Abstract
Protein evolution is crucial for organismal adaptation and fitness. This process takes place by shaping a given 3-dimensional fold for its particular biochemical function within the metabolic requirements and constraints of the environment. The complex interplay between sequence, structure, functionality, and stability that gives rise to a particular phenotype has limited the identification of traits acquired through evolution. This is further complicated by the fact that mutations are pleiotropic, and interactions between mutations are not always understood. Antibiotic resistance mediated by β-lactamases represents an evolutionary paradigm in which organismal fitness depends on the catalytic efficiency of a single enzyme. Based on this, we have dissected the structural and mechanistic features acquired by an optimized metallo-β-lactamase (MβL) obtained by directed evolution. We show that antibiotic resistance mediated by this enzyme is driven by 2 mutations with sign epistasis. One mutation stabilizes a catalytically relevant intermediate by fine tuning the position of 1 metal ion; whereas the other acts by augmenting the protein flexibility. We found that enzyme evolution (and the associated antibiotic resistance) occurred at the expense of the protein stability, revealing that MβLs have not exhausted their stability threshold. Our results demonstrate that flexibility is an essential trait that can be acquired during evolution on stable protein scaffolds. Directed evolution aided by a thorough characterization of the selected proteins can be successfully used to predict future evolutionary events and design inhibitors with an evolutionary perspective.
Keywords: antibiotic resistance, enzyme, fitness landscape, metalloproteins, epistasis
Protein evolution is crucial for organismal adaptation and fitness (1–8). This process takes place by shaping a given 3-dimensional fold for its particular biochemical function within the metabolic requirements and constraints of the environment. The evolution of a protein relies on a subtle interplay between sequence, structure, functionality, and stability that is manifested in a particular phenotype (1, 2). The dissection of this interplay is not trivial, and it is further complicated by 2 facts. The first complication is that mutations are pleiotropic, and classical approaches to this issue fail in providing a global picture. Thus, although functional studies are focused on the effect of mutations on a phenotype overriding the biochemical rationale, structural or biochemical studies usually disregard the organismic impact of mutations. The second difficulty is to understand the interactions between mutations, particularly when they are distant in the 3-dimensional structure of the protein. Only recent efforts have been conveyed to address the interaction of distant mutations at the biochemical level, i.e., their impact on the in vitro activity of enzymes (6, 9). Extensions of these studies to predict the in vivo effect of these interactions on organismal fitness are scarce.
Adaptive molecular changes have been the subject of many previous studies, mostly relying on statistical descriptions at the sequence level. Only recent efforts have aimed at identifying changes in sequence and structure that affect fitness within a defined protein fold (4, 7, 8, 10, 11). The identification of such structural traits acquired through evolution allows both the reconstruction of ancestral genes (7, 8, 12) and the prediction of future evolutionary events (5, 13). The latter approach could be immensely valuable for anticipating the molecular features that might enhance bacterial resistance to antibiotics, thereby informing strategies to combat this clinical threat.
The principal mechanism of bacterial resistance to β-lactam antibiotics is the expression of β-lactamases, a family of enzymes whose evolution is continuously challenged by the indiscriminate use of antibiotics (5, 13, 14). β-Lactam resistance mediated by β-lactamases provides a unique example in which organismal survival depends on the hydrolytic activity of a protein (14). Accordingly, recent studies in the serine-β-lactamase (SβL) TEM-1 and its variants have allowed significant advances in the understanding of protein evolution. Based on the analysis of different possible evolutionary trajectories, Weinreich and coworkers have posited that only a few of them can be successful, thus implying that directed evolution can be exploited to anticipate natural evolutionary scenarios (4). Related experiments on the same enzyme have evinced a trade-off between activity and stability in the evolution of antibiotic resistance mediated by TEM variants (5, 15, 16).
Antibiotic resistance can also be due to the expression of metallo-β-lactamases (MβLs) (17). Despite catalyzing the same reaction, serine and metal-dependent enzymes are not evolutionarily related, and display different catalytic mechanisms. Metallo-β-lactamases are zinc enzymes able to hydrolyze almost all β-lactam antibiotics (except B2 MβLs, which are exclusive carbapenemases) (17). Because there are no clinically useful inhibitors available for them, MβLs challenge the available therapeutics for bacterial infections (18). From the evolutionary point of view, several authors have considered them as young, still incompletely evolved enzymes, based on their lower catalytic efficiencies compared with SβLs (14, 19, 20). However, the broader substrate portfolio exhibited by MβLs may have been achieved at the expense of lower enzymatic activities.
