Abstract
We describe a novel basic leucine zipper containing type I IFN-inducible early response gene SARI (Suppressor of AP-1, Regulated by IFN). Steady-state SARI mRNA expression was detected in multiple lineage-specific normal cells, but not in their transformed/tumorigenic counterparts. In normal and cancer cells, SARI expression was induced 2 h after fibroblast IFN (IFN-β) treatment with 1 U/ml of IFN-β. Antisense inhibition of SARI protected HeLa cells from IFN-β-mediated growth inhibition. As a corollary, overexpression of SARI inhibited growth and induced apoptosis in cancer cells, but not in normal cells. SARI interacted with c-Jun via its leucine zipper, resulting in inhibition of DNA binding of activator protein (AP-1) complex and consequently AP-1-dependent gene expression. Transformed cells relying on AP-1 activity for proliferative advantage demonstrated increased susceptibility to SARI-mediated growth inhibition. These findings uncover a novel mode of IFN-induced anti-tumor growth suppression and suggest potential gene therapy applications for SARI.
Keywords: IFN-inducible gene, Jun-interacting protein, cancer growth suppressing gene
Although IFNs were originally identified as proteins conferring viral interference, in the past 50 years a variety of effects have been attributed to these molecules, such as inhibition of viral, bacterial, and parasitic pathogenesis; inhibition of cell growth; induction of apoptosis; inflammation; immunomodulation; and anti-angiogenesis (1–6). IFNs exert their diverse effects by stimulating the expression of a plethora of IFN-stimulated genes in target cells (2). Both type I (IFN-α, IFN-β, IFN-ω, and IFN-τ) and type II (IFN-γ) IFNs exert potent anti-tumor effects and are used clinically either as a monotherapy or as an adjuvant to chemotherapy or radiotherapy for a number of solid tumors and hematological malignancies that include melanoma, renal cell carcinoma, Kaposi sarcoma, malignant glioma, lymphomas, and leukemias (1). The anti-tumor effects of IFNs are mediated by direct inhibition of proliferation of cancer cells as well as by indirect effects such as inhibition of tumor angiogenesis and up-regulation of tumor-specific antigens and adhesion molecules.
In an in vitro system, treatment of human melanoma cells with type I IFN (IFN-β) and the protein kinase C activator mezerein (MEZ) induces irreversible growth arrest and “terminal differentiation” (7, 8). Differential gene expression analysis between IFN-β/MEZ-treated terminally differentiated versus control melanoma cells identified a variety of novel “melanoma differentiation-associated” (mda) genes, the functional characterization of which has revealed their central role in various IFN-regulated events, such as viral interference, growth inhibition, and induction of apoptosis (9–16).
Activator protein-1 (AP-1), one of the first identified mammalian transcription factors, plays an essential role in regulating cell proliferation and oncogenic transformation (17). AP-1 comprises a variety of dimeric basic region-leucine zipper (bZIP) proteins that belong to the Jun (c-Jun, JunB, JunD), Fos (c-Fos, FosB, Fra-1, and Fra-2), Maf, and ATF subfamilies that recognize either the 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (5′-TGAG/CTCA-3′) or the cAMP response element (5′-TGACGTCA-3′) (18). Among the Jun proteins, c-Jun is the most potent transcriptional activator, and the c-Fos-c-Jun heterodimer positively regulates cell proliferation and transformation (17, 19, 20). c-Jun co-operates with Ha-ras in rat embryonic fibroblast transformation and c-jun−/− cells are refractory to the transforming activity of oncogenic Ras (21, 22). Additionally, overexpression of c-Jun itself can immortalize rodent fibroblasts in culture (21). Augmented AP-1 activity has been observed in more than 90% of human cancers, indicating its seminal importance in human carcinogenesis (23).
The present manuscript describes the cloning and characterization of a novel type I IFN-inducible early response gene. The gene product is a bZIP containing protein that inhibits cell growth by interacting with c-Jun and inhibiting AP-1 activity. The gene was named SARI (Suppressor of AP-1, Regulated by IFN) and its functional analysis reveals a perviously unrecognized molecular pathway underlying the anti-tumor action of IFNs.
Results
Cloning of SARI.
In an effort to unravel the molecular basis of terminal differentiation, we performed differential gene expression analysis by rapid subtraction hybridization employing temporal RNAs isolated from solvent-treated (i.e., control) and IFN-β/MEZ-treated (i.e., terminally differentiated) HO-1 human melanoma cells (9). This allowed us to clone a broad spectrum of mda genes. One of the mda genes, later named SARI based on its functional properties, was originally identified as a novel gene, mda-D-74. The cDNA consists of 2,140 bp and codes for a putative protein of 274 aa residues with a predicted molecular mass of 29.4 kDa and pI 7.2 (Fig. 1A). Genomic BLAST search indicated that the SARI gene is located in the long arm of chromosome 11 between 11q12 and 11q13. The gene consists of three exons. Translation starts in exon 1 and the stop codon is located in exon 3 (Fig. 1B). Sequence analysis using InterPro Scan identified an L-X6-L-X6-L-X6-L motif between 45 and 66 aa residues preceded by a highly basic region, suggesting that SARI might be a basic-region leucine zipper containing transcription factor (Fig. 1A).
