Abstract
The immune response to infection includes activation of the blood clotting system, leading to extravascular fibrin deposition to limit the spread of invasive microorganisms. Some bacteria have evolved mechanisms to counteract this host response. Pla, a member of the omptin family of Gram-negative bacterial proteases, promotes the invasiveness of the plague bacterium, Yersinia pestis, by activating plasminogen to plasmin to digest fibrin. We now show that the endogenous anticoagulant tissue factor pathway inhibitor (TFPI) is also highly sensitive to proteolysis by Pla and its orthologs OmpT in Escherichia coli and PgtE in Salmonella enterica serovar Typhimurium. Using gene deletions, we demonstrate that bacterial inactivation of TFPI requires omptin expression. TFPI inactivation is mediated by proteolysis since Western blot analysis showed that TFPI cleavage correlated with loss of anticoagulant function in clotting assays. Rates of TFPI inactivation were much higher than rates of plasminogen activation, indicating that TFPI is a better substrate for omptins. We hypothesize that TFPI has evolved sensitivity to proteolytic inactivation by bacterial omptins to potentiate procoagulant responses to bacterial infection. This may contribute to the hemostatic imbalance in disseminated intravascular coagulation and other coagulopathies accompanying severe sepsis.
Introduction
Aside from its role in minimizing blood loss, the clotting system is also an effector arm of the immune system that modulates release of inflammatory mediators and prevents the spread of invasive microorganisms through extravascular fibrin deposition. This “quarantine through coagulation” strategy is evolutionarily primitive, as evidenced by the sensitivity of the horseshoe crab clotting system to activation by bacterial endotoxin.1 Indeed, fibrin deposition is one of the hallmarks of inflammation in humans, and the induration accompanying delayed-type hypersensitivity skin reactions is caused by extravascular fibrin deposition.2 Inflammatory responses to infection include expression of tissue factor on activated monocytes as well as increased acute phase proteins like fibrinogen and PAI-I, which contribute to hypercoagulable states.3
Some bacteria modulate the host coagulation system to evade immune responses or facilitate dissemination through extravascular tissues. For example, staphylocoagulase and streptokinase form complexes with prothrombin or plasminogen, converting them into prothrombotic (thrombin) or profibrinolytic (plasmin) enzymes.4 Likewise, plasminogen activator (Pla), a member of the omptin family of bacterial proteases, promotes dissemination of the plague agent, Yersinia pestis, from subcutaneous sites to the lymphatic and circulatory systems.5 Proteolytic activation of plasminogen by Pla is thought to facilitate Y pestis migration through tissue barriers, because plasmin degrades fibrin clots and extracellular matrix components, activates procollagenases, and inactivates collagenase inhibitors.6–8 Infection of plasminogen-knockout mice with wild-type Y pestis and of wild-type mice with Pla-deficient Y pestis both resulted in greater survival outcomes than infection of wild-type mice with wild-type Y pestis. Eliminating fibrinogen expression subverted the survival benefit of knocking out host plasminogen or pathogen Pla.9 The blood clotting system is therefore a main pathophysiologic target of Pla.
Omptins are a family of outer membrane proteases expressed in Gram-negative bacteria that share approximately 50% sequence homology and preferentially proteolyze substrates between 2 basic residues.10 Structurally, they are composed of 10 antiparallel β-strands forming a vase-shaped barrel embedded in the outer membrane. Lipopolysaccharide (LPS) is required for omptin enzymatic activity, and furthermore short O-antigen side chains (rough LPS) are required for omptin activity toward many exogenous macromolecular substrates.11,12 The cocrystal structure of LPS complexed to FhuA, an outer membrane protein with the same overall structure as the Escherichia coli omptin, OmpT, revealed a binding site for LPS and suggested that extended O-antigen side chains (smooth LPS) could sterically interfere with substrate binding.13
Mechanisms commonly cited for activation of clotting in sepsis center on release of inflammatory signals and up-regulation of tissue factor expression.14 The present study defines a mechanism by which Gram-negative bacteria interact directly with the clotting system to induce a hypercoagulable state. We show that bacterial omptins proteolytically inactivate an essential coagulation inhibitor, tissue factor pathway inhibitor (TFPI). The initiation phase of blood clotting involves the formation of an enzyme complex between tissue factor and factor VIIa (TF:FVIIa), which then proteolytically activates factors X (FX),15 IX (FIX),16 and VII (FVII).17 TFPI functions initially by forming stable TFPI-FXa complexes, which are potent inhibitors of TF:FVIIa, blocking further initiation of blood clotting.18 We hypothesize that sensitivity of TFPI to proteolytic inactivation by bacterial omptins has evolved in mammalian hosts to potentiate protective, procoagulant responses to bacterial infection. With the heavy bacterial loads commonly present in the circulation during the late stages of severe septicemia, this interaction may unfortunately accelerate the development of disseminated intravascular coagulation (DIC) and end-organ failure.
Methods
Materials
Monoclonal antibodies targeting the Kunitz-1 and Kunitz-2 domains of TFPI, and recombinant human TFPI expressed and purified from E coli and from SK-hepatoma cells, were generous gifts from Dr George Broze (Washington University, St Louis, MO). Thromboplastin reagents were either Innovin (Dade-Behring, Marburg, Germany) or 3 to 4 ng/mL recombinant human tissue factor relipidated into 80% phosphatidylcholine/20% phosphatidylserine vesicles22 in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.4, 25 mM CaCl2. Human Glu-plasminogen and plasmin were from Hematologic Technologies (Essex Junction, VT). Human pooled normal plasma and plasmas deficient in prothrombin, factor V (FV), FVII, or FX were from George King Bio-Medical (Overland Park, KS). Human FVII and FVIIa were from Enzyme Research Laboratories (South Bend, IN). Chromogenic substrates, Spectrozyme PL, and Spectrozyme FVIIa, were from American Diagnostica (Stamford, CT).
Bacterial strains, growth conditions, and reagents
Bacterial strains are described in Table 1. E coli and Salmonella enterica serovar Typhimurium were grown in Luria-Bertani (LB) medium, while Y pestis was grown on heart infusion or LB media. E coli and S Typhimurium were grown at 37°C except for strains containing temperature-sensitive plasmids pCP20 and pKD46, which were grown at 30°C. Y pestis was grown at room temperature, except when curing temperature-sensitive plasmids at 37°C. Antibiotic concentrations were 100 μg/mL ampicillin and 20 μg/mL chloramphenicol (Cm).
Table 1.
Bacterial strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MC4100 | E coli K-12 F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flhD5301 deoC1 ptsF25 rbsR | 19 |
| 14028 | Wild-type S enterica serovar Typhimurium | ATCC* |
| KIM5 | Y pestis Δpgm | 20 |
| JS83 | 14028 galE496 | 21 |
| JEC51 | MC4100 ΔompT::Cm | This study |
| JS814 | 14028 ΔgalETK2918::Cm | This study |
| JS815 | 14028 ΔgalETK2918::Cm ΔpgtE | This study |
| JEC182 | KIM5 Δpla::Cm | This study |
ATCC, Manassas, VA.
