Mesenchymal stem cells are self-renewing cells with the ability to differentiate into osteocytes, chondrocytes and adipocytes. This article describes a subset of mesenchymal stem cells with distinct phenotypic and functional properties.
Keywords: mesenchymal stem cells, CD56, MSCA-1
Abstract
Background
Conventionally, mesenchymal stem cells are functionally isolated from primary tissue based on their capacity to adhere to a plastic surface. This isolation procedure is hampered by the unpredictable influence of co-cultured hematopoietic and/or other unrelated cells and/or by the elimination of a late adhering mesenchymal stem cells subset during removal of undesired cells. To circumvent these limitations, several antibodies have been developed to facilitate the prospective isolation of mesenchymal stem cells. Recently, we described a panel of monoclonal antibodies with superior selectivity for mesenchymal stem cells, including the monoclonal antibodies W8B2 against human mesenchymal stem cell antigen-1 (MSCA-1) and 39D5 against a CD56 epitope, which is not expressed on natural killer cells.
Design and Methods
Bone marrow derived mesenchymal stem cells from healthy donors were analyzed and isolated by flow cytometry using a large panel of antibodies against surface antigens including CD271, MSCA-1, and CD56. The growth of mesenchymal stem cells was monitored by colony formation unit fibroblast (CFU-F) assays. The differentiation of mesenchymal stem cells into defined lineages was induced by culture in appropriate media and verified by immunostaining.
Results
Multicolor cell sorting and CFU-F assays showed that mesenchymal stem cells were ~90-fold enriched in the MSCA-1+CD56− fraction and ~180-fold in the MSCA-1+CD56+ fraction. Phenotype analysis revealed that the expression of CD10, CD26, CD106, and CD146 was restricted to the MSCA-1+CD56− mesenchymal stem cells subset and CD166 to MSCA-1+CD56± mesenchymal stem cells. Further differentiation of these subsets showed that chondrocytes and pancreatic-like islets were predominantly derived from MSCA-1+CD56± cells whereas adipocytes emerged exclusively from MSCA-1+CD56− cells. The culture of single sorted MSCA-1+CD56+ cells resulted in the appearance of phenotypically heterogeneous clones with distinct proliferation and differentiation capacities.
Conclusions
Novel mesenchymal stem cells subsets with distinct phenotypic and functional properties were identified. Our data suggest that the MSCA-1+CD56+ subset is an attractive starting population for autologous chondrocyte transplantation.
Introduction
Mesenchymal stem/stromal cells (MSC) are self-renewing cells with the ability to differentiate into osteocytes, chondrocytes and adipocytes, but also neuron-like cells, hepatocytes and pancreatic-like cells.1–4 These multipotent cells are found in various adult and fetal tissues including bone marrow, umbilical cord blood, liver, dental pulp, and term placenta.5–13 In culture, expanded MSC express a panel of key markers including CD105 (endoglin, SH2), CD73 (ecto-5′ nucleotidase, SH3, SH4), CD166 (ALCAM), CD29 (β1-integrin), CD44 (H-CAM), and CD90 (Thy-1).1–3,7,14 In contrast to hematopoietic stem cells they lack CD45, CD34, and CD133 expression.1,2,15 Because of their multi-lineage differentiation potential, MSC represent an attractive cell type for replacement therapy of damaged tissues.16–18
MSC can be identified by their ability to form colony forming units-fibroblast (CFU-F) in vitro.2,7 However, these cells are heterogeneous with respect to their proliferation and differentiation capacity. At least two morphologically distinct MSC populations have been identified that differ not only in size but also in their cell division rate and differentiation capacity.4 In addition, analysis of single cell-derived MSC colonies from adult bone marrow revealed differential capacity of colonies to undergo osteogenic, adipogenic, and chondrogenic differentiation.4,19
In most cases, unfractionated bone marrow-derived cells are used as the starting population for the culture of MSC. This isolation method relies on the adherence of fibroblast-like cells to a plastic surface and the removal of non-adherent hematopoietic cells.1–3,7,15,17 The resulting cells are poorly defined and give rise not only to heterogeneous MSC populations but also to osteoblasts and/or osteoprogenitor cells, fat cells, reticular cells, macrophages, and endothelial cells.20,21 To define the starting population more precisely, surface markers such as SH2 (CD105), SH3/SH4 (CD73), SSEA-4, STRO-1 and the low affinity nerve growth factor receptor (CD271), which enrich for MSC, have been employed.21–25 Very recently, platelet derived growth factor receptor-β (PDGF-RB; CD140b) was identified as a selective marker for the isolation of clonogenic MSC.26 Other reports demonstrated a 9.5-fold enrichment of MSC in bone marrow cells with prominent aldehyde dehydrogenase activity.27
Recently, we have shown that CD56, a marker for natural killer, neural, and muscle cells,28–30 is additionally expressed on a small subset of bone marrow CD271bright cells.26 We also demonstrated that W8B2 antigen31 (here designated mesenchymal stem cell antigen-1; MSCA-1) is selectively expressed on CD271bright bone marrow cells and hypothesized that W8B2 antibody reacts with CFU-F, as these clonogenic cells are known to reside in the CD271bright population.26 In this work we investigated the composite phenotype and gene expression profile, as well as the clonogenic, differentiation, and immunomodulation potential of MSCA-1+ and CD56+ MSC populations in bone marrow.
Design and Methods
Isolation of bone marrow and peripheral blood mononuclear cells
Bone marrow was harvested at the Hospital for Workers Compensation from the femoral shafts of patients undergoing total hip replacement. Peripheral blood from healthy volunteers was obtained from the Transfusion Department, Tübingen. Cells were collected in 5000 U heparin (Sigma-Aldrich, Taufkirchen, Germany) after informed consent and approval of the ethics committee of the University of Tübingen. Bone marrow mononuclear cells and peripheral blood mononuclear cells were isolated by Ficoll Histopac density gradient fractionation and remaining erythrocytes lysed in ammonium chloride solution.5
Culture of primary cells
Ficoll-separated and FACS-enriched bone marrow mononuclear cells were cultured as previously described.5 In brief, 2×107 unfractionated or 1×104 sorted MSCA-1+CD56+ and MSCA-1+CD56− bone marrow cells were cultured in gelatine-coated T-75 or T-25 culture flasks in the presence of Knockout™ medium (Invitrogen, Karlsruhe, Germany) and 5 ng/mL recombinant human basic fibroblast growth factor (rh-bFGF; CellSystems, Remagen, Germany).32 After 3 days of culture, non-adherent cells were removed and fresh medium was added. Adherent cells were cultured until they reached 90% confluence.
Colony forming unit-fibroblast assay
CFU-F assays were performed by plating either 1×105 unselected or 500–5,000 FACS-selected bone marrow mononuclear cells in gelatine-coated T-25 flasks containing Knockout™ medium and 5 ng/mL rh-bFGF. After 12 days of culture, adherent cells were washed twice with phosphate-buffered saline, fixed with methanol (Sigma-Aldrich) for 5 min at room temperature, air-dried, and stained with Giemsa solution (Merck, Darmstadt, Germany). CFU-F colonies were macroscopically enumerated. Colony sizes ranged between 1 and 8 mm in diameter.