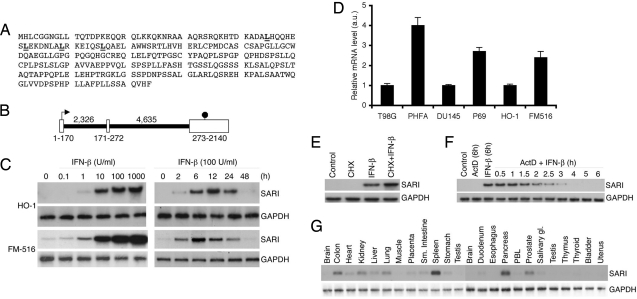
Fig. 1.
SARI is a type I IFN-inducible early response gene down-regulated in cancers. (A) Amino acid sequence of SARI protein. The leucine residues in the leucine zipper are underlined. (B) Genomic structure of the SARI gene. The white boxes represent the exons and the black lines represent the introns. The numbers at the bottom of the white boxes represent the size of the exons and those on top of the black lines represent the size of the introns in bp. The arrow indicates the translation start site and the octagon indicates the stop codon. (C) Dose- (Left) and time-dependent (Right) induction of SARI mRNA by IFN-β in HO-1 and FM516-SV cells analyzed by Northern blotting. GAPDH mRNA expression was used as loading control. (D) Quantitative SARI mRNA expression analysis in cancer (T98G, DU145, and HO-1) and normal cells (PHFA, P69, and FM516-SV; in arbitrary units [a.u.]). Data represent mean ± SD of three independent experiments. (E) IFN-β induction of SARI mRNA occurs at a transcriptional level. HeLa cells were treated with cycloheximide (10 μM) followed by IFN-β (100 U/ml). SARI and GAPDH mRNA expression was analyzed by Northern blotting. (F) HeLa cells were treated with IFN-β (100 U/ml) for 6 h followed by actinomycin D (ActD, 0.5 μg/ml) for 0.5 to 6 h. SARI and GAPDH mRNA expression was analyzed by Northern blotting. (D) Analysis of SARI and GAPDH mRNA expression in different tissues using multiple tissue Northern blot (Clontech).
Gene expression analysis identified SARI as a type I IFN (IFN-α/β)-inducible early response gene. In HO-1 human melanoma cells and SV40 T/tAg-immortalized primary human melanocytes (FM516-SV), SARI mRNA expression could be detected as early as 2 h after IFN-β treatment (100 U/ml) with maximum induction at 12 h post-treatment with a gradual decrease thereafter and a return by 48 h to the basal level of expression (Fig. 1C). The induction could be detected with as little as 1 U/ml of IFN-β. Interestingly, in FM516-SV cells, but not in HO-1 melanoma cells, SARI expression could be detected under de novo conditions (Fig. 1C). A similar expression and induction profile could be detected in a series of normal and cancer cells from diverse tissue origins [supporting information (SI) Fig. S1]. More importantly, under basal conditions, SARI mRNA expression could be detected in multiple normal cells, including primary human fetal astrocytes (PHFAs), immortal normal mammary epithelial cells (HBL100), immortal prostate epithelial cells (P69), and immortal pancreatic mesothelial cells (LT2), but not in their malignant counterparts (Fig. S1). Quantitative RT-PCR confirmed that SARI mRNA expression was significantly higher in PHFA versus T98G malignant glioma cells, P69 versus DU145 prostate cancer cells, as well as FM516-SV versus HO-1 melanoma cells (Fig. 1D). Treatment with cycloheximide, a protein translation inhibitor, did not by itself induce SARI mRNA expression and moderately increased IFN-β-mediated induction, suggesting that IFN-β predominantly regulates SARI expression on a transcriptional level (Fig. 1E). The half-life of the IFN-β-inducible transcript was ≈2 h as revealed by actinomycin D treatment, which prevents new RNA transcription (Fig. 1F). Multiple tissue Northern blots revealed SARI expression in diverse tissues, with highest expression in pancreas and spleen and moderate expression in colon, heart, kidney, liver, lung, and prostate (Fig. 1G). Low-level expression could be detected in placenta, stomach, small intestine, and salivary gland. Brain, muscle, and testis were conspicuous by the absence of SARI expression.
SARI Inhibits Growth of Cancer Cells but not Normal Cells.
