Abstract
Background
Completed genome sequences are rapidly increasing for Rickettsia, obligate intracellular α-proteobacteria responsible for various human diseases, including epidemic typhus and Rocky Mountain spotted fever. In light of phylogeny, the establishment of orthologous groups (OGs) of open reading frames (ORFs) will distinguish the core rickettsial genes and other group specific genes (class 1 OGs or C1OGs) from those distributed indiscriminately throughout the rickettsial tree (class 2 OG or C2OGs).
Methodology/Principal Findings
We present 1823 representative (no gene duplications) and 259 non-representative (at least one gene duplication) rickettsial OGs. While the highly reductive (∼1.2 MB) Rickettsia genomes range in predicted ORFs from 872 to 1512, a core of 752 OGs was identified, depicting the essential Rickettsia genes. Unsurprisingly, this core lacks many metabolic genes, reflecting the dependence on host resources for growth and survival. Additionally, we bolster our recent reclassification of Rickettsia by identifying OGs that define the AG (ancestral group), TG (typhus group), TRG (transitional group), and SFG (spotted fever group) rickettsiae. OGs for insect-associated species, tick-associated species and species that harbor plasmids were also predicted. Through superimposition of all OGs over robust phylogeny estimation, we discern between C1OGs and C2OGs, the latter depicting genes either decaying from the conserved C1OGs or acquired laterally. Finally, scrutiny of non-representative OGs revealed high levels of split genes versus gene duplications, with both phenomena confounding gene orthology assignment. Interestingly, non-representative OGs, as well as OGs comprised of several gene families typically involved in microbial pathogenicity and/or the acquisition of virulence factors, fall predominantly within C2OG distributions.
Conclusion/Significance
Collectively, we determined the relative conservation and distribution of 14354 predicted ORFs from 10 rickettsial genomes across robust phylogeny estimation. The data, available at PATRIC (PathoSystems Resource Integration Center), provide novel information for unwinding the intricacies associated with Rickettsia pathogenesis, expanding the range of potential diagnostic, vaccine and therapeutic targets.
Introduction
Rickettsiae are a group of organisms belonging to the class Alphaproteobacteria, a large and metabolically diverse group of gram-negative bacteria [1]–[3]. Within Alphaproteobacteria, the order Rickettsiales comprises three families: Holosporaceae, Anaplasmataceae and Rickettsiaceae [4], of which Rickettsia spp. are grouped in the latter, along with the monotypic genus Orientia, the scrub typhus agent [5]. Robust phylogenetic analysis further suggests that the abundant free-living marine bacterioplankton Pelagibacter ubique and mitochondria are early-branching groups of the order [6]. Species in the genus Rickettsia are obligate intracellular symbionts of plants [7], amoebae [8], [9], arthropods [e.g., 10]–[13], annelids [14], vertebrates [15] and likely many other organisms [16]. Most Rickettsia-containing vertebrates are secondary hosts that acquired these bacteria via blood-feeding arthropods or the transdermal inoculation or inhalation of the feces of infected arthropods. Rickettsia spp. are often parasitic in the secondary vertebrate host [e.g., 17], and their pathogenicity to some extent has been well studied. In particular, human rickettsial infections are known to cause many diseases, including epidemic typhus (R. prowazekii), murine typhus (R. typhi), murine typhus-like (R. felis), rickettsial pox (R. akari), Rocky Mountain spotted fever (R. rickettsii), Boutonneuse fever (R. conorii), and North Asian tick typhus (R. sibirica). These virulent species of rickettsiae are of great interest both as emerging infectious diseases [18] and for their potential deployment as bioterrorism agents [19], [20].
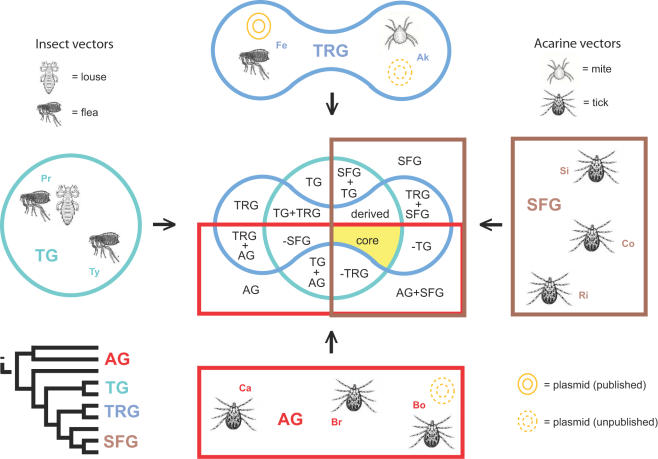
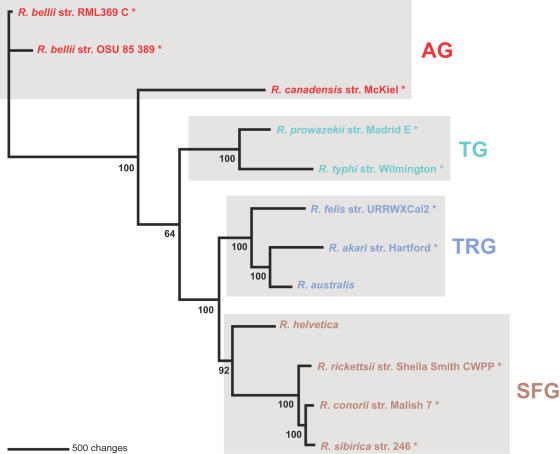
Due to both small genome size and medical importance, ten genome sequences from Rickettsia spp. have been published and annotated in the last decade [9], [21]–[27], providing a foundation to study the evolutionary history of these lineages through comparative genomics. Recently, Gillespie et al. [28] proposed a revision to the long-standing classification of Rickettsia by erecting the transitional group (TRG) as a distinct lineage that shares immediate ancestry with the members of the spotted fever group (SFG) rickettsiae. Coupled with the typhus group (TG) and ancestral group (AG) rickettsiae, these four rickettsial lineages comprising 10 sequenced genomes present an opportunity to create a database that encompasses the distribution of the predicted open reading frames (ORFs) across all ten annotated genomes ( Figure 1 ).