Differentiation of mesenchymal stem cells
Osteoblast and adipocyte differentiation
MSC derived from sorted MSCA-1+CD56+ or unfractionated bone marrow cells were cultured in NH OsteoDiff or NH AdipoDiff medium (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively.5 In brief, 2×104 (osteogenesis) or 4×104 (adipogenesis) MSC were cultured in 24-well Falcon plates (Becton Dickinson, Heidelberg, Germany). After 12 days of culture in NH OsteoDiff medium, cells were fixed with methanol (−20°C, 5 min). Alkaline phosphatase activity in osteoblasts was determined using FAST™ BCIP/NBT substrate (Sigma-Aldrich). Calcium deposition in fixed cells (4% paraformaldehyde, 15 min) was analyzed after staining with 2% alizarin red S (Merck) for 10 min at room temperature. The formation of adipocytes was evaluated after 25 days of culture in NH AdipoDiff medium and staining of methanol-fixed cells with oil red O dye (Sigma-Aldrich) for 45 min at room temperature. Pictures were taken using an Axiovert 40C light microscope (Carl Zeiss GmbH, Göttingen, Germany).
Chondrogenic differentiation
MSC (4×105) were cultured as a pellet for 4 hours in 15 mL Falcon tubes (Becton Dickinson) at 37°C in 20 μL of Incomplete Chondrogenic Induction Medium (PAA, Pasching, Austria) containing 1% ITS-supplements (Sigma-Aldrich), 175 μM L-ascorbic acid (Sigma-Aldrich), 350 μM L-proline (Sigma-Aldrich), and 100 nM dexamethasone (Sigma-Aldrich). After incubation, 400 μL of Complete Chondrogenic Induction Medium supplemented with 10 ng/mL of transforming growth factor β (Sigma-Aldrich) were added. The resulting cell pellets were cultured for 3 weeks, fixed with 4% phosphoformic acid, embedded in paraffin, and cut into 5 μm thick sections. The dried and deparaffinized sections were incubated with Alcian blue solution (Merck) for 45 min at room temperature, washed in 3% acetic acid, embedded and photographed on a Zeiss Axiovert 200 microscope.
Myogenic differentiation
MSC (5×105) were cultured for 7 days on ultra low adherent dishes in Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Invitrogen), supplemented with 100 μM β-mercaptoethanol. The resulting embryoid body-like clusters were incubated in gelatine-coated 24-well dishes for 21–28 days, the resulting cells fixed with 4% phosphoformic acid for 45 mins at room temperature and permeabilized with 0.1% Triton X-100/phosphate-buffered saline for 20 min. Cells were labeled over night at 4°C with rabbit anti-human smooth muscle actin antibody (Spring Bioscience, Freemont, CA, USA) and mouse anti-human sarcomeric actinin (anti α-actinin) antibody (Sigma-Aldrich). After washing, cells were stained with Cy3-conjugated goat anti-rabbit IgG (Jackson Immuno Research), or Alexa Fluor488-conjugated goat anti-mouse IgG (Invitrogen), and 0.4 μg/mL 4′6 diamidino-2 phenylindole (DAPI).
Neuronal differentiation
MSC (3.5×104) were cultured for 6 days in 800 μL NeuroCult® NS-A proliferation medium (CellSystems) and then for 7 days in NeuroCult® NS-A differentiation medium (CellSystems). Cells were fixed with 4% phosphoformic acid and permeabilized with 0.3% Triton-X-100/phosphate-buffered saline (Sigma-Aldrich) prior to incubation overnight with rabbit anti-human glial fibrillary acidic protein antibody or mouse anti-human neuronal class III-β-tubulin antibody (both reagents from CellSystems).
After washing with 0.1% BSA/TBS/Tween-20 (Sigma-Aldrich), cells were stained with Cy3-conjugated porcine anti-rabbit secondary antibody (Jackson Immuno Research, Cambridge, UK), for 30 min at room temperature or Alexa Fluor® 488-conjugated goat anti-mouse IgG secondary antibody (Invitrogen), and 0.4 μg/mL DAPI.
Pancreatic differentiation
MSC (5×105) were plated into 6-well ultra low adherent dishes (Costar; CellSystems) and cultured for 4 days in minimal essential medium containing 1 mM mono-thioglycerol, 15% ES-Cult fetal bovine serum, and 4.5 g/L DMEM high glucose (CellSystems). The resulting cell clusters were then cultured for 6 days in 6-well adherence plates (Falcon) in ITS supplemented serum-free medium (CellSystems). After transfer into poly-L-ornithine coated 24-well plates, cells were cultured for 6 days in pancreatic proliferation medium (CellSystems) containing N2-A and B27 supplements and 25 ng/mL rh-bFGF, and then for 6 more days in 10 mM nicotinamide containing rh-bFGF-free pancreatic differentiation medium (CellSystems). After washing, cells were fixed with 4% phosphoformic acid, permeabilized with 70% ethyl alcohol, and incubated with blocking buffer containing 0.25% Triton X-100 and 2% fetal bovine serum, labeled with polyclonal rabbit anti-human glucagon antibody (1:75 dilution; Dako Cytomations, Glostrup, Denmark) or polyclonal rabbit anti-human insulin antibody (1:200 dilution; Santa Cruz Biotechnology) overnight and stained with secondary goat anti-rabbit IgG-Cy3 (Millipore, Schwalbach, Germany), and 0.4 μg/mL DAPI.
Generation of mesenchymal stem cell-reactive monoclonal antibodies, W8B2 and 39D5
The W8B2 monoclonal antibody (IgG1; specificity for hMSCA-1) was raised by immunization of a 6–8 week-old female Balb/c mouse (Charles River WIGA, Sulzfeld, Germany) with the retinoblastoma cell line WERI-RB-1, as previously described.31 The 39D5 monoclonal antibody (IgG1, CD56) was raised by immunization with the hematopoietic cell line KG-1a.33
Immunofluorescence analysis and cell sorting
Antibodies
The following proprietary antibodies were used: 97C5 (CD10),34 46A11 (CD13),35 39D5 (CD56),33 1G2C2 (CD105),31 104D2 (CD117),34 W6B3C1 (CD133),31 28D4 (CD140b),26 67D2 (CD164),37,38 CUB1 (CD318; CDCP1),39 24D2 (CD340; HER-2),26 W3C4E11 (CD349; frizzled-9),5 HEK-3D6 (unknown),26 W1C3 (unknown),26 W5C4 (unknown),26 W5C5 (unknown),26 W3D5 (unknown),26 and W8B2B10 (MSCA-1).31 CD34-PE (clone 8G12), CD45-PE (clone HI30), CD56-FITC (clone NCAM16.2), CD56-PE (clone NCAM16.2), CD90-APC (5E10), CD63-PE (clone H5C6), CD73-PE (clone AD2), and HLA-DR-PE (clone TÜ36) were purchased from Becton Dickinson (Heidelberg, Germany). The SSEA-4 reactive antibody MC-813-70 was purchased from Chemicon (Hampshire, UK). CD271-APC (clone ME20.4-1.H4) was obtained from Miltenyi Biotec. CD105-PE (clone SN6) was purchased from eBioscience Inc. (San Diego, CA, USA). CD166-PE was a kind gift from Dr. Gene Lay (BioLegend, San Diego, CA, USA).