Strain construction
Gene deletions were made using λ Red-mediated recombination as previously described.23,24 In S Typhimurium, constructs were moved into a clean wild-type background or recipient strains with other known deletion(s) by P22 HT105/1 int-201 (P22)–mediated transduction.25 In some cases, antibiotic resistance cassettes were removed using the temperature-sensitive plasmid, pCP20, carrying FLP recombinase.26 In all cases, appropriate insertion of antibiotic resistance markers was confirmed by PCR.
TFPI incubations/pulldowns
Overnight bacterial cultures were washed once with HBS (20 mM HEPES, pH 7.4, 140 mM NaCl, 0.1% polyethylene glycol [PEG]–8000, 0.1% bovine serum albumin [BSA]), resuspended in HBS or pooled normal plasma containing various concentrations of TFPI, then rotated at room temperature for various times. To test for binding to bacteria, TFPI incubation mixtures were centrifuged at 16 000g for 4 minutes. Supernatants were removed, and the bacterial pellet was washed with HBS and resuspended in HBS. For sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis, samples in nonreducing SDS sample buffer were resolved on 15% Tris-glycine SDS gels (Bio-Rad Laboratories, Hercules, CA). For Western blot analysis, proteins were blotted onto polyvinylidene fluoride (PVDF) membranes and probed with 1 μg/mL anti-TFPI monoclonal antibodies (targeting Kunitz-1 or Kunitz-2 domains) followed by peroxidase-conjugated anti–mouse immunoglobulin G (IgG) and visualization using enhanced chemiluminescence (ECL)–plus Western blotting kits (Invitrogen, Carlsbad, CA).
Preparation of bacterial lysates/cell envelope fractions
Overnight E coli MC4100 cultures at 109 colony forming units (cfu)/mL were washed once, resuspended in HBS, and lysed by sonication. To isolate cell envelope fractions, lysates were centrifuged at 150 000g for 1.5 hours at 4°C. Supernatants were removed, and pellets were resuspended in HBS.
Clotting assays
Samples (50 μL) of TFPI/bacterial incubation mixtures or pulldown samples were combined with 50 μL pooled normal plasma and 50 μL thromboplastin reagent at 37°C to initiate clotting. Time to clot formation was measured using a STart4 coagulometer (Diagnostica Stago, Parsippany, NJ). TFPI anticoagulant activities were quantified by reference to a standard curve generated with purified TFPI.
Measurement of TFPI inactivation rates
Mixtures of 30 nM TFPI and bacteria (107 cfu/mL E coli MC4100 or Y pestis KIM5, or 109 cfu/mL S Typhimurium ΔgalETK) were incubated at 37°C for various times, after which clotting activities were measured. In all instances, the time to clot formation constituted less than 10% of the total incubation time. Linear regression of plots of TFPI activity versus incubation time yielded a line (R2 > 0.95) whose slope represented the rate of TFPI inactivation.
Clotting factor assays
Bacteria (2 × 108 cfu/mL) were incubated with pooled normal plasma at room temperature for 1 hour. Mixtures were then diluted 20-fold into imidazole buffer (50 mM imidazole, pH 7.4, 150 mM NaCl, 1% BSA). Time to clot formation was measured after mixture of 50 μL of this dilution plus 50 μL factor-deficient plasma and 100 μL thromboplastin reagent. Clotting factor activities were determined by reference to standard curves generated using mixtures of varying proportions of pooled normal plasma and factor-deficient plasma.
Mass spectrometry of TFPI cleavage fragment
Determination of TFPI cleavage fragment mass was performed by the University of Illinois at Urbana-Champaign Protein Sciences Facility. Briefly, 24-hour incubations of 1010 cfu/mL E coli MC4100 and 100 μg/mL TFPI were centrifuged at 16 000g for 4 minutes. TFPI fragments in the supernatant fraction were resolved on a C18 reverse-phase high-pressure liquid chromatography (HPLC) column and the major peak was analyzed by electrospray ionization mass spectrometry.
Results
An outer membrane component of E coli interacts directly with TFPI to abrogate its function
We investigated whether bacteria could modulate the function of TFPI, the major endogenous inhibitor of the initiation phase of blood clotting. Adding purified TFPI to plasma prolongs the clotting time initiated by tissue factor, which is a convenient and sensitive way to evaluate its anticoagulant function. In initial experiments, we added E coli K-12 strain, MC4100, to plasma containing sufficient TFPI to greatly prolong the clotting time; inactivation of TFPI will therefore reduce the clotting time back to the TFPI-free value. E coli abrogated the anticoagulant effect of1 μg/mL TFPI (Figure 1A), and the TFPI dose-response was much shallower in the presence of E coli (Figure 1B). The ability of E coli to counteract TFPI anticoagulant function could be due to a direct or indirect action: (1) bacteria could inactivate TFPI (via binding, proteolysis, etc) or (2) bacteria could modify another plasma protein to override TFPI's anticoagulant effect. For example, converting FV to FVa abrogates TFPI's anticoagulant function because FVa protects FXa within the prothrombinase complex from inhibition by TFPI.27
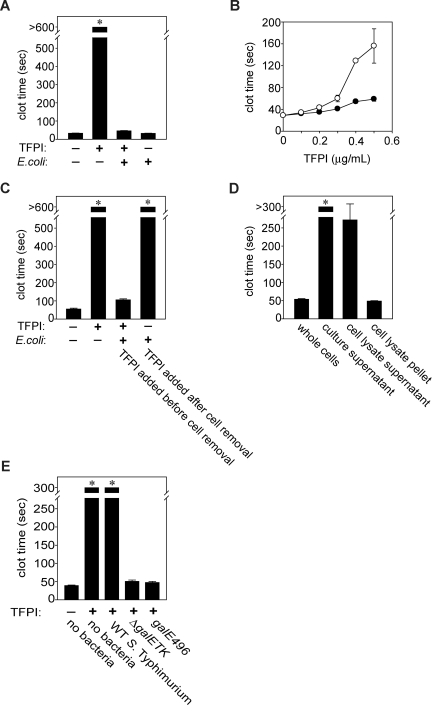
Figure 1.
