at a rate of 130 mV/s. For each event we identified the probe exit voltage, Vexit, as the voltage at which we observed a large and abrupt increase in the conductance while ramping (Fig. 4 C and D).
Histograms of Vexit from experiments with three different probe/target combinations are shown in Fig. 4. When the probe DNA is complementary to the target DNA (Fig. 4E) a significant number of Vexit are negative, indicating probe/target hybridization. In six repeated experiments with complementary probe/target molecules we observed similar populations of negative Vexit. In five repeated experiments where the ssDNA 3′ end was not complementary to the target molecules (Fig. 4F) and in one experiment without target DNA (Fig. 4G), negative Vexit values were rarely observed. On two different pores both complementary and noncomplementary probe/target combinations were used. The data of one of those pores is shown in Fig. 5 E and F. These data provide clear and direct evidence that ssDNA can thread through M1MspA, confirming our hypothesis that the deep blockades observed in Fig. 2 are indeed caused by translocation of ssDNA through the M1MspA nanopore.
Fig. 5.
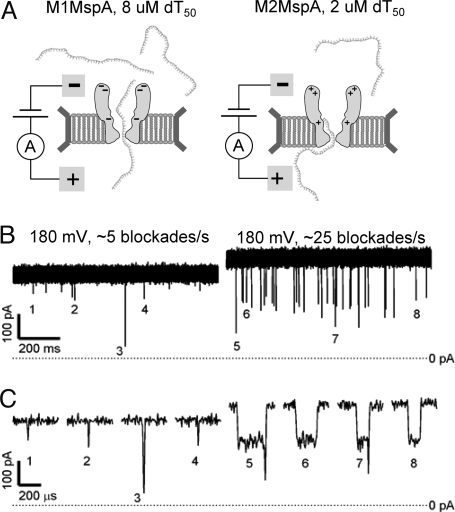
Comparison of dT50 homopolymer blockades for M1MspA and M2MspA. (A) Schematic diagram of experiments. (B) Representative ionic current signals observed for M1MspA with 8 μM dT50 (Left) and M2MspA with 2 μM dT50 (Right). (C) Numbered blockades from traces in B shown at expanded time scales.
We also investigated the interaction between M1MspA and linear, homogeneous ssDNA 50-mers. At 180 mV, the addition of ≈8 μM dT50 into the cis compartment caused ≈5 blockades per second (Fig. 5), a factor of ≈20 increase over the blockade rate in the absence of dT50. Most of these blockades were shorter than 30 μs, which is too brief to resolve internal structure or estimate the depth of the blockade. Experiments with dA50 and dC50 gave similar results. The short duration of the observed blockades suggests that translocation of these linear, homogeneous ssDNA 50-mers is typically shorter than 30 μs. The blockades are also consistent with brief excursions of the polymers into the vestibule that end with escape back into the cis compartment. Although both translocation and escape likely occur in our experiments with linear ssDNA 50-mers, we were unable to estimate the relative frequency of the two processes. The example homopolymer data presented here were obtained with a different long-lived M1MspA pore than the hairpin data described above, but there is quantitative agreement between extensive hairpin datasets taken on the two pores.
To further examine the effect of charges in MspA on its DNA analysis capabilities, we made three additional mutations to M1MspA and replaced negatively-charged residues in the vestibule and around the entrance with positively charged residues (Fig. 1A). The resulting mutant D90N/D91N/D93N/D118R/D134R/E139K (M2MspA) demonstrated expression levels (Fig. 1B) and channel-forming activity similar to WTMspA (Fig. S1). Like M1MspA, M2MspA had smaller conductance than WTMspA (Fig. S1) and exhibited minimal spontaneous blockades for voltages up to and above 180 mV (data not shown). At 180 mV, the addition of 2 μM dT50 to the cis compartment led to blockade rates of ≈25 blockades per second (Fig. 5B). A ≈100-μs partial blockade ending with a clear downward spike was a common blockade pattern (Fig. 5C). The partial blockade durations and their tendency to end with a downward spike both increased with voltage (Fig. S5). These trends are consistent with a process where a polymer enters the vestibule and is held there, producing a partial blockade until one end enters the high-field constriction and initiates translocation. This mechanism has accurately explained a similar partial-to-deep blockade pattern observed with αHL (16). The short duration of the downward spikes suggests that translocation of linear ssDNA 50-mers through M2MspA is shorter than ≈30 μs. We interpret partial blockades that do not end with downward spikes as either escape back into the cis compartment or as translocation that is shorter than ≈10 μs, which is too brief to be observed in our experiments.