Immunofluorescence staining
After blocking of non-specific binding with 10 mg/mL polyglobin (10 min, 4°C), cells were incubated for 15 min with either 20 μL of proprietary antibodies or 10 μL of fluorochrome-conjugated antibodies.5 Cells stained with conjugates were washed twice, suspended in 200 μL FACS buffer and used for flow cytometry. Cells labeled with proprietary antibodies were stained with 20 μL of a F(ab)2 fragment of a R-phycoerythrin (PE) conjugated goat anti-mouse antibody (Dako Cytomations, Glostrup, Denmark) for 15 min, washed twice and analyzed by flow cytometry. For multi-color staining, cells were incubated for 15 min with 10 μL of anti-CD56-FITC and anti-CD271-APC and/or the indicated PE-conjugate. Mouse IgG1 antibodies conjugated with FITC, PE, or APC (Becton Dickinson) were used as isotype controls. After washing, cells were used for flow cytometry. For combined indirect and direct staining, cells were first labeled with the non-conjugated antibody and then stained with 20 μL of 1:25 diluted goat anti-mouse secondary antibody for 15 min. Free binding sites of the secondary antibody were blocked by incubating cells for 25 min with 20 μL mouse IgG polyclonal antibody (0.05 μg/mL; Southern Biotech, Birmingham, AL, USA) prior to counter-staining with CD271-APC and/or CD56-FITC. For controls, secondary step antibody conjugates were used in addition to IgG1-FITC and IgG1-PE. After washing, cells were analyzed by flow cytometry.
Flow cytometric analysis and cell sorting
Cells were sorted on a FACSAria cell sorter (Becton Dickinson), or analyzed on a FACSCantoII flow cytometer (Becton Dickinson). Files were analyzed using the FCS express software (De Novo Software, Ontario, Canada). Single cell sorting in 96-well plates was performed using the ACDU device.
Magnetic-activated cell-separation
For some experiments, bone marrow cells were preselected by MACS (Miltenyi Biotec) using CD271-APC and anti-APC beads. The cells were separated according to the manufacturer’s recommendations.
Inhibition of T-cell proliferation
HLA-mismatched peripheral blood mononuclear cells were labeled with 0.8 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen) and cultured at 75,000 cells/well for 4 days in 96-well plates (Greiner, Frickenhausen, Germany) coated with either 10,000, 20,000 or 30,000 unfractionated or sorted MSC. Cells were cultured in RPMI-1640 (Biochrom) supplemented with either 100 U/mL interleukin-2 (R&D Systems, Minneapolis, MN, USA) and 1 μg/mL OKT3 antibody (Janssen-Cilag, Neuss, Germany), or with 3 μg/mL phytohemogglutinin (Sigma-Aldrich). After incubation, cells were analyzed by flow cytometry.
Inhibition of dendritic cell differentiation
One million freshly isolated monocytes (CD14 MicroBeads, Miltenyi Biotec) were cultured with interleukin-4 (10 ng/mL) and granulocyte-monocyte colony-stimulating factor (5 ng/mL) in the presence or absence of 105 MSC. After 6 days of culture, cells were assayed by flow cytometry for CD14 (Becton Dickinson) and CD1a (Becton Dickinson) expression.40
Gene chip analysis of sorted cells
Ten thousand MSCA-1+CD56− and MSCA-1+CD56+ cells were used for commercial gene chip analysis (Miltenyi Biotec) to perform human whole genome oligo microarray (Agilent Technologies, Böblingen, Germany). Amplified cDNA were quantified using a ND-1000 spectrophotometer (NanoDrop Technologies Inc, Wilmington, DE, USA). Two hundred and fifty nanograms of the library polymerase chain reactions were used as the template for Cy3 and Cy5 labeling. The samples were hybridized for 17 h at 65°C on Agilent’s microarray according to the manufacturer’s instructions. Gene chip scanning and data analysis were carried out using Luminator software (Miltenyi Biotec).
Statistics
Statistical analysis was performed using Student’s t-test and a p-value less than 0.05 was considered statistically significant.
Results
The 39D5 monoclonal antibody detects a rare bone marrow CD271+ subset
Recently, we showed that the CD56-reactive 39D5 monoclonal antibody recognizes a rare subset of CD271bright cells.26 Notably, 39D5 recognizes an epitope of CD56 not expressed on the surface of peripheral blood-derived natural killer cells.33 Indeed, comparative flow cytometric analysis showed that only the commercially available CD56-specific antibody NCAM16.2 but not the 39D5 monoclonal antibody reacted with 20±10% of peripheral blood cells (Figure 1A). In contrast to the situation with peripheral blood cells, both monoclonal antibodies reacted with a small subpopulation (0.5–5.5%) of bone marrow CD271bright cells (Figure 1B). Simultaneous staining of bone marrow cells with 39D5 and NCAM16.2 revealed that both antibodies detected the same CD271bright population (data not shown).
Figure 1.
Characterization of CD56+ bone marrow cells. (A) CD56 epitope NCAM16.2 but not 39D5 is expressed on peripheral blood natural killer cells. (B) CD56 epitopes NCAM16.2 and 39D5 are expressed on a rare CD271+ bone marrow subset. (C) CD271+CD56+ and CD271+CD56− bone marrow cells are clonogenic. CFU-F derived from 500 FACS-sorted cells were stained and scored as described. Data represent the mean CFU-F numbers of three different experiments (*=p<0.01). (D) Expression of selected markers on cultured CD271+CD56− and CD271+CD56+ derived mesenchymal stem cells.
The CD271brightCD56+ population is enriched for CFU-F
To determine the clonogenic potential of sorted CD56+ and CD56− subsets, CFU-F assays were performed. Figure 1C shows a 3 (±0.8)-fold enriched cloning efficiency of CD271brightCD56+ cells compared to CD271brightCD56− cells and a 180 (±52) fold enrichment of CFU-F compared to unfractionated bone marrow cells. The enrichment was independent of the analyzed CD56 epitope (data not shown). Interestingly, CD271brightCD56+ cells did not only give rise to higher colony numbers (38/500 versus 12/500 plated cells) but were also 2 to 4-fold enriched in very large colonies (>100 cells/colony; data not shown).
Phenotype of mesenchymal stem cells derived from sorted CD271brightCD56+/− bone marrow cells
CD271brightCD56+ and CD271brightCD56− bone marrow cells were separated by FACS, cultured in gelatine-coated flasks in the presence of serum replacement medium (n=3),5 stained with the indicated antibodies and analyzed by flow cytometry. Figure 1D shows that CD10, CD140b, CD318, HER2 (CD340), frizzled-9 (CD349), as well as the antibody-defined antigens W1C3, W5C4, W5C5, and W3D5 (and CD166; data not shown) were similarly expressed on MSC derived from both fractions. CD271, SSEA-4 and CD56 were more densely expressed on CD271brightCD56+ derived MSC, whereas MSCA-1 (W8B2 antigen) expression was more pronounced on CD271brightCD56− derived MSC. In contrast to primary MSC (Figure 2B), cultured MSC de novo expressed CD166 and CD318 (and CD109; data not shown) and downregulated CD271 expression.
Figure 2.