Gram-negative bacteria abrogate TFPI anticoagulant activity by interacting with an outer membrane component that is dependent on LPS O-antigen length. (A) Loss of TFPI activity. E coli MC4100 (109 cfu/mL) were incubated with or without 1 μg/mL TFPI in plasma for 1 hour at room temperature, after which clotting times were measured. (B) Dependence on TFPI concentration. E coli MC4100 (109 cfu/mL) were incubated with varying TFPI concentrations in human plasma for 30 minutes at room temperature, after which clotting times were quantified (●), compared with samples containing TFPI but no bacteria (○). (C) Requirement for physical contact with TFPI. E coli MC4100 (109 cfu/mL) were incubated with 1 μg/mL TFPI for 30 minutes, then bacteria were removed by centrifugation and TFPI activity tested in clotted assays. In parallel experiments, E coli MC4100 were incubated in TFPI-free plasma for 30 minutes, removed by centrifugation, and 1 μg/mL TFPI was added to the plasma supernatants, after which clotting assays were performed. (D) TFPI inactivation by cell fractions containing the E coli envelope. E coli MC4100 (109 cfu/mL) were lysed and fractionated, and the indicated cell fractions were incubated with TFPI (3 μg/mL) in HBS for 30 minutes at room temperature, then tested in clotting assays as before. (E) Only S Typhimurium strains expressing truncated O-antigen reduced clotting times. S Typhimurium (109 cfu/mL) were incubated with 3 μg/mL TFPI in HBS for 30 minutes at room temperature, after which bacteria were removed by centrifugation and supernatants tested in clotting assays. The wild-type strain, 14028, expresses extended O-antigen side chains, while strains harboring a deletion (ΔgalETK) or mutation (galE496) in the UDP-4-galactose-epimerase gene express truncated O-antigen. Samples that did not clot by 600 (A,C) or 300 seconds (D,E) are indicated by asterisks. Data are mean plus SEM (n = 3).
To investigate these 2 possibilities, bacteria were incubated with plasma for 30 minutes, then removed by centrifugation. Clotting times exceeded 600 seconds when 1 μg/mL TFPI was subsequently added to the bacteria-treated plasma, indicating no detectable reduction in TFPI activity (Figure 1C). On the other hand, when TFPI and bacteria were incubated together in plasma and then the bacteria removed, the clotting times were drastically shortened. This indicates that E coli must be in physical contact with TFPI to shorten the clotting time, suggesting they antagonize TFPI anticoagulant activity not through FV activation or otherwise “conditioning” the plasma, but rather by direct interaction with TFPI.
The above experiments used whole E coli. To identify the subcellular location of the procoagulant agent, bacterial lysates were ultracentrifuged to separate the cell envelope from cytoplasm/periplasm contents. Particulate-free supernatants containing soluble cytoplasmic/periplasmic material failed to reverse the prolongation of clotting time by TFPI, while the resuspended pellet fraction potently reversed TFPI anticoagulant activity (Figure 1D). Because the pellet contained insoluble cell envelope debris, this suggested that the anti-TFPI activity of the bacteria was attributable to an outer membrane component. The putative procoagulant agent was not secreted, because incubating E coli–conditioned medium with TFPI failed to reverse TFPI's anticoagulant effect (Figure 1D). Furthermore, washing bacteria with buffer before incubating them with TFPI did not remove the procoagulant effect (not shown).
Rough LPS is required for S Typhimurium to abrogate TFPI anticoagulant function
We examined the ability of another Gram-negative enteric bacterium, S Typhimurium, to antagonize TFPI function. In this case, we observed ablation of TFPI anticoagulant function only with strains genetically modified to express rough LPS. LPS O-antigen is missing in E coli K-12 strains,28 whereas the wild-type S Typhimurium strain 14028 produces O-antigen composed of polysaccharide repeats. S Typhimurium strains that are mutant in galE, which encodes UDP-galactose-4′-epimerase, do not synthesize the LPS outer core or O-antigen. Accordingly, we incubated a galE deletion mutant, ΔgalETK, with TFPI for 30 minutes, removed the bacteria and tested the supernatant. We found clot times similar to those without added TFPI and dramatically shorter than clot times obtained using the wild-type strain (Figure 1E). We also tested a S Typhimurium galE point mutant, galE496, which expresses limited O-antigen (< 1% wild-type), again observing clotting times consistent with nearly total abrogation of TFPI function. O-antigen therefore inhibits the ability of S Typhimurium to antagonize TFPI function, probably by blocking access to another outer membrane component.
TFPI inactivation occurs through proteolytic processing by omptins
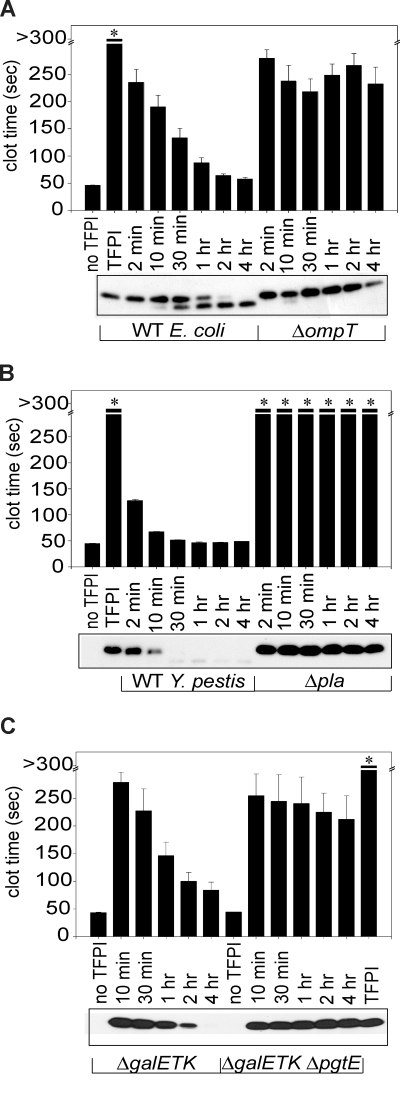
Previous studies have shown that rough LPS is required for proteolytic activation of plasminogen by the omptins, Pla (expressed by Y pestis) and PgtE (expressed by Salmonella).29 Due to the rough LPS requirement for TFPI inactivation, we hypothesized that omptins were responsible for the ability of E coli and S Typhimurium to block TFPI anticoagulant function, via proteolysis. We also hypothesized that Pla expressed on the surface of Y pestis might also abrogate TFPI function via proteolysis. To evaluate TFPI proteolysis, TFPI was incubated with S Typhimurium ΔgalETK and wild-type strains of E coli or Y pestis, resolved on SDS-PAGE, Western blot analyzed, and detected using monoclonal anti-TFPI antibodies. TFPI anticoagulant activity was quantified in parallel samples from these time courses. When TFPI was incubated with omptin-positive cells of all 3 bacteria, we observed a progressive reduction in band intensity for full-length TFPI (Figure 2A-C bottom panels), correlating with loss of TFPI anticoagulant function (Figure 2A-C top panels). In contrast, incubating TFPI with the omptin deletion strains, Y pestis Δpla, E coli ΔompT, or S Typhimurium ΔgalETK ΔpgtE, resulted in almost no loss in band intensity for full-length TFPI even after 4 hours (Figure 2). We also observed little or no loss of TFPI anticoagulant activity with these deletion mutants. Taken together, these results demonstrate that omptins are required for TFPI functional inactivation by bacteria via proteolytic processing of TFPI.
Figure 2.