The observation that basal SARI mRNA expression was detected in normal cells but not in their malignant counterparts suggested that SARI might display tumor suppressor functions that are lost during tumor progression. Based on this consideration, we analyzed the biological consequences of overexpression of this molecule in normal and tumor cells. We constructed a replication-incompetent adenovirus expressing SARI (Ad.SARI), infected PHFA, H4 (malignant glioma), P69, DU-145, FM516-SV, and MeWo (i.e., melanoma) cells with Ad.SARI (100 pfu/cell) and monitored cell growth by viable cell counting. Empty adenovirus (Ad.vec) served as a control. Very interestingly, Ad.SARI profoundly inhibited cancer cell growth (H4, DU145, and MeWo) with little to no effect on normal cells (PHFA, P69, and FM516-SV; Fig. 2A). This finding was confirmed in transformed/tumorigenic cells by clonogenic assays and by anchorage-independent growth assays in soft agar (data not shown). It should be noted that the expression level of SARI upon Ad.SARI infection was equivalent in all cell lines, as confirmed by Western blot analysis using anti-SARI antibody (data not shown). To determine if this inhibition of growth was a result of induction of apoptosis, flow cytometry studies were performed to follow Annexin V staining or propidium iodide staining to identify subG1 (A0) cell populations. Ad.SARI, but not Ad.vec, resulted in a significant induction of apoptosis in cancer cells (H4, DU145, and HeLa), but not in PHFA (Fig. 2B).
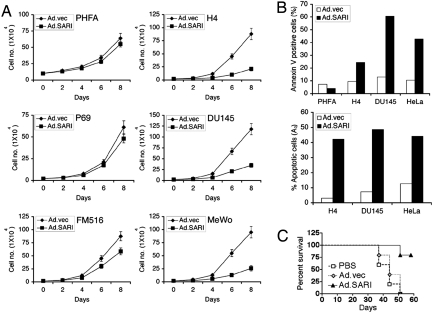
Fig. 2.
SARI suppresses growth of cancer cells in vitro and in vivo. (A) Cancer (H4, DU145, and MeWo) and normal cells (PHFA, P69, and FM-516SV) were infected with either Ad.vec or Ad.SARI at a multiplicity of infection of 100 pfu/cell and cell number was counted at the indicated time points. The data represent mean ± SD of three independent experiments. (B) Cancer (H4, DU145, and HeLa) and normal cells (PHFA) were infected as in A and apoptosis was analyzed by Annexin V binding assay (Upper) and propidium iodide staining assay (Lower), followed by flow cytometry 48 h after infection. (C) Subcutaneous xenografts were established in the flanks of athymic nude mice with DU145 human prostate cancer cells. Established tumors were injected with PBS, Ad.vec, or Ad.SARI as described in Materials and Methods and survival of animals was analyzed by the Kaplan-Meier test.
The growth inhibitory effect of SARI was confirmed by in vivo studies. Subcutaneous xenografts from DU-145 prostate cancer cells were established in the left flank of athymic nude mice. When the tumors reached ≈100 mm3 in size, requiring ≈7 days, intratumoral injections of PBS solution, Ad.vec, or Ad.SARI (108 pfu/injection) were administered three times during the first week and then twice weekly for two more weeks for a total of seven injection. Tumor growth was significantly inhibited by Ad.SARI treatment compared with PBS or Ad.vec treatment (data not shown). Monitoring animal survival demonstrated that Ad.SARI significantly prolonged survival compared with PBS-treated or Ad.vec-infected animals, confirming the tumor-suppressor properties of SARI in vivo (Fig. 2C).
To interrogate the molecular pathway(s) involved in Ad.SARI-mediated growth inhibition, we used normal immortal rat embryonic fibroblasts (CREF) and a clone of CREF transformed by H-ras, v-src, HPV-18, or a specific temperature (i.e., cold)-sensitive mutant of type 5 adenovirus H5hrl (CREF-ras, CREF-src, CREF-HPV, and CREF-H5hrl, respectively) (24, 25). The parental CREF do not grow in soft agar and are non-tumorigenic, whereas transformed clones all grow in soft agar and are tumorigenic. Ad.SARI had no discernible effect on the growth of CREF. Among the tumorigenic clones, a preferential inhibition of growth was observed in CREF-ras and CREF-src clones with a significantly reduced effect on CREF-HPV and CREF-H5hrl clones (Fig. 3A). These findings were bolstered by clonogenic assays as well as by soft agar assays (Fig. 3B and data not shown). These results indicate that signaling pathways, activated by ras and src, but not by nuclear-acting oncogenes such as HPV-18 and H5hrl, make CREF susceptible to Ad.SARI-mediated growth inhibition.
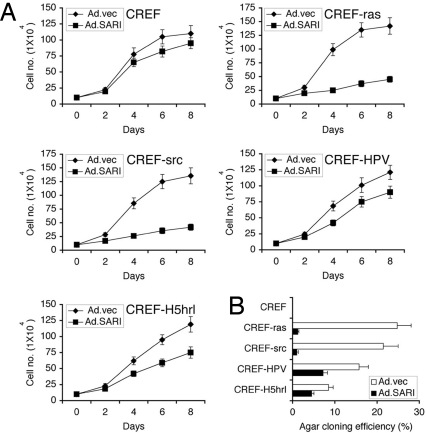
Fig. 3.