Directed evolution on the Bacillus cereus MβL, BcII, by DNA shuffling and selection by cephalexin resistance (a poor substrate of BcII) elicited an evolved variant, M5, displaying an enhanced activity toward cephalexin and others cephalosporins (13). Surprisingly, this enhancement did not take place at the expense of impaired hydrolytic activity toward other substrates, thus generating an enzyme with an even broader substrate spectrum, supporting the suggestion that MβLs can further evolve. Here, we report the structural, biochemical, biophysical, and in vivo functional study of this in vitro evolved metallo-β-lactamase. We conclude that MβLs have a large evolutionary potential that may be still manifested in the clinical environment. From the point of view of protein evolution, we have found that: (i) MβLs can evolve by further expanding their substrate spectrum, i.e., their functional promiscuity; (ii) a mutation enhancing the activity has acted in the rate-determining step of catalysis by stabilizing a key intermediate, suggesting that their active-site chemistry can still evolve; (iii) a mutation gained later in the evolution further enhances the catalytic efficiency by augmenting the flexibility; (iv) both mutations provide not only a more efficient enzyme, but also a higher in vivo fitness at the expense of the enzyme stability. We can conclude that flexibility is an essential trait in evolution provided the stability threshold of the protein has not been exhausted. In addition, these results suggest inhibitory strategies to anticipate further resistance.
Results
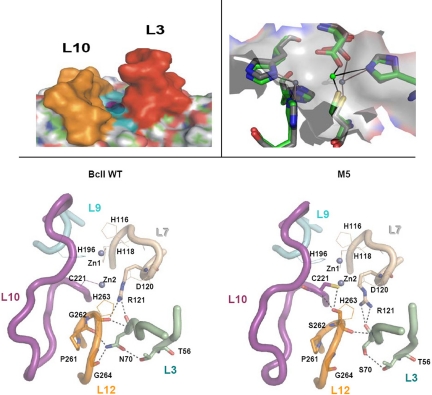
The evolved variant of the metallo-β-lactamase BcII from B. cereus, named M5, contains 4 mutations (N70S, V112A, L250S, and G262S), none of them located in the active site. We solved the X-ray structure of M5 [supporting information (SI) Table S1], which showed that none of these mutations entails changes in the global fold (0.6 Å rmsd on all nonhydrogen atoms vs. wt BcII). The active site of BcII can be described as a shallow groove flanked by loops L3 and L10, hosting 2 Zn(II) ions located in its floor, which is defined by loops L7, L9, and L12 (20, 21) (Fig. 1). Zn1 is bound to 3 His residues (His-116, His-118, and His-196), whereas Zn2 is coordinated to Asp-120, Cys-221, and His-263. Although all mutations are remote, the most significant changes are found in the enzyme active site: Zn2 has moved closer to Zn1 by 0.5–1.0 Å in the mutant, while preserving the same ligand set (Fig. 1). The 2 closest mutations to the active site are N70S and G262S, both located below the active-site floor. The hydroxyl oxygen of Ser-262 is 3.0 Å from the Cys-221 sulfur, and 3.2 Å from the Cys-221 backbone nitrogen atom (Fig. 1). Ser-262 behaves as a second shell ligand of Zn2 and at the same time connects loops L10 and L12. Ser-70 is located in a β-strand 2 residues upstream from Pro-68, which defines the N terminus of a conserved mobile loop (L3). The absence of the Asn-70 side chain results in the removal of 2 H bonds with Pro-261 and Gly-264, which flank the Zn2 ligand His-263 and the mutated residue 262 (located in loop L12; Fig. 1). These are the only H bonds connecting loops L3 and L12. Another notable change in the interloop connections is that Arg-121 (from loop L7) adopts a different conformation in M5, resulting in the loss of a H bond of its guanidinium group with the backbone oxygen of residue 262 (loop L12).
Fig. 1.