Phenotype, clonogenic potential, and morphology of CD271brightMSCA-1+CD56− and CD271brightMSCA-1+CD56+ bone marrow cells. Triple-stained bone marrow cells were gated on the CD271+ subset and analyzed for coexpression of CD56 and selected markers. (A) Display of SSC versus CD271-APC of bone marrow cells. (B) Display of CD56 versus the indicated markers on CD271bright gated cells. (C) Display of CD56 versus MSCA-1 (W8B2) on CD271bright gated cells. Sort windows are indicated as R2 and R3. D ( ) CFU-F numbers derived from 1.000 FACS-sorted bone marrow MSCA-1+CD56− and MSCA-1+ CD56+ cells or 100.000 unfractionated bone marrow cells. The resulting CFU-F were stained and scored 12 days after culture and normalized to 1,000 plated cells (p<0.01). (E) Morphology of bone marrow MSCA-1+CD56− and MSCA-1+CD56+ cells. Subsets were sorted, cytocentrifuged, stained with May-Grünwald-Giemsa solution, and scored on a Zeiss Axiovert 200 microscope (Carl Zeiss Inc, Göttingen, Germany). Note the presence of basophilic-like granules in cells of the triple-positive population.
Gene expression analysis of primary CD271brightCD56− and CD271brightCD56+ bone marrow cells
Whole genome microarray analysis of 10,000 sorted bone marrow cells was performed to compare the gene expression profile of CD271brightCD56− and CD271bright CD56+ bone marrow cells. CD271brightCD56+ cells showed an 11 to 43 fold increased expression of secreted frizzled-related protein 4, esophageal cancer related gene 4 protein, carboxypeptidase E, platelet-derived growth factor A, eukaryotic translation termination factor 1, and CD163 (data not shown). In contrast, genes coding for leukocyte immunoglobulin-like receptor subfamily B, zinc finger protein 212, amphiregulin, HLA class II DM β, spondin 2, and HLA class II DR α were expressed at 62 to 23-fold decreased levels in this subset (data not shown), indicating a high diversity of gene expression profiles in these subsets.
Phenotype of CD271brightCD56+/− bone marrow cells
To compare the expression profiles of surface markers on CD271brightCD56+ and CD271bright CD56− cells, bone marrow cells were triple-stained with anti-CD271, anti-CD56 and a panel of test antibodies and gated on the CD271bright population (Figure 2A; gate R1). Figure 2B shows that CD63, CD73, CD140b, CD164, and W3D5 antigen were expressed at similar levels on both cell subsets, whereas CD45, CD117, CD133, CD318 were negative. In contrast, CD271bright CD56+ cells expressed CD13, CD105, frizzled-9 (CD349), HLA-DR, and MSCA-1 (W8B2 antigen) at reduced levels, whereas CD166 was found exclusively on these cells. The fact that CD166 expression is absent on the majority of primary MSC is surprising because cultured MSC are known to express high levels of CD166. Likewise, the tumor antigen CDCP1 (CD318) was not expressed on primary CD271bright CD56− and CD271brightCD56+ cells (Figure 2B) but highly expressed on cultured MSC (Figure 1D).
Clonogenic capacity of MSCA-1+CD56+/− bone marrow cells
We recently demonstrated that MSCA-1 (W8B2 antigen) expression is restricted to CD271bright cells and that clonogenic MSC reside in the CD271bright but not in the CD271dim population.26 Here we show that MSCA-1 is expressed at high levels on CD271brightCD56− cells and at lower levels on CD271brightCD56+ cell subsets (Figure 2A). The percentage of CD56+ cells in the MSCA-1+ population ranged from 1–20% (21 tested samples; average frequency of CD56+ cells: 8.5%). To analyze the clonogenic capacity of these subsets, cells were fractionated according to sort windows R2 and R3 (Figure 2C). Defined cell numbers were plated into culture flasks and the resulting CFU-F enumerated after 12 days of culture. Figure 2D shows that MSCA-1+CD56+ cells gave rise to 2±0.4 fold higher CFU-F numbers than MSCA-1+CD56− cells. Giemsa staining revealed that MSCA-1+CD56− cells contain a large, bright cytoplasm with vacuoles, whereas MSCA-1+CD56+ cells contain a smaller cytoplasm with basophilic granules (Figure 2E).
Differentiation capacity of mesenchymal stem cells derived from MSCA-1+CD56+/− cells
For differentiation assays, unfractionated or sorted MSCA-1+CD56−, MSCA-1+CD56+ cells were expanded until they had undergone approximately 9–10 cell divisions. Defined numbers of the resulting MSC were then induced to differentiate into cells of the osteogenic, adipogenic, chondrogenic, myogenic, neuronal, and pancreatic lineages.
Osteoblastic differentiation
Culture of MSC (n=5) derived from sorted cell subsets in appropriate medium resulted in the appearance of 95±5% (CD56+) and 70±5% (CD56−) of alkaline phosphatase-positive cells (Figure 3). In contrast, MSC derived from unfractionated bone marrow cells gave rise to only 35±5% alkaline phosphatase-positive cells. Similarly, alizarine red S staining was observed in all osteoblast fractions. However, the number of calcium nodules was 2-fold lower in osteoblasts derived from unfractionated cells.
Figure 3.
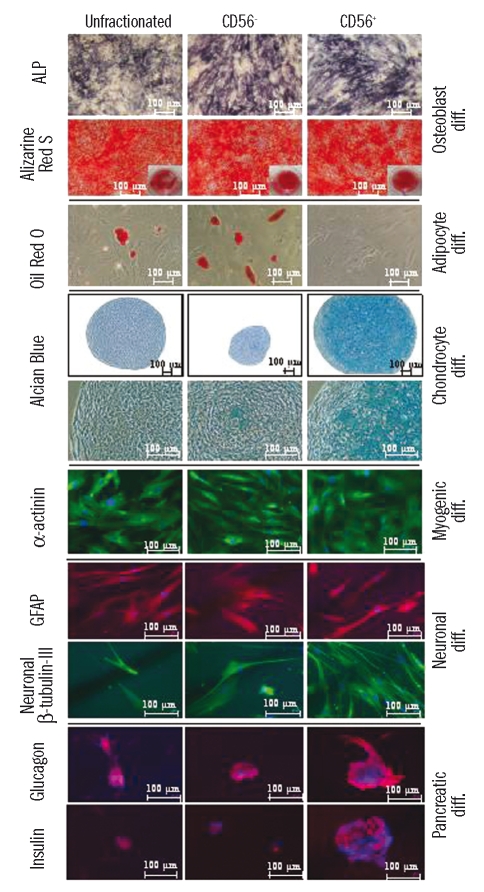
Differentiation potential of mesenchymal stem cells derived from sorted MSCA-1+CD56− and MSCA-1+CD56+ bone marrow cells. Cells triple-stained with CD271, MSCA-1 (W8B2), and CD56 were sorted and cultured as described. Expanded mesenchymal stem cells were induced into osteogenic, adipogenic, chondrogenic, myogenic, neurogenic, and pancreatic differentiation and stained as described. The resulting cells were photographed with a Zeiss Axiovert 200 microscope. Note the exclusive presence of adipocytes in the MSCA-1+CD56 subset and the appearance of chondrocytes and pancreatic-like islets in the MSCA-1+CD56+ fraction.
Adipocyte differentiation
Culture of unfractionated and MSCA-1+CD56− derived MSC (n=5) in adipocyte differentiation medium resulted in the appearance of oil red O incorporating adipocytes. In contrast, MSCA-1+CD56+ derived MSC were unable to generate adipocytes (Figure 3). MSCA-1+CD56− MSC showed a 5±0.5 fold increase of oil red O-positive adipocytes compared to unfractionated cells. Thus, the adipocyte differentiation potential is restricted to the MSCA-1+CD56− subset.