SARI inhibits growth of transformed cells in a Ras- and Src-dependent manner. (A) Normal immortal rat embryonic fibroblasts (CREF) and clones of CREF transformed by H-ras, v-src, HPV-18, or a specific temperature-sensitive mutant of type 5 adenovirus H5hrl (CREF-ras, CREF-src, CREF-HPV, and CREF-H5hrl, respectively) were infected with either Ad.vec or Ad.SARI at a multiplicity of infection of 100 pfu/cell and cell number was counted at the indicated time points. (B) Soft agar colony formation assay of CREF, CREF-ras, CREF-src, CREF-HPV, and CREF-H5hrl cells infected as in A. The data represent mean ± SD of three independent experiments.
SARI Mediates IFN-β-Induced Growth Inhibition.
Bioinformatics analysis of SARI predicted a putative basic leucine zipper motif that might contain a c-JUN-dimerization motif. As c-JUN, a basic leucine zipper containing protein, is a component of the AP-1 transcription factor, and because both ras- and src-mediated signaling result in AP-1 activation, we hypothesized that SARI might interact with c-JUN, thereby interfering with AP-1 function and inhibiting growth of cells dependent on AP-1 for proliferation and survival (e.g., CREF-ras and CREF-src). These assumptions were validated through a series of experiments. We confirmed that CREF were resistant to growth suppression by Ad.SARI, whereas CREF stably overexpressing c-jun became susceptible to Ad.SARI-mediated growth inhibition (Fig. 4A). Stable HeLa cells expressing antisense SARI (HeLa-SARIAs) were established, and we analyzed expression and IFN-β induction of SARI protein in these clones. Cytoplasmic and nuclear extracts were prepared from parental HeLa cells, a control HeLa clone not expressing SARIAs, and HeLa-SARIAs clones untreated or treated with IFN-β. SARI protein was detected exclusively in the nucleus and induction of SARI protein by IFN-β was observed in HeLa cells and in the control clone, but not in HeLa-SARIAs clones, confirming the authenticity of these clones (Fig. 4B). Growth of HeLa and HeLa-SARIAs cells were evaluated upon IFN-β treatment. HeLa-SARIAs cells were more resistant to growth inhibition by IFN-β compared with parental HeLa cells, indicating that SARI plays an important role in mediating IFN-β action (Fig. 4C).
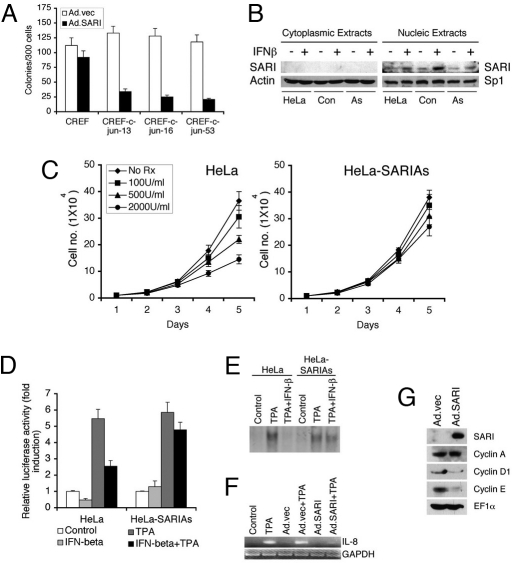
Fig. 4.
SARI inhibits AP-1 activation. (A) CREF and CREF clones stably overexpressing c-jun (CREF-c-jun 13, CREF-c-jun 16, and CREF-c-jun 53) were treated and analyzed as in Fig. 3A. (B) Cytoplasmic and nuclear extracts were prepared from HeLa cells, control HeLa clone not expressing SARIAs (con), and HeLa clones stably expressing antisense SARI (As) treated with IFN-β or untreated, and the expression of SARI was detected using rabbit anti-SARI antibody. The purity of the extracts was confirmed by analyzing Actin (for cytoplasm) and Sp1 (nucleus) expression. (C) HeLa cells and HeLa cells stably expressing antisense SARI (HeLa-SARIAs) were treated with IFN-β (100, 500, and 2,000 U/ml) and cell numbers were counted at the indicated time points. (D) HeLa and HeLa-SARIAs cells were transfected with AP-1-luc, treated with TPA or IFN-β or a combination, and luciferase activity was monitored by a luminometer. Luciferase activity was normalized by β-galactosidase activity. For A–C, data represent mean ± SD of three independent experiments. (E) HeLa and HeLa-SARIAs cells were treated with TPA or IFN-β and nuclear extracts were used for EMSA using a radiolabeled consensus AP-1 probe. (F) HeLa cells were infected with Ad.vec or Ad.SARI at 100 pfu/cell and then treated with TPA. Expression of IL-8 and GAPDH was analyzed by RT-PCR. (G) HeLa cells were infected with Ad.vec or Ad.SARI at 50 pfu/cell for 16 h and the expression of the indicated proteins in total cell lysates were analyzed by Western blot analysis.