Active sites of WT BcII and mutant M5. (Upper Left) Side view of a representation of the active site of BcII, in which the protruding loops L3 and L10 are highlighted in orange and yellow, respectively. (Upper Right) Comparison of the active sites of WT BcII and mutant M5. The Zn2 ion in M5 (in green) is more solvent exposed than the Zn2 ion in WT BcII (in gray). The molecular surface of WT BcII is depicted in transparent gray. (Lower) Upper views of the active sites of WT BcII (Left) and M5 (Right) depicting the metal ions, metal ligands, and loops. The long loops flanking the active site (L3 and L10) as well as the loops containing the metal ligands (L7, L9, and L12) are depicted, together with relevant residues. Loops are indicated in diagram tubes. Zn(II) ions are shown as blue spheres. Metal ligands are shown in lines, residues involved in H bonds are shown in sticks, and H bonds are indicated by black dashed lines. The figures were rendered by using PyMol (http://pymol.sourceforge.net/).
To examine the influence of each of these mutations on the active-site structure, mutants N70S, G262S, and N70S/G262S were produced, and the metal site probed by substitution of the native Zn(II) ion by Co(II) (22). The electronic spectra of the Co(II) derivatives (Fig. S1) show that the ligand field bands between 450 and 660 nm corresponding to the Zn1 site are identical in all variants, revealing that this metal site is unaltered by either of these mutations (22). However, a Cys-221-Co(II) charge transfer band at 343 nm (a reporter of the Zn2 site) is perturbed only when Ser-262 is present, pointing to a key role of the Ser-262-Cys-221 H bond. These experiments reveal that: (i) G262S is the only mutation that has a direct effect on the metal site structure (via the H bond), and (ii) only the Zn2 site is perturbed, whereas the Zn1 site does not show significant changes on this mutation (Fig. 1).
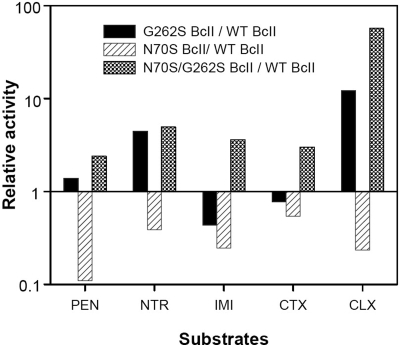
The catalytic performances of the single mutants N70S and G262S, and the double mutant N70S/G262S, were assayed toward 5 different β-lactam substrates including a penicillin, a carbapenem, and 3 cephalosporins (Fig. 2 and Table S2). The G262S mutant shows a selectively enhanced catalytic efficiency toward nitrocefin and cephalexin (the substrate used in the directed evolution experiment) compared with wt BcII, but the catalytic efficiency of the single mutant N70S is compromised for all substrates tested. However, the catalytic efficiency of the double mutant was enhanced for all substrates, with a notable 56-fold increase in activity toward cephalexin. These observations suggest a synergistic effect between these 2 mutations.
Fig. 2.
Catalytic efficiencies of different BcII forms. Relative catalytic efficiencies of G262S BcII, N70S BcII, and N70S/G262S BcII compared with WT BcII against benzylpenicillin (PEN), nitrocefin (NTR), imipenem (IMI), cefotaxime (CTX), and cephalexin (CLX). The relative activity is the ratio of the mutant's kcat/KM value to that of WT. y axis is in log scale.
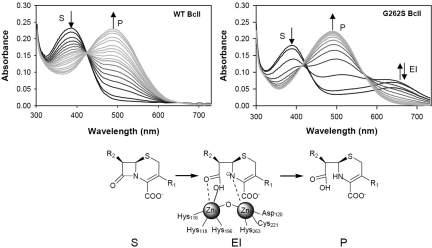
G262S, the only mutation that has a direct impact on the structure of the metal site induces the greatest activity enhancement on cephalexin hydrolysis, a 20-fold increase on kcat (Table S2). This suggests that evolution may have acted by stabilizing the enzyme–transition state complex. This hypothesis was tested by studying the hydrolysis of nitrocefin, a chromophoric cephalosporin that is a useful probe of the catalytic mechanism of MβLs (23). Whereas nitrocefin hydrolysis by wt BcII under single-turnover conditions proceeds through a direct conversion of substrate to product, the same reaction catalyzed by G262S BcII reveals the accumulation of an intermediate, as indicated by the rise of an absorption band with a maximum at 665 nm that decays within 10 ms (Fig. 3 and Fig. S2). This spectral feature is identical to the feature reported for nitrocefin hydrolysis by the MβL CcrA from Bacillus fragilis, and is attributed to an anionic intermediate in which a negatively charged nitrogen is bound to the metal ion at the Zn2 site (Fig. 3) (23). In terms of steady-state parameters, the G262S mutation has increased kcat from 30 to 680 s−1, even larger than that displayed by CcrA (250 s−1). This confirms that the G262S mutation has led to stabilization of a catalytically relevant intermediate, thus acting on the rate-limiting step of the reaction.
