Abstract
Tauopathies are sporadic and genetic neurodegenerative diseases characterized by aggregation of the microtubule-associated protein Tau. Tau pathology occurs in over 20 phenotypically distinct neurodegenerative diseases, including Alzheimer disease and frontotemporal dementia. The molecular basis of this diversity among sporadic tauopathies is unknown, but distinct fibrillar wild-type (WT) Tau conformations could play a role. Using Fourier transform infrared spectroscopy, circular dichroism, and electron microscopy, we show that WT Tau fibrils and P301L/V337M Tau fibrils have distinct secondary structures, fragilities, and morphologies. Furthermore, P301L/V337M fibrillar seeds induce WT Tau monomer to form a novel fibrillar conformation, termed WT*, that is maintained over multiple seeding reactions. WT* has secondary structure, fragility, and morphology that are similar to P301L/V337M fibrils and distinct from WT fibrils. WT Tau is thus capable of conformational diversity that arises via templated conformation change, as has been described for amyloid β, β2-microglobulin, and prion proteins.
Tau filament deposition in Alzheimer disease, frontotemporal dementia, and other tauopathies correlates closely with cognitive dysfunction and cell death (1). About 10% of tauopathies are due to dominant mutations in the Tau gene. These diseases are collectively termed frontotemporal dementia with parkinsonism linked to chromosome 17, FTDP-17 (2–4). Most of the mutations occur in the microtubule-binding region of the Tau protein, which is thought to be both its functional (5) and pathogenic (6) “core.” Approximately 90% of tauopathies occur sporadically and involve only wild-type (WT)2 Tau. Both familial and sporadic tauopathies vary by regional involvement, disease duration, age of onset, Tau isoform expression, and fibril morphology (7). It is unknown how the pathology of WT Tau might generate distinct disease phenotypes in sporadic tauopathies, and whether conformational diversity of the protein could potentially play a role in disease, as it does in prion disorders (8, 9).
Mutations in the Tau gene can generate conformationally distinct Tau species. Structural differences between in vitro prepared WT, G272V, N279K, P301L, V337M, and ΔK280 Tau fibrils have been observed using Fourier transform infrared spectroscopy (FTIR) (10), and differential susceptibilities to protease cleavage in vitro have been described for WT and P301L Tau fibrils (11). Furthermore, Tau filaments extracted from diseased brain are often morphologically distinct, consisting of straight or paired helical filaments of various periodicities and widths (12). It is unknown whether WT Tau can assume self-propagating, structurally distinct fibrillar conformations, as has been described for amyloid β peptide (13), β2-microglobulin (14), and the prion protein (15). In this study, we have used biochemical and biophysical methods to test the hypothesis that WT Tau fibrils exhibit conformational diversity that is maintained by templated conformation change.
EXPERIMENTAL PROCEDURES
Tau Expression and Purification—The microtubule-binding region (amino acids 242–364 of the 441-amino acid Tau isoform) of WT Tau and the P301L/V337M Tau double mutation (designated as MUT) used in all fibrillization reactions were subcloned into pRK172 (a gift from Dr. Virginia Lee). Recombinant Tau microtubule-binding region was prepared as described previously from Rosetta (DE3)pLacI competent cells (Novagen), exploiting the heat stability of Tau protein, followed by cation exchange chromatography (16). Single-use aliquots were stored at –80 °C in 10 mm HEPES and 100 mm NaCl (pH 7.4).
Tau Fibrillization and Seeding Assays—4 μm Tau monomer was incubated for the indicated times at room temperature without agitation in 5 mm dithiothreitol, 10 mm HEPES (pH 7.4), 100 mm NaCl, and 150 μm arachidonic acid (Sigma-Aldrich) (17). Tau appears in the insoluble fraction after a 15-h fibrillization reaction and 100,000 × g ultracentrifugation. Alternatively, seeded reactions were carried out using a 10% aliquot from a fibrillization reaction (i.e. 0.4 μm total Tau protein) and 4 μm monomer in three successive incubations. The primary fibrillization reaction was initiated with 150 μm arachidonic acid. After 15 h, the secondary fibrillization was initiated by exposure of 4 μm monomer to 10% of the primary reaction. After 3 h, the tertiary reaction was initiated by exposure of 4 μm monomer to 10% of the secondary reaction. After 3 h, the quaternary reaction was initiated by exposure of 4 μm monomer to 10% of the tertiary reaction, resulting in a 1,000-fold dilution of the primary seed. The quaternary reaction was allowed to proceed for 96 h before further analysis. WT* was produced by using primary MUT Tau as a seed for WT monomer in serial seeding reactions (MUT seed WT seed WT seed WT).