Parental HeLa and HeLa-SARIAs cells were transfected with an AP-1 reporter plasmid, in which the luciferase gene is regulated by consensus AP-1-binding sites (AP-1-luc), and then treated with TPA, IFN-β, or both. HeLa and HeLa-SARIAs cells responded equally to TPA for AP-1-luc induction (Fig. 4D). However, IFN-β significantly reduced both basal and TPA-induced activation of AP-1-luc only in HeLa cells. HeLa-SARIAs cells displayed significant resistance to the inhibitory effect of IFN-β (Fig. 4D). These observations suggest that under basal conditions TPA efficiently activates AP-1-luc activity. However, SARI, upon induction by IFN-β, interfered with TPA-mediated AP-1-luc activation that could be rescued by antisense inhibition of SARI induction. These findings were further confirmed by electrophoretic mobility shift assay (EMSA) using nuclear extracts from TPA- or TPA/IFN-β-treated HeLa and HeLa-SARIAs cells and a radiolabeled consensus AP-1 probe. In HeLa cells, TPA-induced augmentation of AP-1-binding activity was markedly reduced by IFN-β treatment (Fig. 4E). This inhibition by IFN-β was profoundly abrogated in HeLa-SARIAs cells. These findings indicate that SARI plays a key role in mediating IFN-β-mediated down-regulation of AP-1 activity. The inhibition of AP-1 activity by SARI was reflected in the expression level of an AP-1 downstream gene, IL-8. TPA treatment resulted in IL-8 mRNA induction in control and Ad.vec-infected HeLa cells. However, upon infection with Ad.SARI, TPA-mediated induction of IL-8 mRNA was markedly reduced (Fig. 4F). The steady-state expressions of cell cycle regulatory proteins such as cyclin D1 and cyclin E, which are downstream of AP-1, were also significantly down-regulated upon Ad.SARI infection (Fig. 4G).
SARI Interacts with c-JUN.
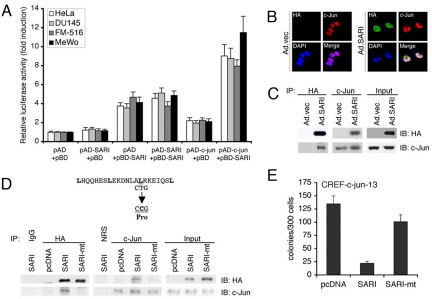
Potential interactions between c-JUN and SARI were evaluated using multiple approaches. We used a mammalian two-hybrid assay in which plasmid pCMV-AD was used for cloning the c-jun cDNA to be expressed as a fusion with the activation domain of VP16 (pAD-c-jun) and plasmid pCMV-BD was used to clone the SARI cDNA to be expressed as a fusion with the DNA-binding domain of GAL4 (pBD-SARI). These constructs were transfected along with a reporter vector that contains five GAL4 binding sites (GAL4UAS) upstream of a minimal TATA box promoter driving the expression of a luciferase reporter gene in HeLa, DU145, FM516-SV, and MeWo cells. Interaction between SARI and c-JUN resulted in the association of the GAL4 DNA binding domain with the VP16 transcription activation domain and a significant induction of luciferase activity was observed only when pAD-c-jun and pBD-SARI were transfected together (Fig. 5A). HeLa cells were infected with Ad.SARI and the cells were processed for dual immunofluorescence analysis using anti-HA antibody to detect HA-tagged SARI and anti-c-Jun antibody to detect c-Jun. Both SARI (green) and c-JUN (red) were detected in the nucleus and the merged images showed overlapping distribution of green, red, and blue (DAPI detecting nucleus), producing a white color (Fig. 5B). Co-immunoprecipitation analysis using SARI-overexpressing HeLa cell lysates and anti-HA and anti-c-JUN antibodies confirmed their interaction (Fig. 5C). Anti-HA and anti-c-JUN antibodies could effectively pull down c-JUN and SARI, respectively. It should be noted that similar assays did not find any interaction between SARI and either c-Fos or ATF-2, indicating that the interaction of SARI with c-Jun is specific (data not shown). Mutation of leucine59 in the bZIP domain of SARI into proline completely inhibited interaction of c-JUN and SARI (Fig. 5D), indicating that the leucine zipper motif mediates the interaction between c-JUN and SARI. Whereas WT SARI could significantly inhibit colony formation of CREF-c-Jun-13 cells, the mutant SARI almost completely lost this activity, indicating that growth inhibition by SARI is mediated by its interaction with c-JUN (Fig. 5E).
Fig. 5.