An important similarity between M1MspA and M2MspA is that translocation of linear ssDNA 50-mers appears to be too fast to produce deep blockades with resolvable structure. This observation suggests that the constriction, which is the same for both mutants, is the region that primarily determines the speed of a linear ssDNA molecule translocating through MspA. Comparing these ≈2–10 base/μs MspA translocation speeds to the ≈0.5–1 base/μs translocation speeds observed with αHL (8, 16) supports the notion that the details of the channel geometry and composition play a leading role in determining translocation speed. In the case of MspA and αHL, the large difference in translocation speed could result from the width of the channel regions flanking the constrictions. If interaction between DNA and the channel walls slows DNA passage (32), then slower translocation would be expected in αHL where the 10–20 bases that are highly confined in the constriction and transmembrane region are forced to interact with the channel walls. In MspA, only the 2–4 bases in the constriction are forced to be in contact with the protein. The charge distribution within the constriction is another significant difference between αHL and the MspA mutants we have investigated. The αHL constriction is formed by the side chains of E111, K147, and M113 (33), forcing the negatively charged ssDNA backbone into extremely close proximity with seven positively-charged and seven negatively-charged residues. The lack of charged residues in the constriction of our MspA mutants could also be responsible for the faster translocation speeds compared with αHL. Future comparative experiments elucidating the origin of these differences will give much needed insight into the factors determining translocation speed.
Further comparison of the homopolymer blockade characteristics between the two MspA mutants gives insight into how the arrangement of charged residues in the channel influences its interactions with DNA. Blockade rates for M2MspA were ≈20 times higher than M1MspA rates for a given ssDNA concentration (Fig. 5B). We also found that M2MspA demonstrated easily observable blockades down to ≈80 mV, whereas almost no blockades were visible for M1MspA below ≈140 mV (data not shown). Finally, partial blockades for M2MspA were at least ≈100 times longer than for M1MspA (Fig. 4C). These trends are consistent with a simple electrostatic model wherein the positively-charged residues in M2MspA both facilitate ssDNA entry into the vestibule and inhibit the escape of ssDNA molecules from the vestibule back into the cis compartment. These observations demonstrate that the appropriate placement of charged residues offers a simple means to substantially tailor the interaction between MspA and DNA.
Conclusions
Our results introduce MspA as a nanopore for nucleic acid analysis. The geometry, stability, and engineerability of this pore make it a very promising candidate for single-molecule analytical applications. Our demonstrations that ssDNA can be electrophoretically driven through MspA mutants and that MspA can be rationally tailored based on its crystal structure represent important steps toward the realization of the potential inherent in the unique geometry of MspA. We anticipate that future mutagenic studies exploring and optimizing the factors governing the interaction between MspA and ssDNA will be of widespread benefit to nanopore DNA sequencing and other nanosensor applications and biophysical investigations.
Materials and Methods
Bacterial Strains and Growth Conditions.
All bacterial strains used in this study are listed in Table S1. Mycobacteria were grown at 37 °C in Middlebrook 7H9 liquid medium (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80 or on Middlebrook 7H10 agar (Difco) supplemented with 0.2% glycerol. Escherichia coli DH5α was used for all cloning experiments and was routinely grown in Luria–Bertani (LB) medium at 37 °C. Hygromycin was used at concentrations of 200 μg/mL for E. coli and 50 μg/mL for M. smegmatis.
Site-Directed Mutagenesis of mspA.
The M1MspA and M2MspA mutants were constructed in a stepwise fashion by site-directed mutagenesis using the combined chain reaction (CCR) (34). The plasmid pMN016 carries a psmyc-mspA transcriptional fusion (31) and was used as a template. The oligonucleotides psmyc1 and pMS-seq1 as forward and reverse primers, respectively, and an appropriate mutagenesis primer (Table S1) were used in CCR. Three subsequent mutations were introduced into mspA to construct the m1mspA gene. Three further mutations were introduced into m1mspA to yield m2mspA (Table S1). All plasmids were verified by sequencing the entire mspA gene before they were transformed into the triple porin mutant M. smegmatis ML16 (31) for protein production.