Thioflavin T—When measuring the fibrillization rate of primary reactions, 12.5 μm thioflavin T (ThT; Sigma-Aldrich) was included in the reaction (18). Measurements were recorded at 455 nm excitation/485 nm emission in a Safire plate reader (Tecan) every 10 min for 1 h and every 30 min for the next 14 h. When measuring the fibrillization rate of quaternary reactions, 12.5 mm ThT was mixed with aliquots of the quaternary reaction every 24 h and measured as described above.
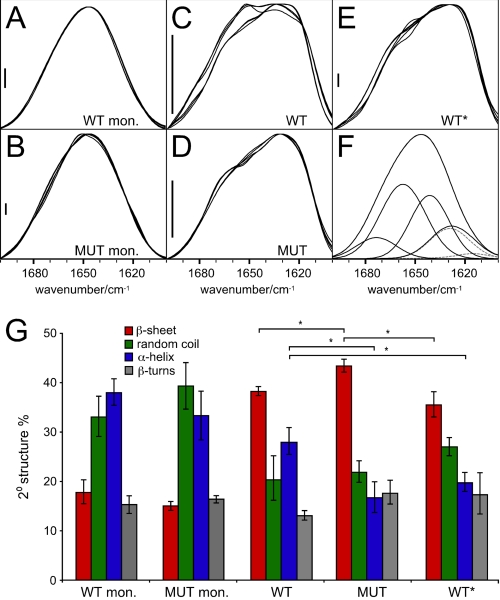
Fourier Transform Infrared Spectroscopy—WT and MUT Tau were polymerized through serially seeded reactions as described above. The polymerized Tau from quaternary reactions was pelleted by ultracentrifugation, resuspended in water, and lyophilized for 48 h. The lyophilisates were resuspended in D2O. Infrared absorbance spectra were recorded against pure D2O. Buffers and salts did not interfere with the amide I absorption band (1700-1600 cm–1). For each sample, 64 double-sided interferograms were scanned bidirectionally and averaged on a PerkinElmer System 2000 FTIR spectrometer, equipped with an MCT detector and purged with dry N2. Spectra were recorded with a 2 cm–1 instrument resolution, and 4-fold zero filling yielded one data point per 0.5 cm–1. Trace signals from water vapor were eliminated with the suppression algorithm of the software Spectrum GX V5 (PerkinElmer Life Sciences). Additionally, low frequency noise was filtered with a Fourier self-deconvolution algorithm at an 8–10 cm–1 cutoff as necessary, and the amide I band was base line-corrected. Secondary structure information was derived by amide I band decomposition as described previously (19). Cauchy curves were numerically determined to add up to the shape of the amide I band. The Cauchy curve integrals were used to calculate the fraction of secondary structure that is assigned to the peak frequency of the curve. Curve fitting all amide I bands with identical initialization parameters for the number of bands, band position, width, and shape and the automatic determination of each intensity as well as the overall base-line values resulted in a high sensitivity for structural changes. The secondary structure fractions presented in Fig. 2G were averaged from six spectra (MUT aggregate from five) of two separate aggregation assays. Thereby, we were able to suppress the negative effects of residual water vapor and to quantify the quality of our analysis with the error bars presented.
FIGURE 2.
FTIR analysis of WT, MUT, and WT* Tau. A, WT monomer (mon.). B, MUT monomer. C–E, quaternary WT, quaternary MUT, and quaternary WT* aggregates, respectively, prepared as described for FTIR. MUT (D) and WT* (E) exhibit distinct spectra from C, WT. WT* aggregates (E) exhibit a red shift of the amide I maximum to 1630 cm–1 compared with WT aggregates (∼1634 cm–1) and MUT aggregates (1632 cm–1). F, a curve fit example demonstrating the achieved accuracy (the calculated sum of band components superimposes the measured amide I band completely) and the five component bands used. These describe (high to low wave number)β-turns, α-helix, random coil, and β-sheet. For β-sheet, a high and a low frequency band were assumed (dashed lines). In A–C and E, six spectra from two preparations are superimposed, and five are superimposed in D. The main reasons for the large variability of the spectra between 1665 and 1645 cm–1 were residual water vapor and a low protein concentration in the samples, especially of WT fibrils. Scale bars = 5 × 10–3 arbitrary units. G, quantification of A–E. The results presented derive from curve fitting and averaging six spectra of two samples for each condition; error bars represent the S.E. *, p < 0.05.