SARI directly interacts with c-JUN. (A) Mammalian two-hybrid assay was performed using pAD-c-jun and pBD-SARI in HeLa, DU145, FM516-SV, and MeWo cells as described in Materials and Methods. Luciferase activity was normalized by β-galactosidase activity. The data represent mean ± SD of three independent experiments. (B) HeLa cells were infected with Ad.vec or Ad.SARI at 50 pfu/cell. The cells were fixed 24 h later and subjected to immunofluorescence analysis using anti-HA and anti-c-JUN primary antibodies and FITC- and rhodamine-conjugated secondary antibodies, respectively. Mounting medium containing DAPI was used for staining the nucleus. Confocal laser scanning microscopy was used to analyze the images. (C) HeLa cells were infected with Ad.vec or Ad.SARI at 50 pfu/cell. Total cell lysates were used for co-immunoprecipitation analysis using anti-HA antibody for immunoprecipitation and anti-c-JUN antibody for immunoblotting and vice versa. (D) Expression plasmid in which Leucine59 of SARI was mutated to proline was generated (SARI-mt). HeLa cells were transfected with empty vector (pcDNA) or expression plasmids expressing WT SARI or SARI-mt. The cell lysates were subjected to co-immunoprecipitation analysis as described in C. (E) CREF-c-jun-13 cells were transfected with empty vector (pcDNA) or expression plasmids expressing WT SARI or SARI-mt and subjected to clonogenic assay for 2 weeks upon selection with hygromycin. Data represent mean ± SD of three independent experiments.
Discussion
IFNs induce growth inhibition by multiple pathways that involve many IFN-stimulated genes (2). SARI is one of these genes that, based on a number of unique properties, highlights its significance in IFN-signaling. SARI expression is induced as early as 2 h after IFN-β treatment with as little as 1 U/ml of IFN-β, pointing to a role as an early mediator of IFN-β action. Inhibition of SARI by an antisense approach in HeLa cells significantly abrogated IFN-β-induced growth inhibition, thus establishing SARI in mediating IFN anti-proliferative action. Whereas steady-state expression of SARI mRNA was detected in normal cells of diverse lineages, such as melanocytes, astrocytes, breast and prostate epithelial cells, and pancreatic mesothelial cells, expression was not detected in multiple cancer cell lines of the same tissue of origin. These findings indicate that, although normal cell survival is compatible with SARI expression, continued proliferation of cancer cells requires that SARI expression be suppressed, a phenomenon characteristic of many tumor suppressor proteins. The most intriguing finding is that, even when using an adenovirus-mediated delivery approach that ensures high-level gene expression in >90% of cells, SARI overexpression did not adversely affect normal cell survival while inducing profound growth inhibition and apoptosis in cancer cells. Accordingly, targeted overexpression of SARI might provide an effective gene for cancer therapy. The molecular mechanism in which steady-state SARI expression is suppressed in cancer cells remains to be determined. However, IFN-induction of SARI expression is retained by cancer cells, indicating a potential separation of regulatory control between steady-state and inducible expression of SARI. Experimental evidences indicate that IFN-β regulates SARI expression at the transcriptional level and characterization of its promoter region would provide further insights into the regulation of this interesting gene.
We demonstrate that, whereas CREF are resistant to SARI-induced growth inhibition, ras, src, and c-jun transformed CREF clones acquire sensitivity to SARI. Both ras and src signaling induce c-jun activation indicating that cells that employ AP-1 for proliferative advantage become susceptible to growth inhibition by SARI. This is a significant observation for using SARI for potential cancer gene therapy purposes. Ras, src, and AP-1 activations are extremely common events in the process of carcinogenesis and SARI would therefore have significant potential to exert broad-spectrum anti-tumor activity (23, 26, 27).
SARI contains a bZIP domain, and our mutational studies confirm that the leucine zipper in SARI mediates its interaction with c-Jun. EMSA using a consensus AP-1 probe and in vitro translated SARI or c-Jun proteins demonstrated that SARI did not bind to the consensus AP-1 site or the AP-1 site in the IL-8 promoter either alone or as a heterodimer with c-Jun (data not shown). Chromatin immunoprecipitation (CHIP) assay using the AP-1 binding site in the IL-8 promoter and anti-SARI antibody also failed to demonstrate DNA binding by SARI (data not shown). SARI is a nuclear protein and, in the nucleus, SARI binds with c-Jun in solution and squelches the DNA-binding activity of AP-1 complexes, thus inhibiting AP-1-mediated gene transcription. A relevant question is why SARI, which contains a putative bZIP basic region domain, fails to bind DNA. One possible explanation could be the lack of stretches of three or more basic amino acid residues in the putative basic region of SARI that is found in other bZIP domain-containing proteins that do bind DNA (28).
SARI is comparable to the IFN-inducible p200 family of proteins (IFI-200) (29, 30). IFI-200 family proteins are induced by IFN, inhibit cell proliferation, and interact with a plethora of proteins such as retinoblastoma protein, p50/p65 nuclear factor-κB, c-Fos, c-Jun, c-Myc, and p53. Thus, these proteins display diverse and pleiotropic effects in target cells. The presence of the bZIP structural motif in SARI indicates that its interaction activities might be more focused on bZIP domain containing proteins. Current studies are focused on identifying other interacting partners of SARI as well as defining its involvement in regulation of relevant transcription factor functions.