Omptin gene deletion prevents E coli and S Typhimurium from inactivating TFPI. Incubation of TFPI with omptin-positive bacterial strains resulted in time-dependent reductions of clotting times (bar graphs), which coincided with reductions in band intensity of full-length TFPI on Western blots (photographs). TFPI (1 μg/mL) was incubated with (A) E coli MC4100 or ΔompT (107 cfu/mL) or (B) Y pestis KIM5 or Δpla (108 cfu/mL) in HBS at room temperature for varying times, then tested in clotting assays. Parallel samples were subjected to Western blot analysis and detected with an anti-TFPI monoclonal antibody reactive with the Kunitz-2 domain. (C) This procedure was repeated using S Typhimurium ΔgalETK or ΔgalETK ΔpgtE (109 cfu/mL) and 3 μg/mL TFPI (3 μg/mL) and an anti-TFPI monoclonal antibody reactive with the Kunitz-1 domain. Samples that did not clot by 300 seconds are indicated by an asterisk. Clotting data are mean plus SEM (n = 3).
Ability of omptins to modify other clotting factors
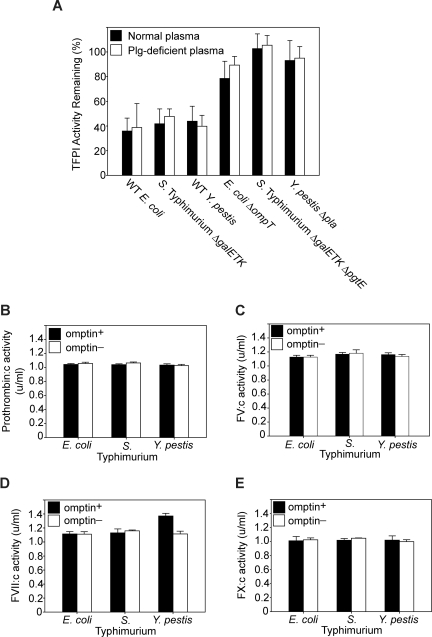
Plasmin has been shown to proteolytically inactivate TFPI.30 To rule out the possibility that omptin activation of plasminogen to plasmin indirectly contributes to TFPI inactivation, we compared TFPI activity levels for bacteria/TFPI incubations conducted in normal plasma versus those conducted in plasminogen-deficient plasma. With all 3 omptin-positive strains tested, no differences in TFPI activity levels were observed between the 2 plasma treatments (Figure 3A). In addition, all the omptin-negative strains caused little or no decrease in TFPI activity, whichever plasma was used. Thus, omptin-mediated plasminogen activation had a negligible effect on TFPI inactivation in these plasma clotting assays.
Figure 3.
Omptin procoagulant activity is mediated through direct TFPI inactivation. (A) Plasmin does not contribute to TFPI inactivation. Clotting times (measured as in Figure 1B) were obtained for mixtures of TFPI and various bacterial strains (108 cfu/mL) incubated in normal plasma (■) as well as in plasminogen-deficient plasma (□). TFPI activities were determined by reference to a standard curve of clot times versus TFPI concentration. (B-D) Omptin activity toward extrinsic clotting factors. Omptin-positive (■, E coli MC4100, S Typhimurium ΔgalETK, and Y pestis KIM5) and omptin-negative (□, E coli ΔompT, S Typhimurium ΔgalETK ΔpgtE, and Y pestis Δpla) bacterial strains were tested in clotting factor assays to measure changes in the specific coagulant activity of (B) prothrombin, (C) FV, (D) FVII, or (E) FX. Data are mean plus SEM (n = 3).
The possibility remained that modification of other clotting factors in plasma may have contributed to the procoagulant effect of omptins. To eliminate this possibility, omptin-positive and omptin-knockout bacterial strains were tested for their ability to enhance or degrade the coagulant activity of prothrombin, FV, FVII, or FX, in clotting assays designed to quantify the specific activities of individual plasma clotting factors. As seen in Figure 3B, C, and E, there was no observable change in activity levels of prothrombin, FV, or FX when plasma was incubated with omptin-positive versus omptin-negative strains. For FVII coagulant activity, however, incubation with wild-type Y pestis caused a modest 30% increase compared with the Δpla strain (Figure 3D). Quantitation of FVIIa levels after a one-hour incubation of wild-type Y pestis with plasma demonstrated a FVIIa concentration of 3.5 ng/mL versus approximately 1 ng/mL when plasma was incubated with the Δpla strain or no bacteria (data not shown). This modest increase in FVIIa concentration would not significantly change the observed clotting time in a typical tissue factor-initiated test of plasma clotting time. Importantly, in these experiments, we never observed any decrease in specific clotting factor activities for FVII, FV, FX, or prothrombin, ruling out nonspecific proteolytic degradation of blood clotting factors by these bacteria.
TFPI is a better substrate than plasminogen for omptin-positive bacteria
To determine if TFPI was a better substrate for omptins than plasminogen,we compared TFPI inactivation rates versus plasminogen activation rates by omptin-expressing bacteria (Table 2). TFPI inactivation was detected by loss of TFPI anticoagulant activity as quantified in Figure 1B. Plasminogen activation was determined by incubating plasminogen with bacteria and monitoring the parabolic increase in plasmin activity (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Using this assay, we calculated a plasminogen activation rate by whole Y pestis KIM5 of 900 molecules/cell per minute (Table 2). Sodeinde et al5 reported a kcat of 0.21/minute Pla activation of human Glu-plasminogen based on experiments with outer membrane preparations. Extrapolating this value by their estimate of 10 000 Pla molecules per Y pestis cell would yield a maximal rate, at saturating plasminogen concentrations, of 2100 molecules/cell per minute. Using an assay similar to the plasminogen activation assay, we obtained a rate of FVII activation by wild-type Y pestis of 1980 molecules/cell per minute. (The Δpla strain as well as all omptin-positive and negative strains of E coli and S Typhimurium failed to generate enough plasmin or FVIIa to allow for accurate determination of activation rates.) Interestingly, we obtained rates of TFPI inactivation by wild-type Y pestis KIM5 that were more than 2 orders of magnitude higher than the rates of plasminogen or FVII activation. Omptin-positive strains of E coli and S Typhimurium inactivated enough TFPI for determination of reaction rates: wild-type E coli MC4100 exhibited a rate of TFPI inactivation similar to that of wild-type Y pestis, while S Typhimurium ΔgalETK exhibited a rate of TFPI inactivation 2-fold higher than the rate of plasminogen activation by wild-type Y pestis. Thus, for all 3 omptin-positive strains tested, the rate of TFPI inactivation is higher than the corresponding rate of plasminogen activation.
Table 2.