Circular Dichroism—Primary and quaternary reactions were ultracentrifuged at 100,000 × g for 1 h at room temperature, and pellets were resuspended in phosphate-buffered saline to a final concentration of ∼4 μm prior to CD. Spectra were recorded on a Jasco spectrometer and reflect four accumulations.
Atomic Force Microscopy—Reactions were adsorbed onto mica chips (Ted Pella, Inc.) for 2 min, washed twice with 200 μl of water, and allowed to dry for at least 1 h prior to tapping mode atomic force microscopy (Veeco).
Fiber Fragility Assays—Whole primary and quaternary fibrillization reactions of WT, MUT, and WT* were dialyzed into phosphate-buffered saline for 15 h. CD spectra were measured from 250 to 200 nm, after which the reactions were sonicated with a titanium probe on a Branson Sonifier 250 at 50% duty and 10% intensity for 1 min, and CD spectra were again measured. The changes in WT, MUT, and WT* signals were compared using Wilcoxon signed rank statistical analysis in fragility assays. The data set (n = 6) presented meets all requirements for this type of one-sided, two-tailed nonparametric analysis (α < 0.025 and *, p = 0.0156 for all fragility assays).
Electron Microscopy—Quaternary fibrillization reactions were adsorbed onto glow-discharged Formvar/carbon copper grids (200 mesh; Ted Pella, Inc.). The grids were washed with ammonium acetate buffer, stained with uranyl acetate, dried, and viewed in an FEI Tecnai F20 electron microscope at 80 kV and a standard magnification of ×25,000 (20). Electron micrographs were recorded with a Gatan UltraScan CCD camera. The magnification was calibrated using negatively stained catalase crystals and ferritin. The percentage of paired helical filaments was compared between WT, MUT, and WT* quaternary reactions using Student's t test. The data set (n = 3) presented meets all requirements for this type of two-tailed, two-sample equal variance analysis (*, p < 10–5).
RESULTS
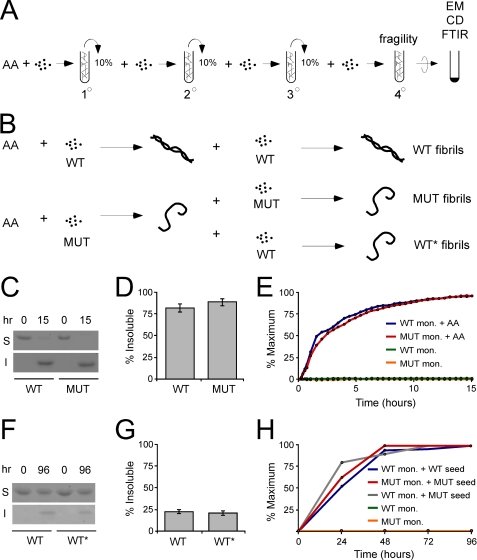
Fibril Seeding Strategy—We established a protocol to create pools of fibrillar protein under uniform conditions (Fig. 1, A and B). The primary WT and MUT fibrillization reactions were induced by arachidonic acid (17). The secondary reaction was a seeded reaction in which 4 μm monomer was incubated with 10% of the primary reaction in the absence of arachidonic acid for 3 h. The secondary seeding reaction was followed by two additional seeding reactions to produce the tertiary and quaternary reactions (Fig. 1A). To induce a novel fibrillar conformation of WT Tau, termed WT*, we performed a cross-seeding reaction in which the primary MUT reaction was incubated with WT monomer. Three serial seeding reactions were then performed using WT monomer (Fig. 1B). After 96 h, quaternary reactions were ultracentrifuged, and the insoluble fractions were compared using FTIR, CD, and EM. Whole reactions were compared for fragility assays. We first determined the extent of fibrillization and the fibrillization rate for each reaction. In primary reactions, 81% of WT monomer and 89% of MUT monomer were present in the insoluble fraction after 15 h (Fig. 1, C and D), indicating that most of the protein was in an aggregated form. Primary WT and MUT reactions had similar rates of fibrillization based on ThT fluorescence (Fig. 1E) (18). In quaternary seeded reactions, WT and WT* also had similar degrees of aggregation. 22% of WT and 21% of WT* quaternary reactions were insoluble after 96 h (Fig. 1, F and G). Monitoring ThT fluorescence of the quaternary reactions produced a t½ of 24 h for WT, 18 h for MUT, and 13 h for WT* (Fig. 1H). Despite different t½ values, all reactions reached a plateau by ∼48 h.