Fig. 3.
Stabilization of a reaction intermediate by mutation G262S. Reaction of BcII WT and G262S with nitrocefin, measured by using a photodiode array detector in a stopped-flow system under single-turnover conditions. All spectra are shown until the reaction completion time. The absorbance changes at 390 (S), 490 (P), and 665 (EI) nm are indicated with arrows. Nitrocefin hydrolysis by WT BcII proceeds by direct conversion of S to P (Left). The G262S mutation stabilizes the anionic intermediate EI (Right), depicted in the reaction scheme.
These data provide a rationale for the effect of the G262S mutation, but do not account for the selection of the N70S mutation, nor explain the broader substrate spectrum displayed by the double N70S/G262S mutant. The N70S mutation is deleterious for the activity of wt BcII, but beneficial when acting on G262S BcII (Fig. 2 and Table S2). Mutations improving the catalytic performance of enzymes (like SβLs) usually have a detrimental effect on the protein stability that is restored by compensatory, stabilizing mutations (1, 2, 15). We therefore studied the stability of the different BcII mutants by guanidine hydrochloride-induced protein unfolding. BcII is an extremely stable protein (ΔGu = 18.4 kcal/mol, see Fig. S3), and the G262S substitution has a large impact on its stability (ΔGu = 4.56 kcal/mol). The N70S mutation induces a smaller destabilization on wt BcII (ΔGu = 12.9 kcal/mol), and the double mutant N70S/G262S is even less stable than either of the single mutants (ΔGu = 3.28 kcal/mol). Thus, N70S is not a compensatory mutation for stability.
The crystal structure of M5 shows that the principal effect of removing the Asn-70 side chain is the elimination of the 2 H bonds connecting loops L3 and L12 (Fig. 1). Loop L3 is flexible in the free enzyme, and it was shown to adopt different conformations on substrate and inhibitor binding (17). Therefore, we decided to explore the effect of this mutation on the protein's flexibility by running molecular dynamics (MD) simulations for WT BcII, G262S, and N70S/G262S BcII in the free state and in the Michaelis complexes with cephalexin. These simulations show that the H bonds involving Asn-70 in wt BcII and Ser-262 in M5 are stable both in the free and in the bound forms. During the dynamics runs of the unbound forms, L3 moves closer to L10 both in wt and G262S BcII. Instead, in N70S/G262S BcII, loop L3 tears apart from L10, resulting in a more accessible active-site groove. The MD simulations of the 3 enzymes with cephalexin docked onto the active site provide a different picture. The conformation of loop L10 in the WT BcII–cephalexin complex shows a loose interaction between the carboxylate moiety of the substrate and the conserved Lys-224 (Fig. S4). This interaction is favored in the 2 mutants, accounting for the enhanced activity in both cases, which is driven by the introduction of Ser-262, which connects loops L12 and L10. Furthermore, in the double mutant, while showing a more open active site in the unbound form, loops L3 and L10 move closer to each other when the substrate is bound (Fig. S4). Thus, the N70S mutation bestows a more flexible scaffold by removing the 2 hydrogen bonds that connect L12 and L3. Because the spectroscopic data (Fig. S1) indicate that this mutation does not alter the metal site structure, it is evident that its acts exclusively on the loop structure and dynamics.
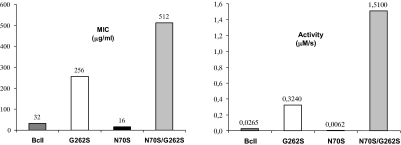
We analyzed the in vivo effect of these mutations by determining the antibiotic resistance in Escherichia coli toward cephalexin and cefotaxime. The expression level of all BcII variants in the periplasm was similar (as revealed by Western blot analysis of the periplasmic extracts), as well as the minimum inhibitory concentration (MIC) toward cefotaxime, used as a reference substrate. Instead, the MIC of cephalexin was enhanced from 32 μg/ml (wt BcII) to 256 μg/ml by the G262S mutation. Mutation N70S was deleterious on a wt background, reducing the MIC by 50%. Instead, the MIC of the double mutant was 512 μg/ml, i.e., when this mutation took place on G262S BcII, it induced high resistance levels. These data closely parallel the in vitro data: the effect of each mutation in organismal fitness is determined exclusively by the catalytic efficiencies (Fig. 4).