Rates of TFPI inactivation versus plasminogen and FVII activation by omptin-positive bacteria
| Strain | TFPI inactivation rate (molecules/minute per cfu) | Plasminogen activation rate (molecules/minute per cfu) | FVII activation rate (molecules/minute per cfu) |
|---|---|---|---|
| Y pestis KIM5 | 217 000 + 42 000 | 900 + 17 | 1980 + 540 |
| S Typhimurium ΔgalETK | 2100 + 170 | ND | ND |
| E coli MC4100 | 204 000 + 30 000 | ND | ND |
Tissue pathway factor inhibitor (TFPI) inactivation rates were determined by incubating TFPI and bacteria for various times at 37°C, then quantifying TFPI activity in clotting assays. Remaining TFPI activities were determined by reference to standard curves using purified TFPI. Experimental conditions resulting in insufficient plasmin or FVIIa for accurate determination of activation rates are listed as not detectable (ND). The lower limit of detection for both assays corresponded to an activation rate of approximately 50 molecules/minute per cfu. Results are mean plus SEM (n = 3).
Comparison of glycosylated and nonglycosylated forms of TFPI as omptin substrates
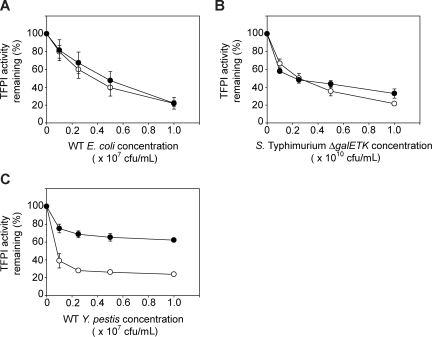
Up to this point, all experiments used recombinant human TFPI purified from E coli, which lacks glycosylation and any other eukaryotic posttranslational modifications. TFPI purified from plasma and from various mammalian cell lines, including SK-hepatoma cells, which are known to be glycosylated and exhibit a higher relative molecular weight (Mr) than TFPI expressed in bacteria. Thus, we compared the sensitivities of E coli–derived TFPI versus SK-hepatoma cell–derived TFPI (SK-TFPI) to omptin-mediated inactivation. Using a clotting assay to monitor changes in TFPI activity levels at increasing bacterial concentrations, we found almost no difference in the ability of omptin-positive E coli and S Typhimurium strains to inactivate the 2 forms of TFPI (Figure 4A,B). Y pestis, however, exhibited differences in rates of inactivation between the 2 TFPI forms (Figure 4C). The rate of SK-TFPI inactivation by Y pestis was calculated as in Table 2 and was found to be 10-fold lower than the rate of bacterially derived TFPI inactivation (20 200 + 990 vs 217 000 + 42 000 molecules/minute/cfu). Nevertheless, this lower inactivation rate is still more than 20 times faster than the corresponding rate of plasminogen activation by Y pestis.
Figure 4.
Comparison of omptin activity toward glycosylated and nonglycosylated forms of TFPI. The omptin-positive bacterial strains: (A) E coli MC4100, (B) S Typhimurium ΔgalETK, or (C) Y pestis Kim5 were incubated with 30 nM glycosylated TFPI (SK-TFPI) derived from SK-Hepatoma cells (●) or nonglycosylated TFPI derived from E coli (○) for 30 minutes at room temperature at the indicated bacterial concentrations (x-axis). Samples were tested in clotting assays using normal plasma, with TFPI activities determined by reference to standard curves as in Figure 3A. Data are mean plus SEM (n = 3).
Analysis of TFPI cleavage fragments generated by OmpT, PgtE, and Pla
Omptins preferentially cleave substrates between consecutive basic residues31,32; such candidate cleavage sites appear 4 times in the primary sequence of TFPI, not including a stretch of 6 consecutive basic residues in the C-terminal basic tail region. For E coli–mediated TFPI proteolysis, this basic tail region seemed a likely cleavage target for OmpT, because SDS-PAGE and Western blot analysis revealed a TFPI fragment between 1 to 2 kDa smaller than the 31 948 Da full-length species (Figure 5A,B fragment 1). Cleavage in the C-terminal basic region would result in a fragment of approximately 30 kDa, corresponding to TFPI fragment 1. Mass spectrometric analysis of this fragment yielded a molecular weight of 28 733 Da, corresponding to cleavage between residues K249 and G250 near the C terminus or between residues F28 and K29 near the N terminus. This latter scissile bond location is highly unlikely, because cleavage by omptins occurs C-terminal, not N-terminal to a basic P1 residue. Wild-type E coli appears to generate only this single TFPI fragment, even after an extended (24-hour) incubation (Figure 5A lane 5).
Figure 5.
Analysis of TFPI cleavage by whole bacteria. (A) Omptin-positive bacterial strains, E coli MC4100, S Typhimurium ΔgalETK, or Y pestis KIM5 (109 cfu/mL) were incubated with TFPI (100 μg/mL) in HBS without BSA for 24 hours at room temperature, after which mixtures were resolved on SDS-PAGE and stained with Coomassie blue. (B) TFPI (3 μg/mL) was incubated with omptin-positive bacterial strains (109 cfu/mL) in HBS for varying times, after which the mixtures were Western blot analyzed and detected with an anti-TFPI monoclonal antibody reactive with the Kunitz-2 domain. Discrete fragments are indicated by arrows and numbered. (C) S Typhimurium, (D) E coli, or (E) Y pestis at 109 cfu/mL were incubated with TFPI (3 μg/mL) for 30 minutes at room temperature in HBS after which bacteria were removed by centrifugation at 16 000g for 4 minutes. Supernatants were transferred to separate tubes while the bacterial pellets were washed once with HBS, pelleted, and resuspended in HBS. Western blot analysis of the resulting supernatant and bacterial pellet fractions were probed with monoclonal antibodies reactive with the Kunitz-2 domain of TFPI. (F) Amino acid sequence following the C-terminal ends of the Kunitz-3 domains of human (Homo sapiens), rabbit (Oryctolagus cuniculus), mouse (Mus musculus), and rat (Rattus norvegicus) TFPI. Residues identical in all 4 species are highlighted in gray.
The S Typhimurium ΔgalETK strain also generates a fragment of similar Mr after a 24-hour incubation with TFPI (Figure 5A lane 4). In addition, the ΔgalETK strain also generates a TFPI fragment of 20 to 25 kDa (Figure 5B fragment 2). This band comigrates with the fragment detected in Western blot analysis of bacterial pellet samples from pull-down experiments using a Kunitz-2 domain specific monoclonal antibody (Figure 5C). This fragment is not seen in the supernatants, suggesting that the fragment contains regions of TFPI that facilitate binding to the bacterial outer membrane (most likely the Kunitz-3 domain and/or C-terminal basic region). A third cleavage fragment generated by S Typhimurium ΔgalETK is visible on Coomassie-stained SDS-PAGE of supernatant samples from 24-hour incubations (Figure 5A fragment 4). This band corresponds to one seen on Western blot analysis of supernatant samples probed with a Kunitz-1 domain-specific monoclonal antibody (data not shown). It also corresponds to the lowest Mr band generated by wild-type Y pestis (Figure 5A), with an Mr of 10 to 15 kDa. Y pestis generates a second band of approximately 18 kDa (Figure 5B fragment 3). In addition, Y pestis also generates a TFPI fragment that comigrates with the highest Mr fragment generated by S Typhimurium ΔgalETK and wild-type E coli strains (Figure 5B fragment 1). After 24 hours, this band is nonexistent in the Y pestis incubation samples as demonstrated by SDS-PAGE (Figure 5A lane 3). The fact that omptin-positive strains generated discrete TFPI bands on SDS-PAGE rather than a smear indicates that omptins target specific residues on TFPI for cleavage.