In summary, we identified a novel IFN-inducible nuclear protein that, by direct protein-protein interaction, interferes with the function of the AP-1 transcription factor. Apart from mediating IFN-induced growth inhibition, it would be intriguing to identify whether SARI plays any role in other IFN actions, such as anti-viral or anti-angiogenic activity. As a nuclear protein in normal cells, does SARI play any role in regulation of cell cycle activity? Is it part of the normal cell cycle checkpoint control? Inhibition of SARI in normal cells as well as generation of a SARI-KO mouse would provide insights into these questions. These studies are being actively pursued to better understand the molecular function of this important molecule, which will provide valuable insights into the mechanism of action of IFNs and their role as cancer-suppressing molecules.
Materials and Methods
Cell Lines and Culture Conditions.
A detailed description of the cell lines used in this study is provided in SI Text.
Cell Growth and Apoptosis Studies.
Cell growth was monitored by trypan blue dye exclusion assay. Apoptosis was monitored by Annexin V binding assays and propidium staining followed by flow cytometry as described (31, 32).
Transient Transfections and Luciferase Assays.
Transient transfections and luciferase assays were performed as described (33). Luciferase activity was normalized by β-galactosidase activity.
5′ and 3′ Rapid Amplification of cDNA Ends.
5′ and 3′ rapid amplification of cDNA ends was performed by using a GeneRacer kit (Invitrogen) according to the manufacturer's instructions.
RNA Extraction, Northern Blot Analysis, RT-PCR, Quantitative RT-PCR, and Multiple Tissue Northern Blot.
Total RNA extraction and Northern blotting were performed as described (33). The primers used for RT-PCR were as follows: IL-8 sense, 5′ GGTGCAGAGGGTTGTGGAGAA 3′; IL-8 antisense, 5′ GCAGACTAGGGTTGCCAGATT 3′; GAPDH sense, 5′ ATGGGGAAGGTGAAGGTCGGAGTC 3′; GAPDH antisense, 5′ GCTGATGATCTTGAGGCTGTTGTC 3′. Quantitative RT-PCR was performed using a kit from Stratagene based on SYBR Green I DNA-binding dye. Multiple tissue Northern blot was obtained from Clontech and hybridization was performed according to the manufacturer's instructions.
Preparation of Whole-Cell Lysates, Western Blotting, and Co-Immunoprecipitation and Immunofluorescence Analysis.
Western blotting was performed as previously described (33). The primary antibodies used were anti-HA (Covance), anti-Cyclin A, anti-Cyclin D1, anti-Cyclin E and anti-EF1α (Upstate). Co-immunoprecipitation and immunofluorescence analyses were performed as described (34).
EMSA.
Nuclear extracts were prepared from 2 to 5 × 108 cells and EMSA was performed using consensus AP-1 probe (Santa Cruz Biotechnology) as described (33).
ChIP Assays.
ChIP assays were performed using a commercially available kit from Active Motif (35). PCR was performed using IL-8 promoter-specific primers: sense, 5′ ATGTCAGCTCTCGACGAAAATAGA 3′; and antisense, 5′ GGAGGGATTGCAAGGTTTAGC 3′.
Generation of Anti-SARI Antibody.
His-tagged recombinant SARI protein was generated in Escherichia coli and purified using Ni-NTA metal chelate affinity chromatography. The purified protein was used to raise polyclonal antibodies in the rabbits according to the manufacturer's protocol (Invitrogen).
Mammalian Two-Hybrid Assay.
Plasmid pCMV-AD was used for cloning the c-jun cDNA to be expressed as a fusion with the activation domain of VP16 (Clontech). Plasmid pCMV-BD was used to clone the SARI cDNA to be expressed as a fusion with DNA-binding domain of GAL4. These constructs were transfected along with a reporter vector that contains five GAL4 binding sites (GAL4UAS) upstream of a minimal TATA box promoter driving the expression of a luciferase reporter gene. Luciferase activity was monitored by a luminometer and normalized by β-galactosidase activity.
Statistical Analysis.
All of the experiments were performed at least three times. The results are expressed as mean ± SD. Statistical comparisons were made using an unpaired two-tailed Student t test. A P value <0.05 was considered as significant.
Supplementary Material
Acknowledgments.
We thank Dr. Tina Cirman for initial IFN studies with SARI and Nichollaq Vozhilla for technical assistance with the animal studies. The present study was supported in part by National Institutes of Health Grants CA035675 and CA097318; the Samuel Waxman Cancer Research Foundation (SWCRF); and by the National Foundation for Cancer Research. D.S. is the Harrison Endowed Scholar in Cancer Research. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research and is an SWCRF Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807975106/DCSupplemental.