Purification of MspA.
MspA porins were selectively extracted from M. smegmatis and purified by subsequent anion exchange and gel filtration chromatography as described (35, 36).
Single-Channel Experiments.
Bilayers were made with diphytanoyl-PA and diphytanoyl-PC lipids prepared in equal proportion and were formed across a horizontal, ≈20-μm-diameter aperture in Teflon as described (7). MspA was added to one side of the bilayer (cis side) at a concentration of ≈2.5 ng/mL. The cis side was grounded, and positive voltage was applied to the trans side of the bilayer. We used an Axopatch-1B patch–clamp amplifier (Axon Instruments) to apply voltage across the bilayer and measure the ionic current flowing through the pore. The analog signal was low-pass-filtered at 50 kHz with a 4-pole Bessel filter. The amplified, filtered signal was digitized at 250 kHz. Data acquisition was controlled with custom software written in LabWindows/CVI (National Instruments). All experiments were performed at 21 ± 2 °C in 1 M KCl, 10 mM Hepes/KOH buffered at pH 8. Homogeneous ssDNA oligonucleotides dA50, dC50, and dT50 and hairpin constructs hp08 (5′-GCTGTTGC TCTCTC GCAACAGC A50-3′), hp10 (5′-GCTCTGTTGC TCTCTC GCAACAGAGC A50-3′), and hp12 (5′-GCTGTCTGTTGC TCTCTC GCAACAGACAGC A50-3′) were synthesized by Integrated DNA Technologies (IDT).
Trans-Bilayer Probe Experiments.
nA was obtained from Invitrogen. Two different 5′-biotynlated ssDNA constructs, 5′-bt-dC6dA54 d(CTCTATTCTTATCTC)-3′ and 5′-bt-dC6dA54 d(CACACACACACACAC)-3′, were synthesized by IDT. nA and the ssDNA constructs were mixed at a concentration of 50 μM in a 1:1 ratio in our experimental 1 M KCl buffer and stored in at −20 °C until immediately before use. The 15-nt-long target DNA, 3′-GAGATAAGAATAGAG-5′ was synthesized by IDT, suspended in our experimental buffer, and stored at −20 °C until immediately before use. The trans compartment was preloaded with ≈100 μM target DNA and the cis compartment was filled with DNA-free buffer. After a bilayer was formed, the cis compartment was perfused to remove any target DNA that diffused through the aperture. Once a stable M1MspA pore was established, the nA–ssDNA complexes were added to the trans compartment to a final concentration of ≈1 μM. Custom experimental control software written in LabWindows was used to continuously monitor the current and apply the appropriate voltages.
Data Analysis.
Data analysis was implemented with custom software written in Matlab (The MathWorks). Blockades were identified as intervals where the ionic current dropped below a threshold of 80% of the unblocked current level, remained there for at least 12 μs, and returned spontaneously to the unblocked level. Blockades were rejected if the unblocked signal directly preceding or following it had an average current that deviated from the typical unblocked level by more than twice the rms noise of the unblocked signal. Blockades were also rejected if they occurred within 26 μs of another blockade. Deep blockades were identified as intervals where the ionic current dropped <50% of the unblocked level. Intervals where the current remained between 80% and 50% of the unblocked level were identified as partial blockades. We parameterized each event by the dwell times and average currents of its constituent partial and deep subintervals. The tD values used to parameterize the hairpin deep blockade dwell time distributions were estimated as the peak of the probability density distribution of the log10 of the dwell times (Fig. S4). This distribution was estimated with the Matlab Kernel smoothing density estimator using a normal kernel function and a width of 0.15. Transbilayer data were analyzed by detecting abrupt changes in the conductance from <1 to >1 nS. The voltage at which these changes occurred was recorded and then summarized in the histograms shown in Fig. 4 E–G.
Supplementary Material
Acknowledgments.