FIGURE 1.
Fibril seeding. A, arachidonic acid (AA) is used to stimulate monomer fibrillization in the primary reaction. In the secondary reaction, 10% of the primary reaction is used to seed the fibrillization of Tau monomer. In the tertiary reaction, 10% of the secondary reaction is used to seed the fibrillization of Tau monomer. In the quaternary reaction, 10% of the tertiary reaction is used to seed the fibrillization of Tau monomer. For fragility studies, the whole quaternary reaction was used, uncentrifuged. For FTIR, CD, and EM, the quaternary reaction was ultracentrifuged for 1 h at 100,000 × g, and the pellet was used for measurements. B, arachidonic acid is used to stimulate WT or MUT fibrillization in the primary reaction. To generate WT fibrils, 10% of the primary WT reaction is incubated with WT monomer, followed by three serial seeding reactions as described in A. To generate MUT fibrils, 10% of the primary MUT reaction is incubated with MUT monomer, followed by three serial seeding reactions as described in A. To generate WT* fibrils, 10% of the primary MUT reaction is incubated with WT monomer, followed by three serial seeding reactions as described in A. C, after 15 h, primary WT and MUT have comparable degrees of fibrillization as determined by solubility. Primary reactions were ultracentrifuged for 1 h at 100,000 × g. The soluble (S) and insoluble (I) fractions were compared by Coomassie stain. D, shown is quantification of three separate experiments. After 15 h, 81% of WT Tau monomer is insoluble versus 89% of MUT Tau, indicating comparable degrees of fibrillization. E, primary WT and MUT fibrillization reactions were initiated with arachidonic acid and measured over time using ThT fluorescence at ex455/em485. Both reactions proceeded at a similar rate. mon., monomer. F, after 96 h, WT and WT* quaternary reactions have comparable degrees of fibrillization. Quaternary reactions were ultracentrifuged for 1 h at 100,000 × g. The soluble and insoluble fractions were compared by Coomassie stain. G, shown is quantification of three separate experiments. After 96 h, 22% of WT Tau monomer is insoluble versus 21% of WT* Tau, indicating comparable degrees of fibrillization. H, WT monomer, MUT monomer, WT, MUT, and WT* quaternary reactions were monitored for 96 h using ThT fluorescence at ex455/em485. The t½ values for WT, MUT, and WT* are 24, 18, and 13 h, respectively. All reactions reach a plateau by ∼48 h.
WT and WT* Fibrils Have Different Secondary Structures Based on Fourier Transform Infrared Spectroscopy—We used FTIR to compare monomer and quaternary fibril structures. The applied data processing is extremely sensitive for structural differences, which enabled a detailed comparison of the selected Tau conformations. WT and MUT monomer showed no appreciable differences in their secondary structure (Fig. 2, A, B, and G). In contrast, quaternary WT and MUT Tau fibrils exhibited distinct FTIR spectra. WT Tau had relatively more heterogeneity in signal, partially because of lower signal intensity, but also possibly representing a larger diversity of structures, whereas MUT Tau was quite homogeneous (Fig. 2, C and D). Spectral deconvolution indicated that, with a significance level of 5% in a Student's t test, WT fibrils contained significantly more α-helix, significantly less β-sheet, and relatively less β-turn than MUT (Fig. 2G). Likewise, quaternary WT* spectra were clearly distinct from WT spectra, with significantly less α-helical and apparently greater random coil content (Fig. 2, E and G). The absolute values for the secondary structure fractions are only estimates (approximately ±10–15%) because a structural template for calibration of Tau secondary structure analysis does not exist (19). Still, the applied method is highly sensitive for spectral differences between samples of a similar origin (19, 21, 22). Thus, although we cannot define with absolute assurance the structure of the fibrils, FTIR spectral differences between WT and WT* fibrils are clear.