References
- 1.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Zhang SY, et al. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–236. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 5.Barber GN. The interferons and cell death: guardians of the cell or accomplices of apoptosis? Semin Cancer Biol. 2000;10:103–111. doi: 10.1006/scbi.2000.0313. [DOI] [PubMed] [Google Scholar]
- 6.Fisher PB, Grant S. Effects of interferon on differentiation of normal and tumor cells. Pharmacol Ther. 1985;27:143–166. doi: 10.1016/0163-7258(85)90067-1. [DOI] [PubMed] [Google Scholar]
- 7.Fisher PB, et al. Effect of recombinant human fibroblast interferon and mezerein on growth, differentiation, immune interferon binding and tumor associated antigen expression in human melanoma cells. Anticancer Res. 1986;6:765–774. [PubMed] [Google Scholar]
- 8.Fisher PB, Prignoli DR, Hermo H, Jr, Weinstein IB, Pestka S. Effects of combined treatment with interferon and mezerein on melanogenesis and growth in human melanoma cells. J Interferon Res. 1985;5:11–22. doi: 10.1089/jir.1985.5.11. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Kang DC, Alexandre D, Fisher PB. RaSH, a rapid subtraction hybridization approach for identifying and cloning differentially expressed genes. Proc Natl Acad Sci USA. 2000;97(23):12684–12689. doi: 10.1073/pnas.220431297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Lin J, Fisher PB. A molecular definition of terminal cell differentiation in human melanoma cells. Mol Cell Differ. 1994;2:221–239. [Google Scholar]
- 11.Jiang H, et al. The melanoma differentiation-associated gene mda-6, which encodes the cyclin-dependent kinase inhibitor p21, is differentially expressed during growth, differentiation and progression in human melanoma cells. Oncogene. 1995;10:1855–1864. [PubMed] [Google Scholar]
- 12.Jiang H, et al. Cell cycle gene expression and E2F transcription factor complexes in human melanoma cells induced to terminally differentiate. Oncogene. 1995;11:1179–1189. [PubMed] [Google Scholar]
- 13.Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB. Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- 14.Jiang H, Su ZZ, Boyd J, Fisher PB. Gene expression changes associated with reversible growth suppression and the induction of terminal differentiation in human melanoma cells. Mol Cell Differ. 1993;1:41–66. [Google Scholar]
- 15.Kang DC, et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang DC, LaFrance R, Su ZZ, Fisher PB. Reciprocal subtraction differential RNA display: an efficient and rapid procedure for isolating differentially expressed gene sequences. Proc Natl Acad Sci USA. 1998;95:13788–13793. doi: 10.1073/pnas.95.23.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 18.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 19.Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryseck RP, Bravo R. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene. 1991;6:533–542. [PubMed] [Google Scholar]
- 21.Vandel L, et al. Stepwise transformation of rat embryo fibroblasts: c-Jun, JunB, or JunD can cooperate with Ras for focus formation, but a c-Jun-containing heterodimer is required for immortalization. Mol Cell Biol. 1996;16:1881–1888. doi: 10.1128/mcb.16.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 24.Boylan JF, Shih TY, Fisher PB, Zimmer SG. Induction and progression of the transformed phenotype in cloned rat embryo fibroblast cells: studies employing type 5 adenovirus and wild-type and mutant Ha-ras oncogenes. Mol Carcinog. 1992;5:118–128. doi: 10.1002/mc.2940050207. [DOI] [PubMed] [Google Scholar]
- 25.Lin L, et al. Activation of Ras/Raf protects cells from melanoma differentiation-associated gene-5-induced apoptosis. Cell Death Differ. 2006;13:1982–1993. doi: 10.1038/sj.cdd.4401899. [DOI] [PubMed] [Google Scholar]
- 26.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 27.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 28.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Asefa B, et al. The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32:155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: an emerging family of IFN-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 31.Lebedeva IV, et al. Bcl-2 and Bcl-x(L) differentially protect human prostate cancer cells from induction of apoptosis by melanoma differentiation associated gene-7, mda-7/IL-24. Oncogene. 2003;22:8758–8773. doi: 10.1038/sj.onc.1206891. [DOI] [PubMed] [Google Scholar]
- 32.Lebedeva IV, et al. The cancer growth suppressing gene mda-7 induces apoptosis selectively in human melanoma cells. Oncogene. 2002;21:708–718. doi: 10.1038/sj.onc.1205116. [DOI] [PubMed] [Google Scholar]
- 33.Su Z, et al. PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells. Nucleic Acids Res. 2001;29:1661–1671. doi: 10.1093/nar/29.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar D, et al. Molecular basis of NF-kappaB activation by astrocyte elevated gene-1 (AEG-1) Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 35.Su ZZ, Sarkar D, Emdad L, Barral PM, Fisher PB. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007;213:502–510. doi: 10.1002/jcp.21128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.