Discussion
For decades, naturally occurring strains of Y pestis have been known possess a coagulase activity unseen in other Yersinia species.33 Sodeinde and Goguen determined that this activity was dependent on the expression of the omptin gene, pla, which also confers the ability to promote fibrinolysis.34 As a result, the coagulase activity was thought to be an artifact of Pla-mediated conversion of plasminogen into plasmin, an enzyme known to degrade fibrin.5 We now report a biochemical basis for the Y pestis coagulase activity: Pla inactivates TFPI and activates FVII by limited proteolysis. Furthermore, we have established that 2 other members of the omptin family, OmpT and PgtE, also confer the ability to proteolytically inactivate TFPI. Through the use of omptin gene deletions we have established that cleavage of TFPI by bacteria requires the expression of omptins, and this cleavage correlates with loss of TFPI anticoagulant activity. Genetic manipulation of O-antigen expression in S Typhimurium also revealed that TFPI inactivation requires rough LPS expression, a finding that agrees with previous studies showing that rough LPS is required for omptin activity toward exogenous macromolecular substrates.11,35,36
Multiple pools of TFPI exist in vivo.18 This includes soluble TFPI circulating in plasma, soluble TFPI mobilized by heparin injection (apparently loosely bound to endothelial heparan sulfate proteoglycans), and soluble TFPI released from activated platelets. There are tightly membrane-bound forms of TFPI on endothelial cells and also a truncated form of TFPI covalently attached to lipoprotein particles (with very little anticoagulant activity). The specific (patho)physiologic roles for each of these TFPI pools remain to be elucidated, but recent studies have shown that even low nanomolar concentrations of circulating TFPI have potent anticoagulant activity when assisted by protein S.37 Proteolytic inactivation by omptins should antagonize the anticoagulant function of any of the various pools of soluble TFPI.
SDS-PAGE and Western blot analysis of TFPI cleavage fragments generated by omptin-positive E coli, S Typhimurium, and Y pestis strains demonstrated discrete fragments, including at least one 29-kDa fragment generated by all 3 species. This suggests the 3 omptins may target a common cleavage site. Mass spectrometric analysis of the fragment generated by E coli indicated it resulted from the loss of residues C-terminal to K249. This conclusion is established by the fact that omptins require substrates with a basic P1 residue and only cleavage at K249 would fulfill this requirement as well as generate a fragment that matches the Mr of 28 733 Da determined from mass spectrometry. Cleavage after K249 removes the C-terminal basic tail of TFPI, a region that facilitates TFPI binding to membranes38 and membrane receptors.39 Loss of this region would explain the segregation of proteolyzed TFPI to the supernatant fraction of TFPI pulldowns conducted with wild-type E coli (Figure 5D).
The proposed cleavage site between K249 and G250 does not fully conform to the generally observed substrate preference of OmpT for cleaving between consecutive basic residues.31,32 This suggests specific interactions outside the immediate vicinity of the OmpT active site that could enhance its interaction with TFPI as was demonstrated for plasminogen.40 Interestingly, omptin-mediated cleavage of plasminogen also does not occur between dibasic residues, but rather between arginine and valine. Alignment of TFPI primary sequences from human, rabbit, mouse, and rat reveals that the K/G cleavage site is conserved, occurring 10 residues C-terminal to the Kunitz-3 domain in all 4 species (Figure 5F). This is interesting, as rodents are a natural reservoir for Y pestis, and in fact in rat TFPI, the P1′ glycine residue is replaced by an arginine, perhaps making it a better substrate for OmpT. Separation of the C-terminal tail region from the inhibitory Kunitz-1 and -2 domains of TFPI profoundly depresses TFPI's anticoagulant function by preventing its association with FXa.41 And since formation of a FXa-TFPI inhibitory complex is a prerequisite for TFPI's inhibition of TF:FVIIa, loss of the tail region would nullify inhibition of both FXa and TF:FVIIa. Thus, the K249-G250 scissile bond is ideally situated to exert a maximal impact on TFPI function upon its hydrolysis.
Conservation across mammalian species of this TFPI scissile bond raises the possibility that the TFPI-omptin interaction evolved not as a virulence mechanism, but rather a host defense mechanism to promote fibrin entrapment of pathogens expressing cell surface and/or extracellular proteases. The fact that all 3 omptins examined in this report inactivate TFPI faster than the rate at which Y pestis activates plasminogen supports this notion. If it were a virulence mechanism, greater variability would be expected among the omptins in their activities against TFPI, because the evolutionary pressures that produced an enzyme capable of processing TFPI would be expected to be different for each species. This is readily apparent in the unique ability of Pla to activate plasminogen efficiently and in the demonstration that Pla expression increases Y pestis virulence in vivo.5,9 Previous studies have shown that TFPI is sensitive to inactivation by a variety of host-derived proteases, most of which are either released by leukocytes (chymase,42 matrix metalloproteinases,43 cathepsin G,44 and neutrophil elastase45) or up-regulated upon activation of the clotting cascade (thrombin,46 FXa,47 plasmin30). Because activation of clotting is a key component of inflammatory responses, TFPI inactivation by both groups of enzymes may be expected to increase during infection. We therefore hypothesize that TFPI has evolved a generalized sensitivity to proteolytic degradation to potentiate host clotting responses to infection. Furthermore, we propose that TFPI has specifically evolved sensitivity to proteolytic degradation by the conserved omptin protease family as a means of detecting and responding to Gram-negative bacterial pathogens. The ability of Pla to promote fibrinolysis through plasminogen activation is likely an adaptation unique to Y pestis that evolved to facilitate this organism's unusual life cycle.