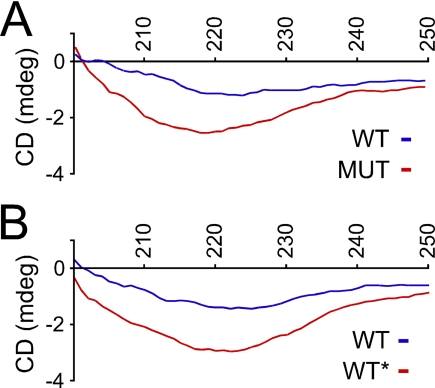
WT and WT* Fibrils Have Distinct Secondary Structures Based on Circular Dichroism—To extend the FTIR studies, we used CD spectroscopy to compare WT, MUT, and WT* fibrils. We prepared primary reactions as described above. After 15 h, we ultracentrifuged the reactions and resuspended the pellets in phosphate-buffered saline. The MUT minimum occurred at 218 nm versus 223 nm for WT fibrils. These minima indicate that MUT fibrils contain more β-sheet structure than WT fibrils and that WT fibrils contain more α-helix, consistent with data obtained from FTIR. The minimum for MUT fibrils was also consistently deeper than that of WT fibrils. We then compared CD spectra of quaternary WT and WT* reactions, prepared as previously described, again measuring only the insoluble material. The CD spectrum of quaternary WT* was clearly different from that of quaternary WT (Fig. 3B), also with a deeper minimum signal. These data supported the conformational differences suggested by FTIR. Some quantitative differences in WT spectra were observed between experiments, possibly from variations in batches of arachidonic acid or Tau protein preparations. Qualitative differences between WT and MUT spectra were consistently observed within each experiment (Fig. 3A). To assure that the spectra observed were due to aggregates arising from the seeding reaction and not simply those carried over from the primary reaction, we serially diluted primary WT and MUT reactions. The resulting spectra from the quaternary dilutions are non-existent (supplemental Fig. 1A); thus, aggregates in the primary seeding reaction (which are diluted 1,000-fold) are not sufficient to produce the signals observed in quaternary reactions. We also ruled out the possibility that minor differences in protein concentration could account for the spectral differences between the samples (supplemental Fig. 1B).
FIGURE 3.
CD analysis of WT, MUT, and WT* Tau fibrils. All compared samples had similar protein concentrations (<10% difference). A, insoluble material from primary WT and MUT fibrillization reactions have distinct CD spectra. B, insoluble material from quaternary WT and WT* fibrillization reactions have distinct CD spectra. Spectra reflect four accumulations each. mdeg, millidegrees.
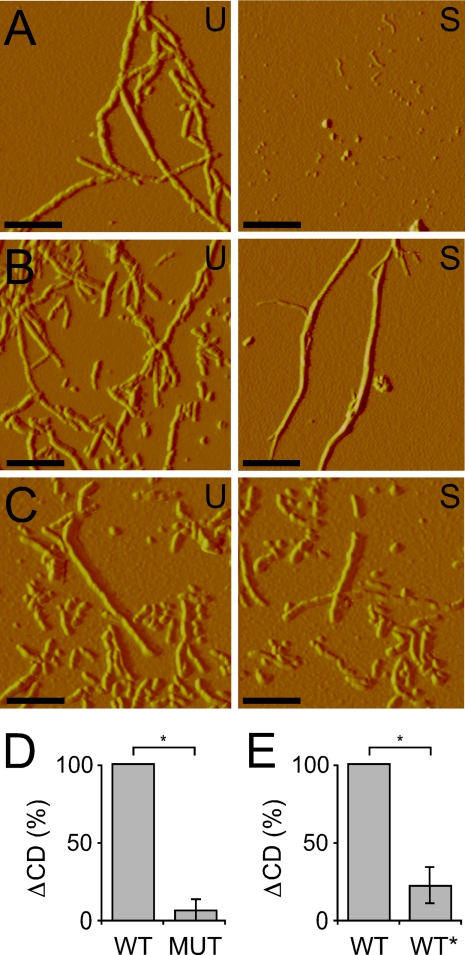
WT and WT* Fibrils Have Distinct Fragilities—Prior reports indicate that Sup35 prion strains are differentially susceptible to sonication-induced shearing (23), and long β2-microglobulin amyloid fibrils exhibit deeper CD spectroscopic minima at 223 nm than short fibrils (24), implying that CD changes before and after sonication can be used to evaluate fibril strength. Consequently, as an additional comparison of structural differences between the fibril types, we developed a method to evaluate the relative sensitivity of WT, MUT, and WT* fibrils to sonication-induced breakage. First, we found that sonication treatment disproportionately disrupted quaternary WT Tau fibrils, but not MUT or WT* fibrils, based on atomic force microscopy (Fig. 4, A–C). To quantify fragility, we compared the CD spectra at 223 nm of the primary reactions before and after sonication at 10% intensity. Like β2-microglobulin fibrils, sonication of Tau fibrils resulted in shorter fibrils that gave rise to a shallower minimum at 223 nm. After six experiments, the relative loss of MUT CD signal was 19% of the loss WT signal (Fig. 4D and supplemental Fig. 2, A and B). MUT fibrils break down to an extent similar to WT fibrils after sonication at 20% intensity (data not shown). WT* fibrils were also stronger than WT fibrils and less disrupted by sonication. Quaternary WT* fibrils exhibited 21% of the loss of signal versus that of quaternary WT fibrils (Fig. 4E and supplemental Fig. 2, C and D). Thus, the differences between WT and WT* observed by FTIR and CD spectroscopy are supported by distinct fibril fragilities.