Fig. 4.
Organismal fitness parallels catalytic efficiency toward cephalexin. Plots of the minimum inhibitory concentrations (MIC) of cephalexin toward E. coli cells expressing the different mutants (Left) compared with the in vitro catalytic efficiencies of the purified enzymes vs. cephalexin (Right).
Discussion
Here, we have been able to dissect the specific roles of 2 mutations in an evolved metallo-β-lactamase in conferring higher resistance levels to antibiotics. We have solved the crystal structure of the evolved lactamase, and performed a kinetic, biophysical, and mechanistic study of single and double mutants, which allowed us to trace a general picture of molecular evolution of this enzyme directly linked to organismal fitness, measured in terms of bacterial survival.
All of the experimental data herein presented allow us to postulate the following scenario for MβL evolution. The G262S mutation is responsible for the enhanced cephalexinase activity through a 2-fold effect: the H bond between Ser-262 and Cys-221 not only allows loop L10 to adopt a less open conformation on cephalexin binding, but it also leads to a better positioning of the Zn2 ion. Recent rational mutagenesis studies on BcII have shown that shifting the position of the Zn2 site into a slightly more buried location (and thereby also increasing the Zn1-Zn2 distance) results in a severe impairment of the lactamase activity (24, 25). Zn2 has been suggested to play an essential role in substrate binding and orientation (in concert with Lys-224) (24–27), and in favoring C–N bond cleavage through the stabilization of a negative charge on the β-lactam nitrogen (Fig. 3) (23). M5 displays the shortest Zn-Zn distance (3.2–3.4 Å) among all native and mutant enzymes from this family (17), with the Zn2 ion much more exposed than in WT BcII (Fig. 1), thus being better suited to stabilize a negatively charged reaction intermediate. This reveals that MβL activity can evolve still further by improving the rate-limiting step by fine tuning the position of Zn2.
The G262S mutation, while enhancing cephalexin hydrolysis, induces a relative tightening of L10 with respect to L12 that does not favor penicillin, cefotaxime, and imipenem hydrolysis (Fig. 2). Instead, the N70S mutation gives rise to an enzyme with a broader substrate spectrum, by augmenting the loop flexibility, as confirmed by the molecular dynamics analysis. This enhanced loop dynamics is manifested in a broader substrate spectrum of the double mutant compared with G262S BcII, which results in a 5-fold enhancement of the cephalexinase activity (Table S2). A thermodynamic double-mutational cycle analysis (Fig. S5) reveals that the effect of these 2 mutations in this activity is not additive, disclosing a strong synergy (or “coupling”) between them (9, 28) (Table S3).
This in vitro synergy manifested in the catalytic efficiencies correlates with the effect of the mutations in organismal fitness, i.e., antibiotic resistance (Fig. 4). The N70S mutation on wt BcII renders bacteria more sensitive to cephalexin. Instead, when this mutation is incorporated on G262S BcII, it induces a considerable increase on resistance. The finding of opposite effects of a single mutation in different genetic backgrounds is known as sign epistasis, a concept of great relevance for the understanding of protein evolution (1, 2, 4). The existence of sign epistasis reveals that antibiotic resistance mediated by MβLs does not increase monotonically in the mutational landscape. Indeed, sign epistasis limits the number of viable evolutionary pathways (4). This is confirmed by analysis of the clones isolated in the different stages of the evolution experiment: the G262S mutation is present in the first round of selection, whereas N70S is only found in the last rounds of evolution.
Metallo-β-lactamases are present in bacteria that also express a serine-β-lactamase. The serine enzymes display an already optimized catalytic machinery, with catalytic efficiencies often close to the diffusional limit. On evolution, the substrate specificity of the SβL TEM-1 can be altered by mutations that widen the active-site entrance, without affecting the reaction mechanism (5). These mutations compromise the enzyme stability, and are usually accompanied by restoring, stabilizing mutations (2, 5, 15, 16). Thus, the fitness landscape of SβLs is determined by a trade-off between the activity and stability features introduced by the mutations (15). In terms of organismal fitness, this trade-off is manifested in sign epistasis, which limits the accessible evolutionary pathways.