Chondrogenic differentiation
To analyze the chondrocyte differentiation potential, MSC (n=3) derived from fractionated and unfractionated cells were cultured in appropriate medium and the resulting cell pellets stained with Alcian blue. Although chondrogenic differentiation was detected in both fractions, pellet slices from MSCA-1+CD56+ cells were 5±1.6 times larger than those from MSCA-1+CD56− cells (Figure 3). In addition, viable chondrocytes were detected almost exclusively in the MSCA-1+CD56+ subset, whereas MSCA-1+CD56− pellets contained mainly apoptotic cells. MSC from unfractionated cells gave rise to heterogeneous pellet sizes but consistently with fewer viable cells. These data suggest that effective chondrogenesis is restricted to the MSCA-1+CD56+ MSC subset.
Myogenic differentiation
Culture of MSCA-1+CD56± derived MSC (n=2) in medium designed for differentiation into striated muscle cells resulted in the appearance of striated muscle-specific β-actinin staining in cells of all fractions (Figure 3). In contrast, the smooth muscle-specific marker smooth muscle actin was negative in all fractions (data not shown). Undifferentiated MSC showed very weak β-actinin staining (data not shown).
Neuronal differentiation
MSC (n=3) cultured in neuronal differentiation medium were stained for glial fibrillary acidic protein and neuronal β-tubulin-III. Figure 3 shows a prominent staining of cells derived from unfractionated as well as from MSCA-1+CD56+ and MSCA-1+CD56− derived MSC. No staining was observed in undifferentiated MSC or in differentiated cells labeled with isotype-matched control antibodies (data not shown).
Pancreatic differentiation
Culture of MSC (n=2) in pancreatic differentiation medium resulted in glucagon and insulin staining of pancreatic-like islets in cells of all fractions (Figure 3). However, islets derived from MSCA-1+CD56+ MSC were larger and the staining intensity of these markers was much more pronounced compared to MSCA-1+CD56− derived or unfractionated MSC. No staining was observed in undifferentiated MSC or in differentiated cells labeled with isotype-matched control antibodies (data not shown).
Inhibition of T-cell proliferation and dendritic cell differentiation
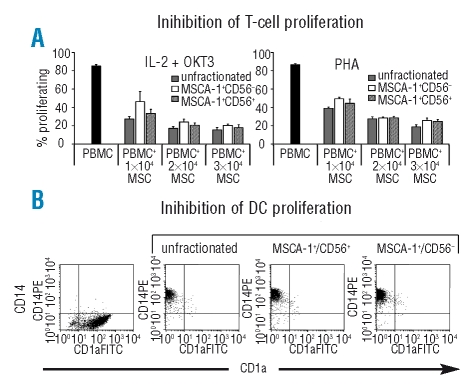
MSC derived from MSCA-1+CD56± cells were analyzed for their capacity to inhibit T-cell proliferation and dendritic cell differentiation (n=3). The anti-proliferative capacity of MSC was analyzed by co-culturing CSFE-labeled peripheral blood mononuclear cells with the indicated MSC subsets and stimulating the mononuclear cell proliferation either with interleukin-2 and OKT3 or with phytohemogglutinin. Figure 4A shows that MSC from all fractions inhibited the proliferation of peripheral blood mononuclear cells without significant differences in a dose-dependent manner and to a similar extent. We also tested the effect of MSC derived from unfractionated and MSCA-1+CD56± populations on the interleukin-4 + granulocyte-monocyte colony-stimulating factor induced differentiation of monocytes into dendritic cells (n=3). Figure 4B shows that all MSC subsets were able to inhibit differentiation from CD14+ monocytes to CD1a+ dendritic cells and we found no evidence for a differential capacity between the different subpopulations.
Figure 4.
Inhibitory effect of MSC subsets on T-cell proliferation and dendritic cell differentiation. (A) Inhibition of T-cell proliferation. MSC derived from MSCA-1+CD56−, MSCA-1+CD56+ and unfractionated bone marrow cells inhibit the interleukin-2 (IL-2) (100 U/mL) + OKT3 (1 μg/mL) or phytohemagglutinin (PHA) (3 μg/mL) induced proliferation of PBMC to a similar extent. (B) Inhibition of dendritic cell differentiation. MSC derived from MSCA-1+CD56, MSCA-1+CD56+, and unfractionated bone marrow cells inhibit the interkeukin-4 (IL-4) (10 ng/mL) + granulocyte-monocyte colony-stimulating factor (5 ng/mL) induced differentiation of CD14+ monocytes into CD1a+ dendritic cells. In the absence of MSC, monocytes differentiated into CD1a+ dendritic cells.
Single cell analysis of MSCA-1+CD56± clones
The growth characteristics, phenotype, and developmental capacity of individual MSCA-1+CD56+ and MSCA-1+CD56− cells were determined by sorting single cells into gelatine-coated 96-well culture plates and culturing them in serum replacement medium until macroscopically visible colonies (>20 cells) appeared.
The cloning efficiency of sorted MSCA-1+CD56+ and MSCA-1+CD56− cells was 11/96 and 5/96, respectively (clone numbers represent the mean of two individual experiments). This approximately 2-fold increased frequency of CD56+ cells is in line with the 2-fold higher colony scores of bulk sorted cells described in Figure 2D.
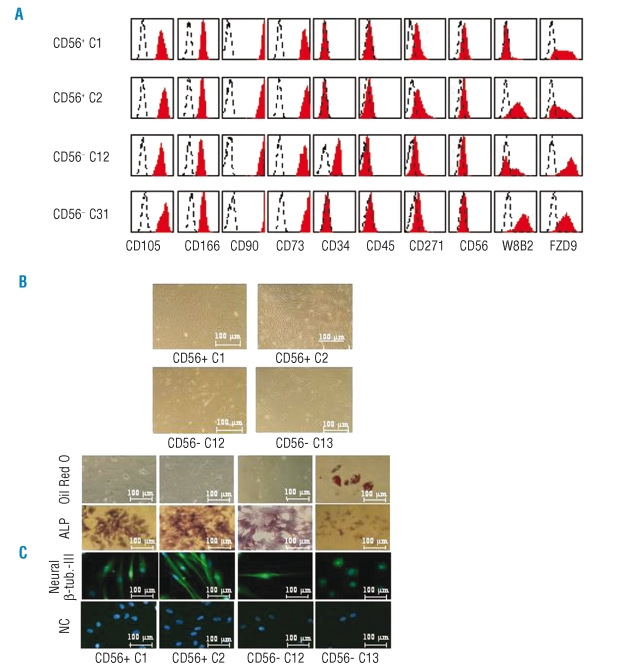
The resulting colonies were transferred into T-25 flasks and expanded until they reached about 18 cell divisions (corresponding to about 2.6×105 cells and 60–70% confluence). Phenotype analysis showed that all 16 clones were negative for CD45 but expressed CD73, CD90, CD105, and CD166 and showed reduced CD271 expression (Figure 5A, Table 1). All CD56+ clones, except clone C3, downregulated CD56 expression. In contrast, CD56 expression was induced in two of four MSC clones derived from CD56− cells (C14, C15).
Figure 5.