FIGURE 4.
WT fibrils are more fragile than MUT and WT* fibrils. A, primary WT fibrils were visualized by atomic force microscopy, untreated (U) and after sonication (S). Scale bars = 0.25 μm. B, identically prepared primary MUT fibrils, untreated and after sonication, demonstrate the resistance of fibrils to sonication. C, quaternary WT* fibrils, untreated and after sonication, demonstrate the resistance of fibrils to sonication. D, quantification of the loss of CD signal at 223 nm for WT and MUT primary Tau fibrillization reactions before and after sonication indicates MUT fibrils are stronger than WT fibrils. Loss of signal for MUT was calculated as a fraction of loss of signal for WT, which was set to 100%. *, p = 0.0156 (n = 6). E, quantification of the loss of CD signal at 223 nm for WT and WT* quaternary Tau fibrillization reactions before and after sonication indicates WT* Tau fibrils are stronger than WT fibrils. *, p = 0.0156 (n = 6). Loss of signal for WT* was calculated as a fraction of loss of signal for WT, which was set to 100%.
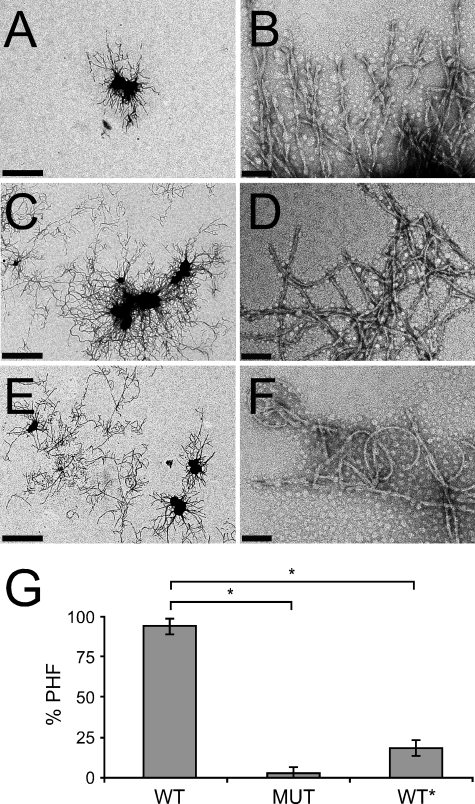
WT and WT* Fibrils Have Different Morphologies Based on Electron Microscopy—As a final comparison of the distinct structures, we used negative stain EM to evaluate the morphologies of the three fibril types. WT fibrils consisted of predominantly paired helical filaments (Fig. 5, A and B), whereas MUT Tau fibrils were more similar to straight filaments. They were thinner, lacked the typical paired helical morphology, and displayed a higher degree of flexibility (Fig. 5, C and D). WT* fibrils were distinct from WT fibrils in their lack of predominant paired helical structure and had similar curvature and width to MUT Tau (Fig. 5, E and F). When three separate experiments were counted and quantified, we found significant differences in the proportion of paired helical filaments in WT fibril preparations versus MUT and in WT versus WT*. 94% of WT fibrils appeared as paired helical filaments, whereas only 2% of MUT fibrils and 18% of WT* fibrils were helical in nature and instead appeared as straight, flexible fibrils (Fig. 5G). Taken together with the preceding experiments, these data confirm our findings that WT Tau fibrils can adopt a distinct, stable, fibrillar structure that is propagated via templated conformation change.
FIGURE 5.
Negative stain electron microscopy of Tau fibrils. High (A) and low (B) magnification EM images show that quaternary WT fibrils have a paired helical filament morphology. In contrast, high (C) and low (D) magnification EM images show that quaternary MUT fibrils have a distinct, curved morphology that lacks the helical appearance of the WT fibrils. High (E) and low (F) magnification EM images show that quaternary WT* fibrils have a curved morphology similar to MUT Tau. Scale bars in A, C, and E = 1 μm; scale bars in B, D, and F = 0.1 μm. G, shown is the quantification of three separate experiments. 94% of quaternary WT fibrils are paired helical filaments (PHF), whereas 2% of MUT fibrils and 18% of WT* are paired helical filaments. *, p < 10–5.