Our finding that Y pestis inactivates the naturally occurring glycosylated form of TFPI at a rate 10-fold slower than that of nonglycosylated TFPI seems to support a profibrinolytic role for Pla in Y pestis virulence. However, the Pla-mediated rate for glycosylated TFPI inactivation is still 20-fold greater than that for plasminogen activation and 10-fold greater than the PgtE-mediated rate of inactivation of both forms of TFPI. One intriguing possibility is that Y pestis initially promotes activation of the clotting system to form a protective fibrin barrier against detection by host inflammatory cells at the regional draining lymph node (bubo), which is the nidus for bubonic plague infections in the body. Then as localized bacterial numbers grow, enough bacteria are present to sustain the infection even without the benefit of the fibrin barrier. Concurrently, the number of Pla enzymes will have increased sufficiently to compensate for the modest plasminogen activating ability of individual Pla molecules, thereby dissolving the fibrin barrier and allowing the bacteria to disseminate to other parts of the body. This strategy would seem to recapitulate the relationship between the clotting and fibrinolytic systems in vivo where plasminogen activation is greatly accelerated by the presence of fibrin, allowing orderly formation and then degradation of a fibrin clot. This scenario is consistent with in vivo studies that have shown that Pla expression generates an ameliorated inflammatory response and an alteration in the morphology of the regional draining lymph node with abundant fibrin deposition.48,49
In contrast to Y pestis, most naturally occurring strains of E coli and Salmonella express smooth LPS, which sterically prevents access of exogenous macromolecular substrates to the omptin active site. This is consistent with the notion that these bacteria have evolved countermeasures to prevent the procoagulant effect of TFPI inactivation. O-antigen length can be quite variable, however, with nearly 50% of E coli strain 0111 LPS moieties possessing O-antigen of less than 5 polysaccharide units.50 In addition, S Typhimurium alters the composition of its LPS to a rough form inside intracellular macrophage vacuoles, thereby revealing full proteolytic activity toward α2-antiplasmin.51 It also up-regulates the expression of PgtE in this environment under the influence of the PhoP/PhoQ regulatory system.52,53 Thus, it is possible that TFPI proteolysis by omptins under pathologic conditions may contribute to the hemostatic imbalance that characterizes DIC and other sepsis-associated coagulopathies, especially considering the heavy bacterial loads, up to 109 cfu/mL in blood, which may be present during the late stages of severe septicemia. Bacterial aggregates found in tissue lesions (ie, the bubo) undoubtedly contain even higher bacterial numbers, well within the concentration range (107-109 cfu/mL) used in the present study to quantify TFPI inactivation. Tang et al recently reported decreased TFPI levels and increased TF-dependent coagulation in the lungs of septic baboons compared with healthy baboons,54 which would be consistent with this premise. Other sepsis model studies have shown little decrease in TFPI activity under septic conditions.55 In light of the failure of recombinant TFPI (Tifacogin) treatment to effect a positive outcome in severe sepsis in a recent phase III clinical trial,55 our findings suggest that the omptin/TFPI interaction may have direct physiologic relevance. With this in mind, it may be worthwhile to examine the therapeutic use of modified forms of TFPI that are engineered to be less susceptible to omptin-mediated proteolytic inactivation.
Supplementary Material
Acknowledgments
We thank Dr George Broze for his generous gifts of recombinant TFPI and anti-TFPI monoclonal antibodies. We also thank Dr Bradford Schwartz for invaluable comments and suggestions on this work and Joshua Turner for his kind assistance in conducting experiments with Yersinia.
This work was supported by National Institutes of Health (NIH) grants R01 HL47014 (J.H.M.), AI052344 and AI056148 (R.I.T.), and AI63230 (J.M.S.), and American Heart Association grant 0810203Z (T.H.Y.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.H.Y. and J.E.C. performed the experiments and contributed figures; R.I.T. and J.M.S. designed experiments and analyzed results; and T.H.Y. and J.H.M. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James H. Morrissey, Department of Biochemistry, College of Medicine, University of Illinois at Urbana-Champaign, 417 Med Sci Bldg MC-714, 506 S Mathews, Urbana, IL 61801; e-mail: jhmorris@illinois.edu.
References
- 1.Ohki M, Nakamura T, Morita T, Iwanaga S. A new endotoxin sensitive factor associated with hemolymph coagulation system of horseshoe crab (Limulidae). FEBS Lett. 1980;120:217–220. doi: 10.1016/0014-5793(80)80301-2. [DOI] [PubMed] [Google Scholar]
- 2.Colvin RB, Johnson RA, Mihm MC, Jr, Dvorak HF. Role of the clotting system in cell-mediated hypersensitivity. I. Fibrin deposition in delayed skin reactions in man. J Exp Med. 1973;138:686–698. doi: 10.1084/jem.138.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levi M, van der PT, Buller HR. Bidirectional relation between inflammation and coagulation. Circulation. 2004;109:2698–2704. doi: 10.1161/01.CIR.0000131660.51520.9A. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich R, Panizzi P, Fuentes-Prior P, et al. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 5.Sodeinde OA, Subrahmanyam YV, Stark K, et al. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 6.Bogenmann E, Jones PA. Role of plasminogen in matrix breakdown by neoplastic cells. J Natl Cancer Inst. 1983;71:1177–1182. [PubMed] [Google Scholar]
- 7.Kucharewicz I, Kowal K, Buczko W, Bodzenta-Lukaszyk A. The plasmin system in airway remodeling. Thromb Res. 2003;112:1–7. doi: 10.1016/j.thromres.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Masson V. [Roles of serine proteases and matrix metalloproteinases in tumor invasion and angiogenesis]. Bull Mem Acad R Med Belg. 2006;161:320–326. [PubMed] [Google Scholar]
- 9.Degen JL, Bugge TH, Goguen JD. Fibrin and fibrinolysis in infection and host defense. J Thromb Haemost. 2007;5(suppl 1):24–31. doi: 10.1111/j.1538-7836.2007.02519.x. [DOI] [PubMed] [Google Scholar]
- 10.Kukkonen M, Korhonen TK. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int J Med Microbiol. 2004;294:7–14. doi: 10.1016/j.ijmm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Kukkonen M, Suomalainen M, Kyllonen P, et al. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol Microbiol. 2004;51:215–225. doi: 10.1046/j.1365-2958.2003.03817.x. [DOI] [PubMed] [Google Scholar]
- 12.Lahteenmaki K, Kukkonen M, Jaatinen S, et al. Yersinia pestis Pla has multiple virulence-associated functions. Adv Exp Med Biol. 2003;529:141–145. doi: 10.1007/0-306-48416-1_28. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 14.Levi M. Disseminated intravascular coagulation: what's new? Crit Care Clin. 2005;21:449–467. doi: 10.1016/j.ccc.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Jesty J, Spencer AK, Nakashima Y, Nemerson Y, Konigsberg W. The activation of coagulation factor X. Identity of cleavage sites in the alternative activation pathways and characterization of the COOH-terminal peptide. J Biol Chem. 1975;250:4497–4504. [PubMed] [Google Scholar]