Phenotype and differentiation capacity of mesenchymal stem cells derived from single cells. (A) Bone marrow cells triple-stained with CD56-FITC, MSCA-1-PE and CD271-APC were gated and sorted as described in Figure 2C. Single cells were sorted in 96-well plates and cultured for 12 days. The resulting mesenchymal stem cells colonies were transferred to T-25 flasks and cultured for 12 more days and stained with the indicated antibodies. Note the heterogeneous expression profiles. (B) Morphology of MSCA-1+CD56+ derived mesenchymal stem cells clones C1 and C2, and MSCA-1+CD56− derived clones C12 and C13. (C) Osteogenic, adipogenic, and neuronal differentiation potential of mesenchymal stem cells derived from single MSCA-1+CD56− and MSCA-1+CD56+ bone marrow cells. Note that the MSCA-1+CD56− clone C13 gives rise to adipocytes but not osteoblasts.
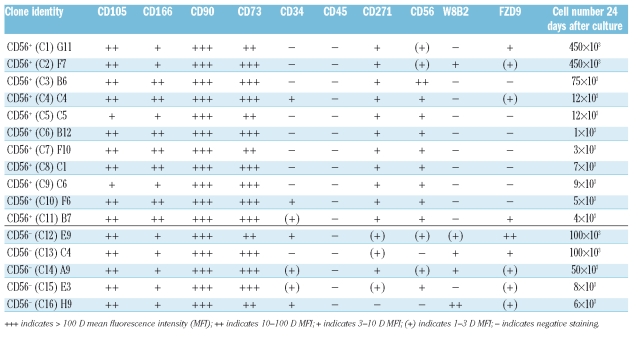
Table 1.
Phenotype and proliferation of mesenchymal stem cells clones (C1-C16) derived from single MSCA-1+CD56− and MSCA-1+CD56+ bone marrow cells.
Interestingly, significant CD34 expression was observed in two of each CD56+ and CD56− clones, whereas MSCA-1 was detected only in one CD56+ clone and in four out of five CD56− clones. Similarly, frizzled-9 expression was observed in only four out of 11 CD56+ clones and in all CD56− clones. These data demonstrate that each single clone has an individual expression profile with preferential expression of MSCA-1 and frizzled-9 in CD56− clones.
Expansion of all 16 individual cells for 24 days resulted in the appearance of spindle-shaped cells with fibroblast-like morphology (Figure 5B). A strong heterogeneity among individual clones with regard to proliferation potential was observed (Table 1). Although the average number of cells derived from CD56+ clones was about double that from CD56− clones (93.5×103 versus 52.8×103 cells after 24 days of culture), no correlation between individual clones or phenotype profiles could be detected (Table 1).
The highly proliferating clones C1 and C2 (MSCA-1+CD56+) and C12 and C13 (MSCA-1+CD56−) were also analyzed for their osteoblast, adipocyte and neuronal differentiation potential. Figure 5C illustrates that – in line with the results of bulk-sorted cells – only one CD56− clone but none of the CD56+ clones gave rise to oil red O dye-incorporating adipocytes. Alkaline phosphatase-positive osteoblasts and neuronal β-tubulin-III expressing neuron-like cells were generated by three of four clones but not by the CD56− clone C13 (Figure 5C). Of note, β-tubulin-III-positive cells were 5- to 10-times more frequent in the CD56+ clones than in the positive CD56− clone. Together, these results underline the high degree of heterogeneity of individual clones derived from MSCA-1+CD56+ and MSCA-1+CD56− cells.
Discussion
We have previously identified novel candidate MSC subsets in bone marrow using proprietary monoclonal antibodies.26 The aim of this study was the characterization of MSC populations recognized by the CD56-specific 39D5 and MSCA-1-specific W8B2 monoclonal antibodies. Phenotypic analysis showed that the targets of these reagents are selectively expressed on CD271bright but no other bone marrow cells. As CFU-F are only found in CD271bright cells, it was likely that the reagents detected MSC. Indeed, CD271brightCD56+ and CD271bright MACA-1+ cells did not only co-express the established MSC markers CD13, CD73, CD105, and CD140b and lacked expression of CD133 and CD45, but they also gave rise to CFU-F. Interestingly, the cloning efficiency was 2 to 4 times higher in the MSCA-1+CD56+ subset than in the MSCA-1+CD56− subset. In addition, effective chondrocyte and pancreatic-like islet differentiation could be induced only in cells of the MSCA-1+CD56+ fraction. In contrast, adipocytes emerged only from MSCA-1+CD56− cells. Sorting of single cells from both subsets not only confirmed the distinct proliferation and differentiation capacities of MSCA-1+CD56+ and MSCA-1+CD56− cells but also showed an unexpected degree of heterogeneity among single clones. In fact, each clone displayed a unique phenotype, proliferation rate, and differentiation capacity.
Recently, Prockop et al. described two morphologically distinct subsets in cultured MSC consisting of either small and rapidly self-renewing (RS) cells or of larger, more mature and slowly replicating cells (mMSC). RS cells gave rise to low forward and side scatter (FSlo/SSlo) and mMSC18,41–43 to large forward and side scatter (FShi/SShi) signals. No suitable surface marker was identified to separate these subsets. When comparing light scatter-defined, culture-derived RS cells and mMSC with the surface marker-defined cell subsets described in this report, a limited analogy between primary MSCA-1+CD56+ cells and RS cells and between MSCA-1+CD56− cells and mMSC can be drawn. Thus, MSCA-1+CD56+ cells reside in the FSlo-medium/SSlo and MSCA-1+CD56− cells in the more heterogeneous FSlo-hi/SSlo-hi population (data not shown), suggesting that MSCA-1+CD56+ cells are more mature. In fact, the number and the average CFU-F size of MSCA-1+CD56+ cells was 2–4 fold increased compared to MSCA-1+CD56− cells. In addition, only MSCA-1+CD56+ cells were able to effectively differentiate into chondrocytes, as revealed by the increased cartilage pellet size and extensive proteoglycan staining. In contrast to RS cells, MSCA-1+CD56+ cells were unable to differentiate into adipocytes. Instead, this capacity was detected in MSCA-1+CD56− cells, indicating that this subset contains lineage-restricted progenitor cells, too. In conclusion, RS cells represent rapidly proliferating immature progenitor cells with multilineage differentiation capacity, whereas MSCA-1+CD56+ cells are enriched for immature MSC with increased proliferation rates but with a more restricted differentiation capacity. In contrast to MSCA-1+CD56+ cells, the MSCA-1+CD56− subset is enriched for more mature and less clonogenic MSC but – unlike mMSC – retains multipotential cells including adipogenic progenitor cells.
Several groups have reported that MSC derived from single cells are heterogeneous with respect to their proliferation and differentiation capacity.4,18 The frequency of individual clones with specific differentiation potentials varied significantly. In one study, almost 100% of colonies underwent osteogenic, 80% adipogenic, but only 30% chondrogenic differentiation.4 In our studies we demonstrated that each individual colony of 16 tested clones displayed discrete expression, proliferation, and differentiation features. Of these, only the large colonies (occurring more frequently in the MSCA-1+CD56+ fraction) could be expanded and used for differentiation assays, indicating that only these colonies are derived from immature MSC. Adipocyte differentiation could be detected only in colonies derived from MSCA-1+CD56− but not MSCA-1+CD56+ cells which is in line with the results obtained from bulk-sorted cells. Although cartilage differentiation of single cells was not analyzed, we anticipate a several log decade enrichment of chondrocyte progenitors in the MSCA-1+CD56+subset because of their almost exclusive presence in this population and because only 1 in 104–105 bone marrow cells express this phenotype. Together, our studies confirm the vast heterogeneity of individual MSC clones detected by other groups and show, for the first time, that even narrowly defined, rare MSC populations are highly heterogeneous with respect to their phenotype, their proliferation capacity, and their differentiation potential.