This study reveals a different situation for the evolution of metallo-β-lactamases: (i) the mutation altering the substrate specificity does not shape the active site cavity; instead, G262S has a direct impact on the active-site chemistry by stabilizing a reaction intermediate; (ii) no stabilizing, compensatory mutations were found; (3) rather, N70S is a destabilizing compensatory mutation for G262S. The fact that the resistance toward cefotaxime (used as a reference substrate) was not decreased by any of the mutations, and that expression levels of the mutants in the periplasm were similar, reveal that the stability threshold of MβLs has not been exhausted (3, 29). Thus, the high stability of the MβL fold (30) compared with that of serine enzymes (15, 16) allows the selection of resistance mutations at the expense of the protein stability. This work also favors the hypothesis put forward by Bloom and coworkers that protein stability promotes evolvability (29) toward other opposite proposals (31).
Protein stability is not necessarily associated with evolutionary fitness (3). DePristo and coworkers have suggested that, because optimum protein stability falls within a narrow range, highly stable proteins lacking the required flexibility to meet their functional needs may not be favored by evolution (2). This work provides a direct evidence for this hypothesis: provided the protein is stable enough (like BcII), mutations acting on the mechanism (G262S) and the protein flexibility (N70S) are able to shape the evolution of MβL-mediated antibiotic resistance.
The G262S mutation has already been observed in enzymes coded by transferable genetic elements in opportunistic and pathogenic bacteria: IMP-1 and its putative ancestor, IMP-6, differ only by this mutation, which expands the enzyme's substrate spectrum (32). This relatively recent event in the natural evolution in MβLs confirms that these enzymes have not yet reached their optimal performance. The effect of this mutation on the rate-limiting step of the reaction allows us to propose a strategy for designing a mechanism-based inhibitor with an evolutionary perspective. Recent mechanistic studies suggest that intermediate stabilization by the Zn2 site is essential in MβL catalysis (33, 34). Our results show that optimization of the chemistry in the active site can still be optimized by better positioning Zn2. We therefore propose the design of anionic transition state analogues targeted to bind the Zn2 site, which would provide better lead compounds for inhibitors, instead of ligands aimed to fit into an oxyanion hole (a strategy for designing SβL inhibitors, that has failed for MβLs).
β-Lactamase-mediated antibiotic resistance is a fascinating evolutionary scenario. Organismal fitness is determined by the expression of 2 nonparalogous types of enzymes displaying functional redundancy. Here, we have shown that the naturally occurring alleles of serine and metallo-β-lactamases lie in different positions of the adaptive landscape. Serine-dependent enzymes seem to display an already optimized catalytic machinery, and mutations enhancing the activity or widening the substrate spectrum require compensatory, stabilizing mutations. Instead, MβL evolution can still occur by acting the active-site chemistry through second-sphere mutations. Given their high stability, flexibility can be acquired by evolution by these enzymes at the expense of stability. These results suggest that MβLs may have been recruited more recently by bacteria to supplement the catalytic profile of SβLs. Indeed, their plasticity (regulated by the 2 loops flanking the active site) seems to be a crucial element in MβL evolution.
The comparative analysis of the evolutionary traits acquired by these 2 types of enzymes provide a detailed description of how subtle changes in the protein structure affect the catalytic mechanism, protein stability and flexibility, with a direct impact on organismal fitness.
Materials and Methods
Materials.
All chemicals were of the highest quality available. Tetracyclin, ampicillin, chloramphenicol, kanamycin, benzylpenicillin, cephalexin, cefotaxime, and 4-(2-pyridylazo)-resorcinol (PAR) were purchased from Sigma; nitrocefin was purchased from Calbiochem and imipenem was a kind gift from Merck, Sharp & Dohme. E. coli XL1 Blue MRF cells (Stratagene) were used as the cloning and recipient strain for plasmids. E. coli BL21 (DE3) pLysS′ cells (Stratagene) were used for protein production. Luria–Bertani (Sigma) was used as growth medium for all bacterial strains. The enzymes used for DNA manipulation were purchased from Promega. Vent DNA polymerase was from New England Biolabs. Oligonucleotides were synthesized by Biosynthesis.