We thank Dr. Bertil Hille for pointing out MspA and for the loan of the patch–clamp amplifiers; Dr. Mark Troll for helping to initiate this research; Risa Wong and Kyle Langford for help in gathering the data; and Marcus Collins for help with data interpretation; Matt Wiggin and Nahid Jetha from the University of British Columbia for technical advice for the transbilayer experiments. This work was supported by the National Institutes of Health, National Human Genome Research Institute, $1000 Genome Program Grant 5R21HG004145.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807514106/DCSupplemental.
References
- 1.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bezrukov SM, Vodyanoy I, Brutyan RA, Kasianowicz JJ. Dynamics and free energy of polymers partitioning into a nanoscale pore. Macromolecules. 1996;29:8517–8522. [Google Scholar]
- 3.Braha O, et al. Designed protein pores as components for biosensors. Chem Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 4.Li J, et al. Ion-beam sculpting at nanometer-length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 5.Heng JB, et al. Sizing DNA using a nanometer-diameter pore. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71 doi: 10.1103/PhysRevE.71.051903. 051903. [DOI] [PubMed] [Google Scholar]
- 7.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 10.Howorka S, Bayley H. Probing distance and electrical potential within a protein pore with tethered DNA. Biophys J. 2002;83:3202–3210. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys Rev Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 12.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Dunning JE, Huang AP, Nyamwanda JA, Branton D. DNA heterogeneity and phosphorylation unveiled by single-molecule electrophoresis. Proc Natl Acad Sci USA. 2004;101:13472–13477. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the α-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler TZ, Gundlach JH, Troll M. Ionic current blockades from DNA and RNA molecules in the α-hemolysin nanopore. Biophys J. 2007;93:3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornblower B, et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nat Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 18.Astier Y, Kainov DE, Bayley H, Tuma R, Howorka S. Stochastic detection of motor protein–RNA complexes by single-channel current recording. Chemphyschem. 2007;8:2189–2194. doi: 10.1002/cphc.200700179. [DOI] [PubMed] [Google Scholar]
- 19.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Khalid S, Sansom MS, Bayley H. Outer membrane protein G: Engineering a quiet pore for biosensing. Proc Natl Acad Sci USA. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores. Angew Chem Int Ed Engl. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Astier Y, Braha O, Bayley H. Toward single-molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 23.Keyser UF, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–477. [Google Scholar]
- 24.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederweis M. Mycobacterial porins: New channel proteins in unique outer membranes. Mol Microbiol. 2003;49:1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x. [DOI] [PubMed] [Google Scholar]
- 26.Stahl C, et al. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol Microbiol. 2001;40:451–464. doi: 10.1046/j.1365-2958.2001.02394.x. [DOI] [PubMed] [Google Scholar]
- 27.Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 28.Heinz C, Engelhardt H, Niederweis M. The core of the tetrameric mycobacterial porin MspA is an extremely stable β-sheet domain. J Biol Chem. 2003;278:8678–8685. doi: 10.1074/jbc.M212280200. [DOI] [PubMed] [Google Scholar]
- 29.Mahfoud M, Sukumaran S, Hulsmann P, Grieger K, Niederweis M. Topology of the porin MspA in the outer membrane of Mycobacterium smegmatis. J Biol Chem. 2006;281:5908–5915. doi: 10.1074/jbc.M511642200. [DOI] [PubMed] [Google Scholar]
- 30.Niederweis M, et al. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol Microbiol. 1999;33:933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 31.Stephan J, et al. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol Microbiol. 2005;58:714–730. doi: 10.1111/j.1365-2958.2005.04878.x. [DOI] [PubMed] [Google Scholar]
- 32.Slonkina E, Kolomeisky AB. Polymer translocation through a long nanopore. J Chem Phys. 2003;118:7112–7118. [Google Scholar]
- 33.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 34.Bi W, Stambrook PJ. CCR: A rapid and simple approach for mutation detection. Nucleic Acids Res. 1997;25:2949–2951. doi: 10.1093/nar/25.14.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz C, Niederweis M. Selective extraction and purification of a mycobacterial outer membrane protein. Anal Biochem. 2000;285:113–120. doi: 10.1006/abio.2000.4728. [DOI] [PubMed] [Google Scholar]
- 36.Heinz C, Roth E, Niederweis M. Purification of porins from Mycobacterium smegmatis. Methods Mol Biol. 2003;228:139–150. doi: 10.1385/1-59259-400-X:139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.