In the past, several surface markers were identified for the prospective isolation of bone marrow-derived MSC. These markers include CD73, CD105, CD271, STRO-1, SSEA-4, and GD2.4,19–26,44 However, none of these markers stringently recognizes CD271brightCD45−/lo cells. In this study, we described two antibodies, W8B2 and 39D5, which fulfil these criteria. Moreover, the CD56-specific 39D5 monoclonal antibody selectively reacts only with a rare MSC subset.26 Although CD56 is also expressed on natural killer cells and activated T cells, the 39D5 epitope is surprisingly absent on these cells. There is no explanation for the differential expression of CD56 epitopes because all known isoforms vary in amino acid sequences of their intra-cellular domain. At present, one might speculate that the 39D5 epitope on natural killer cells is inaccessible due to structural changes in this molecule during membrane assembly, to lipid compositions that inhibit the access of the 39D5 antibody, or to hitherto unknown post-translational modifications of CD56. Further investigations are necessary to study this issue.
Injuries of articular cartilage and spinal disks are major clinical problems because of the limited self-regenerating ability of this tissue. Diseases including rheumatoid arthritis, trauma, acute osteochondral fractures, and spinal disk injuries are directly related to the lack of effective chondrogenesis.45 Despite the progress in orthopedic surgery and increasing success in autologous chondrocyte transplantation,46,47 cell biology-related approaches for cartilage regeneration remain challenging. A major concern is the use of cultured cells for clinical purposes, the initiating cells of which are only poorly characterized. As an attractive alternative, highly enriched and well-defined CD271brightMSCA-1+CD56+ bone marrow cells with potent chondrogenic differentiation capacity may be employed as the starting population. These cells may be either used directly for injections into spinal disk spaces or expanded and differentiated in vitro into chondrocytes prior to use in clinical settings.
In conclusion, we prospectively identified for the first time two phenotypically distinct MSC subpopulations in bone marrow with differential clonogenic and differentiation capacity. We demonstrated that the 39D5 epitope of CD56 is selectively expressed on cells of a minor MSC population and that 39D5+ cells show increased clonogenic and proliferative potential as well as a unique chondrocyte and pancreatic differentiation capacity. We also introduced MSCA-1 as novel and selective pan-MSC marker and showed that only MSCA-1+CD56− but not MSCA-1+CD56+ MSC were able to differentiate into adipocytes. MSCA-1+CD56+ MSC may be used as the starting population of choice for the treatment of several diseases, in particular for rheumatoid arthritis, trauma, acute osteochondral fractures, and spinal disk injuries.
Acknowledgments
The authors would like to thank Flavianna Cerabona for excellent technical assistance.
Footnotes
Authorship and Disclosures
All authors meet the criteria for being contributing authors. VLB and ST contributed to the conception and design of the study, data analysis and interpretation, and manuscript writing; PMB, FG and HR participated in the conception and design of the study, data analysis and interpretation; PdZ and BS participated in provision of study material; IM and TS contributed to data analysis and interpretation; WEF participated in data analysis and interpretation; LK contributed to the final approval of the manuscript; HJB contributed to the conception and design of the study, data analysis and interpretation, and final approval of manuscript. All authors were involved in the discussion and interpretation of data and all approved the final version.
The authors reported no potential conficts of interest.
Funding: this work was supported by the Deutsche Forschungsgemeinschaft (DFG), Sonderforschungsbereich SFB-685 (Immunotherapy: Molecular Basis and Clinical Applications) project Z2: Core Facility Cell Sorting, by the DFG project BU 516/2-1: Identifizierung und funktionelle Untersuchung von MSC-spezifischen Molekülen, and by the Federal Ministry for Education and Research (BMBF), BioProfile Stuttgart/Neckar-Alb; project 0313668B: Entwicklung eines bioartifiziellen Leberreaktors mit allogenen humanen Hepatozyten.
References
- 1.Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–92. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 2.Caplan AI. Mesenchymal stem cells. J Orthop Res. 1991;9:641–50. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 3.Jones EA, Kinsey SE, English A, Jones RA, Straszynski L, Meredith DM, et al. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheum. 2002;46:3349–60. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 5.Battula VL, Bareiss PM, Treml S, Conrad S, Albert I, Hojak S, et al. Human placenta and bone marrow derived MSC cultured in serum-free, b-FGF-containing medium express cell surface frizzled-9 and SSEA-4 and give rise to multilineage differentiation. Differentiation. 2007;75:279–91. doi: 10.1111/j.1432-0436.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 6.Erices A, Conget P, Minguell JJ. Mesenchymal progenitor cells in human umbilical cord blood. Br J Haematol. 2000;109:235–42. doi: 10.1046/j.1365-2141.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 7.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 8.Fukuchi Y, Nakajima H, Sugiyama D, Hirose I, Kitamura T, Tsuji K. Human placenta-derived cells have mesenchymal stem/progenitor cell potential. Stem Cells. 2004;22:649–58. doi: 10.1634/stemcells.22-5-649. [DOI] [PubMed] [Google Scholar]
- 9.Mareschi K, Biasin E, Piacibello W, Aglietta M, Madon E, Fagioli F. Isolation of human mesenchymal stem cells: bone marrow versus umbilical cord blood. Haematologica. 2001;86:1099–100. [PubMed] [Google Scholar]
- 10.Miki T, Lehmann T, Cai H, Stolz DB, Strom SC. Stem cell characteristics of amniotic epithelial cells. Stem Cells. 2005;23:1549–59. doi: 10.1634/stemcells.2004-0357. [DOI] [PubMed] [Google Scholar]
- 11.Yen BL, Huang HI, Chien CC, Jui HY, Ko BS, Yao M, et al. Isolation of multipotent cells from human term placenta. Stem Cells. 2005;23:3–9. doi: 10.1634/stemcells.2004-0098. [DOI] [PubMed] [Google Scholar]
- 12.Zvaifler NJ, Marinova-Mutafchieva L, Adams G, Edwards CJ, Moss J, Burger JA, et al. Mesenchymal precursor cells in the blood of normal individuals. Arthritis Res. 2000;2:477–88. doi: 10.1186/ar130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, et al. Stem cell properties of human periodontal ligament cells. J Periodontal Res. 2006;41:303–10. doi: 10.1111/j.1600-0765.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 14.Huss R. Perspectives on the morphology and biology of CD34-negative stem cells. J Hematother Stem Cell Res. 2000;9:783–93. doi: 10.1089/152581600750062228. [DOI] [PubMed] [Google Scholar]
- 15.Caplan AI. Review: mesenchymal stem cells: cell-based reconstructive therapy in orthopedics. Tissue Eng. 2005;11:1198–211. doi: 10.1089/ten.2005.11.1198. [DOI] [PubMed] [Google Scholar]