DISCUSSION
In this study, we tested the hypothesis that WT Tau can assume distinct fibrillar conformations that are propagated serially by templated conformation change. We have exploited an artificial protein, MUT Tau, containing two disease-causing mutations as a convenient tool with which to drive WT Tau into a distinct fibrillar conformation, termed WT*. We have compared WT, MUT, and WT* fibril conformations using four independent measures: FTIR, CD spectroscopy, sensitivity to sonication-induced breakage, and EM analysis of fibril morphology. Each of these assays demonstrated that WT* represents a novel conformation of WT fibril that is maintained over time through templated conformation change.
Our data are consistent with the idea that sporadic tauopathies, which involve only WT Tau, might derive phenotypic diversity from conformational differences in WT Tau fibrils. In addition to MUT Tau fibrils, we have also observed that ΔK280 Tau fibrils can seed a new conformation of WT Tau (supplemental Fig. 3), suggesting that the ability to induce WT Tau to adopt unique conformations by templated conformation change is not restricted to a particular seed. It remains to be determined to what extent a mutant Tau protein in patients can trigger misfolding of the wild-type and whether other factors might play a role. Indeed, the stimuli that induce WT Tau to form fibrils of a unique conformation in sporadic tauopathy patients could be myriad: splice isoforms, post-translational modifications, a heterologous seed (e.g. amyloid β), oxidation events, etc. Our experiments suggest a theoretical framework with which to consider phenotypic diversity in the sporadic tauopathies. Once WT Tau is sent down a particular conformational path, whatever the inciting stimulus, it maintains this distinct conformation via templated conformation change.
Recent evidence from studies of prion disease suggests a strong link between the conformation of a fibrillar protein and the resultant phenotype (8, 9). According to this model, once a protein adopts a particular fibrillar conformation, that conformation is propagated over time through subsequent seeded fibrillization reactions. The properties of a given fibril type (such as growth rate, fragility, and subsequent toxicity) play an important role in specifying phenotypic features (23). Our results support data reported by others that multiple amyloid-forming proteins have the ability to form distinct conformers based on templated conformation change (13, 14). Indeed, the seeding barrier that has been reported between P301L mutant Tau and WT Tau is reminiscent of prion strains (11, 25) and is consistent with our hypothesis, which would allow for cross-seeding barriers to exist between Tau mutants. Our studies indicate that, like other amyloid proteins such as amyloid β, β2-microglobulin, and prion protein, WT Tau can adopt multiple distinct fibrillar conformations that are maintained over time via templated conformation change. In future experiments, it will be fascinating to determine whether phenotypic diversity in sporadic tauopathies can be linked to unique WT Tau fibril conformations.
Supplementary Material
Acknowledgments
We thank Jonathan Weissman for use of the atomic force microscope and David Agard and Brian Shoichet for use of their Jasco spectrophotometers.
This work was supported, in whole or part, by National Institutes of Health Grant R01 NS50284-03 from NINDS (to M. I. D.) and a National Institutes of Health Training Grant for Neurodegenerative Diseases (to B. F.). This work was also supported by the Sandler Family Supporting Foundation, the Taube Family Foundation Program in Huntington's Disease Research, and the Muscular Dystrophy Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: WT, wild-type; FTIR, Fourier transform infrared spectroscopy; ThT, thioflavin T; EM, electron microscopy; MUT, P301L/V337M double mutation.