DNA Techniques.
DNA preparation and related techniques were performed according to standard protocols (35). Site-directed mutagenesis was performed by using the megaprimer PCR method (36) (see details in SI Materials and Methods). Sequences were determined at the Sequencing Facility of University of Maine (Orono, ME).
Protein Expression and Purification.
All proteins were overexpressed in E. coli BL21(DE3) pLysS′ as fusion proteins with GST, purified and quantified as described in ref. 37 (see details in SI Materials and Methods). The metal content was determined in protein samples dialyzed 4 times against metal-free 10 mM Hepes, pH 7.5, 0.2 M NaCl, at 4 °C, by using the colorimetric reagent 4-(2-pyridylazo)-resorcinol (PAR) under denaturing conditions (38). The apoproteins were obtained as described elsewhere (22, 39).
X-ray Crystallography.
Crystals of M5 were grown by hanging drop vapor diffusion at 18 °C. One microliter of protein solution was mixed with 1 μl of reservoir solution, which contained 1 ml of 24% PEG 3350, 0.1 M sodium tartrate, 1 mM DTT, 1 mM zinc acetate, 0.1 M sodium cacodylate, pH 5.5. Needle-like crystals grew and were cryocooled by using a solution containing 35% PEG 3350, 0.1 M sodium tartrate, 1 mM DTT, 5 mM zinc acetate, 0.1 M sodium cacodylate, pH 6.0. The crystals were taken to the European Synchrotron Radiation Facility (beamline ID29), where they initially diffracted to 6-Å resolution. Annealing 1 crystal for 10 s yielded diffraction to 2.8 Å, and a dataset was collected. The data were processed and refined as described in the SI Materials and Methods. Data collection and refinement statistics are detailed in Table S1.
Kinetic Studies.
Steady-state kinetic parameters were measured and estimated as described in ref. 13. Single-turnover kinetic studies were performed in an SX.18-MVR stopped flow (Applied Photophysis) associated to a photodiode-array detector with some modifications on the already reported protocol (23) (see SI Materials and Methods). The experimental progress curves were analyzed by using the numerical integration algorithm implemented on the software Dynafit (40).
Molecular Dynamics Simulations.
The structure of di-Zn(II) BcII from B. cereus [PDB ID code 1BC2 (20)] was selected as the reference starting structure for all our calculations, as reported in ref. 41. The force-field parameters for the protein frame, the counterions, and water are those of the AMBER PARM98 (42) and TIP3P (43), respectively, and the coordination sphere of the Zn(II) ions was treated as in ref. 41. For the antibiotics, the gaff force field was used (44), as for the charges. Electrostatic interactions were computed by using the Particle Mesh Ewald (PME) algorithm (45). Further details are provided in SI Materials and Methods.
In Vitro Stability.
Equilibrium unfolding induced by guanidine hydrochloride (Gdn-HCl) was monitored by following the changes in tryptophan fluorescence by using a Varian Eclipse spectrofluorometer. Experiments were performed at 25 °C in 50 mM Hepes, pH 7.5, containing 200 mM NaCl. Further details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
P.E.T. received a postdoctoral fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina. This work was supported by grants from ANPCyT and Howard Hughes Medical Institute (A.J.V.). A.J.V. is an International Scholar of Howard Hughes Medical Institute and a staff member of Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3FCZ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807989106/DCSupplemental.