- 16.Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6:230–47. [PubMed] [Google Scholar]
- 17.Friedenstein AJ. Stromal mechanisms of bone marrow: cloning in vitro and retransplantation in vivo. Haematol Blood Transfus. 1980;25:19–29. doi: 10.1007/978-3-642-67319-1_3. [DOI] [PubMed] [Google Scholar]
- 18.Prockop DJ, Sekiya I, Colter DC. Isolation and characterization of rapidly self-renewing stem cells from cultures of human marrow stromal cells. Cytotherapy. 2001;3:393–6. doi: 10.1080/146532401753277229. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz EM, Le BK, Dominici M, Mueller I, Slaper-Cortenbach I, Marini FC, et al. Clarification of the nomenclature for MSC: the International Society for Cellular Therapy position statement. Cytotherapy. 2005;7:393–5. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 20.Seshi B, Kumar S, Sellers D. Human bone marrow stromal cell: coexpression of markers specific for multiple mesenchymal cell lineages. Blood Cells Mol Dis. 2000;26:234–46. doi: 10.1006/bcmd.2000.0301. [DOI] [PubMed] [Google Scholar]
- 21.Simmons PJ, Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55–62. [PubMed] [Google Scholar]
- 22.Conconi MT, Burra P, Di LR, Calore C, Turetta M, Bellini S, et al. CD105(+) cells from Wharton’s jelly show in vitro and in vivo myogenic differentiative potential. Int J Mol Med. 2006;18:1089–96. [PubMed] [Google Scholar]
- 23.Gang EJ, Bosnakovski D, Figueiredo CA, Visser JW, Perlingeiro RC. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–51. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 24.Liu PG, Zhou DB, Shen T. Identification of human bone marrow mesenchymal stem cells: preparation and utilization of two monoclonal antibodies against SH2, SH3. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2005;13:656–9. [PubMed] [Google Scholar]
- 25.Quirici N, Soligo D, Bossolasco P, Servida F, Lumini C, Deliliers GL. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp Hematol. 2002;30:783–91. doi: 10.1016/s0301-472x(02)00812-3. [DOI] [PubMed] [Google Scholar]
- 26.Buhring HJ, Battula VL, Treml S, Schewe B, Kanz L, Vogel W. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–71. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- 27.Gentry T, Foster S, Winstead L, Deibert E, Fiordalisi M, Balber A. Simultaneous isolation of human BM hematopoietic, endothelial and mesenchymal progenitor cells by flow sorting based on aldehyde dehydrogenase activity: implications for cell therapy. Cytotherapy. 2007;9:259–74. doi: 10.1080/14653240701218516. [DOI] [PubMed] [Google Scholar]
- 28.Azizi SA, Stokes D, Augelli BJ, Digirolamo C, Prockop DJ. Engraftment and migration of human bone marrow stromal cells implanted in the brains of albino rats–similarities to astrocyte grafts. Proc Natl Acad Sci USA. 1998;95:3908–13. doi: 10.1073/pnas.95.7.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zola H, Swart B, Cholson I, Voss E. CD56. Leukocyte and Stromal Cell Molecules- The CD markers. New Jersey: Wiley-Liss; 2007. pp. 138–9. [Google Scholar]
- 30.Plappert CF, Schachner M, Pilz PK. Neural cell adhesion molecule (NCAM−/−) null mice show impaired sensitization of the startle response. Genes Brain Behav. 2006;5:46–52. doi: 10.1111/j.1601-183X.2005.00132.x. [DOI] [PubMed] [Google Scholar]
- 31.Vogel W, Grunebach F, Messam CA, Kanz L, Brugger W, Buhring HJ. Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica. 2003;88:126–33. [PubMed] [Google Scholar]
- 32.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–4. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 33.Ritz J, Trinchieri G, Lanier LL. NK-cell antigens: section report. In: Schlossman SF, Boumsell L, Gilks W, Harlan JM, Kishitomoto T, editors. Leucocyte Typing V. White Cell Differentiation Antigens. V. Oxford New York Tokyo: Oxford University Press; 1995. pp. 1367–72. [Google Scholar]
- 34.Jones M, Mason D. CD10 workshop panel report. In: Kishimoto Teal., editor. Leucocyte Typing VI. VI. Garland Publishing, Inc; New York & London: 1998. pp. 131–2. [Google Scholar]
- 35.Todd RF. Myeloid antigens: section report. In: Schlossman SF, Boumsell L, Gilks W, editors. Leucocyte Typing-V. V. Oxford: New York, Tokyo; 1995. pp. 739–70. [Google Scholar]
- 36.Broudy VC, Lin NL, Buhring HJ, Komatsu N, Kavanagh TJ. Analysis of c-kit receptor dimerization by fluorescence resonance energy transfer. Blood. 1998;91:898–906. [PubMed] [Google Scholar]
- 37.Doyonnas R, Yi-Hsin CJ, Butler LH, Rappold I, Lee-Prudhoe JE, Zannettino AC, et al. CD164 monoclonal antibodies that block hemopoietic progenitor cell adhesion and proliferation interact with the first mucin domain of the CD164 receptor. J Immunol. 2000;165:840–51. doi: 10.4049/jimmunol.165.2.840. [DOI] [PubMed] [Google Scholar]
- 38.Hennersdorf F, Florian S, Jakob A, Baumgartner K, Sonneck K, Nordheim A, et al. Identification of CD13, CD107a, and CD164 as novel basophil-activation markers and dissection of two response patterns in time kinetics of IgE-dependent upregulation. Cell Res. 2005;15:325–35. doi: 10.1038/sj.cr.7290301. [DOI] [PubMed] [Google Scholar]
- 39.Buhring HJ, Kuci S, Conze T, Rathke G, Bartolovic K, Grunebach F, et al. CDCP1 identifies a broad spectrum of normal and malignant stem/progenitor cell subsets of hematopoietic and nonhematopoietic origin. Stem Cells. 2004;22:334–43. doi: 10.1634/stemcells.22-3-334. [DOI] [PubMed] [Google Scholar]
- 40.Nauta AJ, Kruisselbrink AB, Lurvink E, Willemze R, Fibbe WE. Mesenchymal stem cells inhibit generation and function of both CD34+-derived and monocyte-derived dendritic cells. J Immunol. 2006;177:2080–7. doi: 10.4049/jimmunol.177.4.2080. [DOI] [PubMed] [Google Scholar]
- 41.Larson BL, Ylostalo J, Prockop DJ. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26:193–201. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 42.Smith JR, Pochampally R, Perry A, Hsu SC, Prockop DJ. Isolation of a highly clonogenic and multipotential subfraction of adult stem cells from bone marrow stroma. Stem Cells. 2004;22:823–31. doi: 10.1634/stemcells.22-5-823. [DOI] [PubMed] [Google Scholar]
- 43.Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841–5. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez C, Hofmann TJ, Marino R, Dominici M, Horwitz EM. Human bone marrow mesenchymal stromal cells express the neural ganglioside GD2: a novel surface marker for the identification of MSCs. Blood. 2007;109:4245–8. doi: 10.1182/blood-2006-08-039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xian CJ, Foster BK. Repair of injured articular and growth plate cartilage using mesenchymal stem cells and chondrogenic gene therapy. Curr Stem Cell Res Ther. 2006;1:213–29. doi: 10.2174/157488806776956904. [DOI] [PubMed] [Google Scholar]
- 46.Scharstuhl A, Schewe B, Benz K, Gaissmaier C, Buhring HJ, Stoop R. Chondrogenic potential of human adult mesenchymal stem cells is independent of age or osteoarthritis etiology. Stem Cells. 2007;25:3244–51. doi: 10.1634/stemcells.2007-0300. [DOI] [PubMed] [Google Scholar]
- 47.Schewe B, Fritz J, Gaissmaier C, Weise K. [Cartilage cell transplant: surgical technique with matrix-associated chondrocytes] Unfallchirurg. 2006;109:577–82. doi: 10.1007/s00113-006-1122-5. [DOI] [PubMed] [Google Scholar]