- 16.Osterud B, Rapaport SI. Activation of factor IX by the reaction product of tissue factor and factor VII: additional pathway for initiating blood coagulation. Proc Natl Acad Sci U S A. 1977;74:5260–5264. doi: 10.1073/pnas.74.12.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuenschwander PF, Fiore MM, Morrissey JH. Factor VII autoactivation proceeds via interaction of distinct protease-cofactor and zymogen-cofactor complexes. Implications of a two-dimensional enzyme kinetic mechanism. J Biol Chem. 1993;268:21489–21492. [PubMed] [Google Scholar]
- 18.Broze GJ., Jr Tissue factor pathway inhibitor and the current concept of blood coagulation. Blood Coagul Fibrinolysis. 1995;6(suppl 1):S7–S13. doi: 10.1097/00001721-199506001-00002. [DOI] [PubMed] [Google Scholar]
- 19.Silhavy T, Berman M, Enquist L. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 20.Perry RD, Fetherston JD. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann BA, Slauch JM. Transduction of low-copy number plasmids by bacteriophage P22. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu D, Ellis HM, Lee EC, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maloy SR, Stewart VJ, Taylor RK. Genetic analysis of pathogenic bacteria a laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 26.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 27.Mast AE, Broze GJ., Jr Physiological concentrations of tissue factor pathway inhibitor do not inhibit prothrombinase. Blood. 1996;87:1845–1850. [PubMed] [Google Scholar]
- 28.Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 29.Kukkonen M, Suomalainen M, Kyllonen P, et al. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol Microbiol. 2004;51:215–225. doi: 10.1046/j.1365-2958.2003.03817.x. [DOI] [PubMed] [Google Scholar]
- 30.Li A, Wun TC. Proteolysis of tissue factor pathway inhibitor (TFPI) by plasmin: effect on TFPI activity. Thromb Haemost. 1998;80:423–427. [PubMed] [Google Scholar]
- 31.Dekker N, Cox RC, Kramer RA, Egmond MR. Substrate specificity of the integral membrane protease OmpT determined by spatially addressed peptide libraries. Biochemistry. 2001;40:1694–1701. doi: 10.1021/bi0014195. [DOI] [PubMed] [Google Scholar]
- 32.McCarter JD, Stephens D, Shoemaker K, et al. Substrate specificity of the Escherichia coli outer membrane protease OmpT. J Bacteriol. 2004;186:5919–5925. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jawetz E, Meyer KF. Studies on plague immunity in experimental animals. II. Some factors of the immunity mechanism in bubonic plague. J Immunol. 1943;49:15–30. [Google Scholar]
- 34.Sodeinde OA, Goguen JD. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect Immun. 1988;56:2743–2748. doi: 10.1128/iai.56.10.2743-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beesley ED, Brubaker RR, Janssen WA, Surgalla MJ. Pesticins. 3. Expression of coagulase and mechanism of fibrinolysis. J Bacteriol. 1967;94:19–26. doi: 10.1128/jb.94.1.19-26.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer RA, Brandenburg K, Vandeputte-Rutten L, et al. Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2002;269:1746–1752. doi: 10.1046/j.1432-1327.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 37.Hackeng TM, Sere KM, Tans G, Rosing J. Protein S stimulates inhibition of the tissue factor pathway by tissue factor pathway inhibitor. Proc Natl Acad Sci U S A. 2006;103:3106–3111. doi: 10.1073/pnas.0504240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warshawsky I, Bu G, Mast A, et al. The carboxy terminus of tissue factor pathway inhibitor is required for interacting with hepatoma cells in vitro and in vivo. J Clin Invest. 1995;95:1773–1781. doi: 10.1172/JCI117855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mast AE, Stadanlick JE, Lockett JM, et al. Tissue factor pathway inhibitor binds to platelet thrombospondin-1. J Biol Chem. 2000;275:31715–31721. doi: 10.1074/jbc.M006595200. [DOI] [PubMed] [Google Scholar]
- 40.Kukkonen M, Lahteenmaki K, Suomalainen M, et al. Protein regions important for plasminogen activation and inactivation of α2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol Microbiol. 2001;40:1097–1111. doi: 10.1046/j.1365-2958.2001.02451.x. [DOI] [PubMed] [Google Scholar]
- 41.Lockett JM, Mast AE. Contribution of regions distal to glycine-160 to the anticoagulant activity of tissue factor pathway inhibitor. Biochemistry. 2002;41:4989–4997. doi: 10.1021/bi016058n. [DOI] [PubMed] [Google Scholar]
- 42.Hamuro T, Kido H, Asada Y, et al. Tissue factor pathway inhibitor is highly susceptible to chymase-mediated proteolysis. FEBS J. 2007;274:3065–3077. doi: 10.1111/j.1742-4658.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- 43.Belaaouaj AA, Li AG, Wun TC, Welgus HG, Shapiro SD. Matrix metalloproteinases cleave tissue factor pathway inhibitor—effects on coagulation. J Biol Chem. 2000;275:27123–27128. doi: 10.1074/jbc.M004218200. [DOI] [PubMed] [Google Scholar]
- 44.Petersen LC, Bjorn SE, Nordfang O. Effect of leukocyte proteinases on tissue factor pathway inhibitor. Thromb Haemost. 1992;67:537–541. [PubMed] [Google Scholar]
- 45.Higuchi DA, Wun T-C, Likert KM, Broze GJ., Jr The effect of leukocyte elastase on tissue factor pathway inhibitor. Blood. 1992;79:1712–1719. [PubMed] [Google Scholar]
- 46.Ohkura N, Enjyoji K, Kamikubo Y, Kato H. A novel degradation pathway of tissue factor pathway inhibitor: incorporation into fibrin clot and degradation by thrombin. Blood. 1997;90:1883–1892. [PubMed] [Google Scholar]
- 47.Salemink I, Franssen J, Willems GM, et al. Factor Xa cleavage of tissue factor pathway inhibitor is associated with loss of anticoagulant activity. Thromb Haemost. 1998;80:273–280. [PubMed] [Google Scholar]
- 48.Butler T. A clinical study of bubonic plague. Observations of the 1970 Vietnam epidemic with emphasis on coagulation studies, skin histology and electrocardiograms. Am J Med. 1972;53:268–276. doi: 10.1016/0002-9343(72)90168-4. [DOI] [PubMed] [Google Scholar]
- 49.Sebbane F, Gardner D, Long D, Gowen BB, Hinnebusch BJ. Kinetics of disease progression and host response in a rat model of bubonic plague. Am J Pathol. 2005;166:1427–1439. doi: 10.1016/S0002-9440(10)62360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldman RC, Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107:145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- 51.Lahteenmaki K, Kyllonen P, Partanen L, Korhonen TK. Antiprotease inactivation by Salmonella enterica released from infected macrophages. Cell Microbiol. 2005;7:529–538. doi: 10.1111/j.1462-5822.2004.00483.x. [DOI] [PubMed] [Google Scholar]
- 52.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to α-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 54.Tang H, Ivanciu L, Popescu N, et al. Sepsis-induced coagulation in the baboon lung is associated with decreased tissue factor pathway inhibitor. Am J Pathol. 2007;171:1066–1077. doi: 10.2353/ajpath.2007.070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham E, Reinhart K, Opal S, et al. Efficacy and safety of tifacogin (recombinant tissue factor pathway inhibitor) in severe sepsis: a randomized controlled trial. JAMA. 2003;290:238–247. doi: 10.1001/jama.290.2.238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.