References
- 1.Braak, H., and Braak, E. (1991) Acta Neuropathol. 82 239–259 [DOI] [PubMed] [Google Scholar]
- 2.Hutton, M., Lendon, C. L., Rizzu, P., Baker, M., Froelich, S., Houlden, H., Pickering-Brown, S., Chakraverty, S., Isaacs, A., Grover, A., Hackett, J., Adamson, J., Lincoln, S., Dickson, D., Davies, P., Petersen, R. C., Stevens, M., de Graaff, E., Wauters, E., van Baren, J., Hillebrand, M., Joosse, M., Kwon, J. M., Nowotny, P., Che, L. K., Norton, J., Morris, J. C., Reed, L. A., Trojanowski, J., Basun, H., Lannfelt, L., Neystat, M., Fahn, S., Dark, F., Tannenberg, T., Dodd, P. R., Hayward, N., Kwok, J. B., Schofield, P. R., Andreadis, A., Snowden, J., Craufurd, D., Neary, D., Owen, F., Oostra, B. A., Hardy, J., Goate, A., van Swieten, J., Mann, D., Lynch, T., and Heutink, P. (1998) Nature 393 702–705 [DOI] [PubMed] [Google Scholar]
- 3.Spillantini, M. G., Murrell, J. R., Goedert, M., Farlow, M. R., Klug, A., and Ghetti, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7737–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poorkaj, P., Bird, T. D., Wijsman, E., Nemens, E., Garruto, R. M., Anderson, L., Andreadis, A., Wiederholt, W. C., Raskind, M., and Schellenberg, G. D. (1998) Ann. Neurol. 43 815–825 [DOI] [PubMed] [Google Scholar]
- 5.Lewis, S. A., Wang, D. H., and Cowan, N. J. (1988) Science 242 936–939 [DOI] [PubMed] [Google Scholar]
- 6.Wischik, C. M., Novak, M., Thogersen, H. C., Edwards, P. C., Runswick, M. J., Jakes, R., Walker, J. E., Milstein, C., Roth, M., and Klug, A. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 4506–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buee, L., Bussiere, T., Buee-Scherrer, V., Delacourte, A., and Hof, P. R. (2000) Brain Res. 33 95–130 [DOI] [PubMed] [Google Scholar]
- 8.Tanaka, M., Chien, P., Naber, N., Cooke, R., and Weissman, J. S. (2004) Nature 428 323–328 [DOI] [PubMed] [Google Scholar]
- 9.Legname, G., Nguyen, H. O., Peretz, D., Cohen, F. E., DeArmond, S. J., and Prusiner, S. B. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19105–19110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Bergen, M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E. M., and Mandelkow, E. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyagi, H., Hasegawa, M., and Tamaoka, A. (2007) J. Biol. Chem. 282 20309–20318 [DOI] [PubMed] [Google Scholar]
- 12.Crowther, R. A., and Goedert, M. (2000) J. Struct. Biol. 130 271–279 [DOI] [PubMed] [Google Scholar]
- 13.Petkova, A. T., Leapman, R. D., Guo, Z., Yau, W. M., Mattson, M. P., and Tycko, R. (2005) Science 307 262–265 [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi, K., Takahashi, S., Kawai, T., Naiki, H., and Goto, Y. (2005) J. Mol. Biol. 352 952–960 [DOI] [PubMed] [Google Scholar]
- 15.Bessen, R. A., Kocisko, D. A., Raymond, G. J., Nandan, S., Lansbury, P. T., and Caughey, B. (1995) Nature 375 698–700 [DOI] [PubMed] [Google Scholar]
- 16.Goedert, M., and Jakes, R. (1990) EMBO J. 9 4225–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson, D. M., and Binder, L. I. (1995) J. Biol. Chem. 270 24306–24314 [DOI] [PubMed] [Google Scholar]
- 18.LeVine, H., III (1993) Protein Sci. 2 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ollesch, J., Kunnemann, E., Glockshuber, R., and Gerwert, K. (2007) Appl. Spectrosc. 61 1025–1031 [DOI] [PubMed] [Google Scholar]
- 20.Wille, H., Govaerts, C., Borovinskiy, A., Latawiec, D., Downing, K. H., Cohen, F. E., and Prusiner, S. B. (2007) Arch. Biochem. Biophys. 467 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ollesch, J., Poschner, B. C., Nikolaus, J., Hofmann, M. W., Herrmann, A., Gerwert, K., and Langosch, D. (2008) Eur. Biophys. J. 37 435–445 [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, M. W., Weise, K., Ollesch, J., Agrawal, P., Stalz, H., Stelzer, W., Hulsbergen, F., de Groot, H., Gerwert, K., Reed, J., and Langosch, D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14776–14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka, M., Collins, S. R., Toyama, B. H., and Weissman, J. S. (2006) Nature 442 585–589 [DOI] [PubMed] [Google Scholar]
- 24.Adachi, R., Yamaguchi, K., Yagi, H., Sakurai, K., Naiki, H., and Goto, Y. (2007) J. Biol. Chem. 282 8978–8983 [DOI] [PubMed] [Google Scholar]
- 25.Miyasaka, T., Morishima-Kawashima, M., Ravid, R., Kamphorst, W., Nagashima, K., and Ihara, Y. (2001) J. Neuropathol. Exp. Neurol. 60 872–884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.