References
- 1.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 2.Depristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 3.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 4.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 5.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 6.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 7.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 8.Zhu G, Golding GB, Dean AM. The selective cause of an ancient adaptation. Science. 2005;307:1279–1282. doi: 10.1126/science.1106974. [DOI] [PubMed] [Google Scholar]
- 9.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 10.Miller SP, Lunzer M, Dean AM. Direct demonstration of an adaptive constraint. Science. 2006;314:458–461. doi: 10.1126/science.1133479. [DOI] [PubMed] [Google Scholar]
- 11.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomatis PE, Rasia RM, Segovia L, Vila AJ. Mimicking natural evolution in metallo-beta-lactamases through second-shell ligand mutations. Proc Natl Acad Sci USA. 2005;102:13761–13766. doi: 10.1073/pnas.0503495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 15.Beadle BM, Shoichet BK. Structural bases of stability-function tradeoffs in enzymes. J Mol Biol. 2002;321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 16.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowder MW, Spencer J, Vila AJ. Metallo-beta-lactamases: Novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 18.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: The quiet before the storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasia RM, Vila AJ. Exploring the role and the binding affinity of a second zinc equivalent in B. cereus metallo-beta-lactamase. Biochemistry. 2002;41:1853–1860. doi: 10.1021/bi010933n. [DOI] [PubMed] [Google Scholar]
- 20.Fabiane SM, et al. Crystal structure of the zinc-dependent beta lactamase from Bacillus cereus at 1.9 A resolution: Binuclear active site with features of a mononuclear enzime. Biochemistry. 1998;37:12404–12411. doi: 10.1021/bi980506i. [DOI] [PubMed] [Google Scholar]
- 21.Carfi A, et al. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orellano EG, Girardini JE, Cricco JA, Ceccarelli EA, Vila AJ. Spectroscopic characterization of a binuclear metal site in Bacillus cereus beta-lactamase II. Biochemistry. 1998;37:10173–10180. doi: 10.1021/bi980309j. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Fast W, Benkovic SJ. Direct observationof an enzime bound intermediate in the catalytic cycle of the metallo-beta-lactamase from Bacteroides fragilis. J Am Chem Soc. 1998;120:10788–10789. [Google Scholar]
- 24.Llarrull LI, et al. Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity. J Biol Chem. 2007;282:18276–18285. doi: 10.1074/jbc.M700742200. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez JM, Medrano Martin FJ, Costello AL, Tierney DL, Vila AJ. The Zn2 position in metallo-beta-lactamases is critical for activity: A study on chimeric metal sites on a conserved protein scaffold. J Mol Biol. 2007;373:1141–1156. doi: 10.1016/j.jmb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Spencer J, et al. Antibiotic recognition by binuclear metallo-beta-lactamases revealed by X-ray crystallography. J Am Chem Soc. 2005;127:14439–14444. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- 27.Moran-Barrio J, et al. The metallo-beta-lactamase GOB is a mono-Zn(II) enzyme with a novel active site. J Biol Chem. 2007;282:18286–18293. doi: 10.1074/jbc.M700467200. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc Natl Acad Sci USA. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oelschlaeger P, Mayo SL, Pleiss J. Impact of remote mutations on metallo-beta-lactamase substrate specificity: Implications for the evolution of antibiotic resistance. Protein Sci. 2005;14:765–774. doi: 10.1110/ps.041093405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oelschlaeger P, Schmid RD, Pleiss J. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-beta-lactamases IMP-1 and IMP-6. Biochemistry. 2003;42:8945–8956. doi: 10.1021/bi0300332. [DOI] [PubMed] [Google Scholar]
- 33.Llarrull LI, Tioni MF, Vila AJ. Metal content and localization during turnover in B. cereus metallo-β Lactamase. J Am Chem Soc. 2008;130:15842–15851. doi: 10.1021/ja801168r. [DOI] [PubMed] [Google Scholar]
- 34.Tioni MF, et al. Trapping and characterization of a reaction intermediate in carbapenem hydrolysis by B. cereus metallo-β-lactamase. J Am Chem Soc. 2008;130:15852–15863. doi: 10.1021/ja801169j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 36.Barik S. In: In Vitro Mutagenesis Protocols. Trower MK, editor. Totowa, NJ: Humana Press; 1996. pp. 203–215. [Google Scholar]
- 37.Paul-Soto R, et al. Mono- and binuclear Zn2+-beta-lactamase. Role of the conserved cysteine in the catalytic mechanism. J Biol Chem. 1999;274:13242–13249. doi: 10.1074/jbc.274.19.13242. [DOI] [PubMed] [Google Scholar]
- 38.Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 39.Llarrull LI, Tioni MF, Kowalski J, Bennett B, Vila AJ. Evidence for a dinuclear active site in the metallo-beta-lactamase BcII with substoichiometric Co(II). A new model for metal uptake. J Biol Chem. 2007;282:30586–30595. doi: 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 40.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 41.Dal Peraro M, Vila AJ, Carloni P. Substrate binding to mononuclear metallo-beta-lactamase from Bacillus cereus. Proteins. 2004;54:412–423. doi: 10.1002/prot.10554. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, et al. AMBER 7. San Francisco: University of California; 2002. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura J, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 44.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 45.Darden T, York D, Pedersen L. Particle mesh Ewald: An N. Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.