Figure 1. Venn diagram depicting 15 intersections for the four rickettsial groups.
Classification scheme based on molecular phylogeny estimation [28], the topology of which is shown in the lower left; AG = ancestral group, TG = typhus group, TRG = transitional group, SFG = spotted fever group. Genome codes are as follows: Br = R. bellii str. RML369-C, Bo = R. bellii str. OSU 85 389, Ca = R. canadensis str. McKiel, Pr = R. prowazekii str. Madrid E, Ty = R. typhi str. Wilmington, Ak = R. akari str. Hartford, Fe = R. felis str. URRWXCal2, Ri = R. rickettsii str. Sheila Smith CWPP, Co = R. conorii str. Malish 7, and Si = R. sibirica str. 246. Arthropod hosts are illustrated for each genome, and strains known to harbor plasmids are depicted.
Establishing orthology across multiple genomes serves not only to identify genes with shared evolutionarily histories, but also facilitates genome annotation [29], [30], and significant attention has focused on algorithms for creating orthologous groups (OGs). Recent work has centered on the following four aspects: i) overall improvement of OG assignment in the face of paralogy, ii) building tools for the cross-querying of taxon-specific databases, iii) creating databases that house specific gene or protein profiles for facilitating the identification of orthologs in novel sequences, and iv) the inclusion of phylogeny estimation into the processes of assigning orthology and detecting paralogy.
At the PathoSystems Resource Integration Center (PATRIC) [31], OGs have been preliminarily established for several groups of organisms, including Rickettsia spp. The advantage of a Rickettsia-specific database lies not only in the ability to query exclusively against the 10 genomes currently annotated in our system, but also to evaluate the results of several algorithmic approaches that create OGs. Furthermore, PATRIC offers continued updates to the annotation of rickettsial genes and proteins, and provides multiple sequence alignments as well as phylogenetic trees, when applicable, for each OG consisting of two to ten rickettsial taxa. The database will continually evolve with the addition of newly sequenced rickettsial genomes, with existing OG assignments driving the curation process of raw genome data.
In the present study, we report the rickettsial OGs (RiOGs) in conjunction with a highly robust phylogeny of the core rickettsial genes, providing an evolutionary framework for interpreting the genomic characteristics of the four main lineages of Rickettsia. These data highlight the genetic anomalies previously characterized for this genus, such as extremely reduced genomes and the high presence of putative pseudogenes, and also reveal novel characteristics including the lack of group-specific virulence factors and high occurrence of lateral transfer between groups that harbor plasmids (AG and TRG rickettsiae). Information on the conserved core genes, as well as those that may be involved in specific functions that define monophyletic groups, host associations, and plasmid-related behavior, will be valuable resources for future laboratory work (e.g., development of vaccines, diagnostics and therapeutics) as well as further evolutionary studies of this intriguing obligate intracellular bacterial group.
Results and Discussion
Synteny and Phylogeny of Rickettsia Genomes
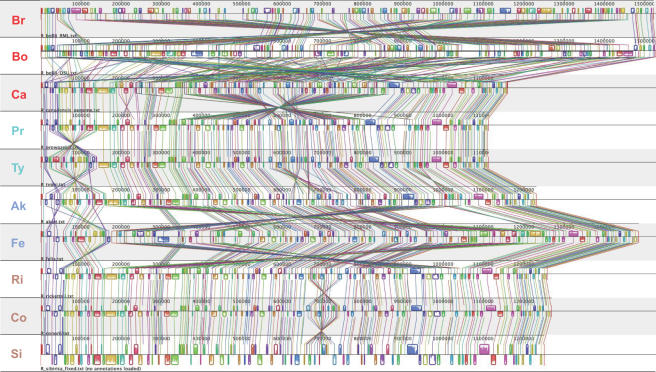
Whole genome alignments for the ten analyzed Rickettsia taxa reveal highly conserved colinearity in six of the seven derived species (sans R. bellii and R. canadensis) with minimal gene rearrangements, most of which occur near the predicted origin of replication termination ( Figure 2 ). However, the R. felis genome contains several long-range symmetrical inversions in the central region of the alignment that are not found in other taxa. Removal of R. felis from the alignment illustrates the highly conserved synteny across the derived rickettsia taxa ( Figure S1-A ). Furthermore, switching the positions of R. akari and R. felis in the alignment ( Figure S1-B ) demonstrates that these central inversions in R. felis, as well as a large genome size, are autapomorphic (uniquely derived) traits within derived rickettsiae. Among the three AG rickettsiae, R. canadensis (formerly R. canada) is more colinear with the derived taxa than it is to either R. bellii strain. Like R. felis, R. canadensis contains several autapomorphic symmetrical inversions in the central region of the alignment, yet they are smaller than the long-range inversions found in R. felis. As previously reported [32], R. bellii str. RML369-C shares little colinearity with other rickettsial genomes, and our analysis of both R. bellii genomes is in agreement with this observation. Despite several long and short range inversions between the R. bellii str. RML369-C and R. bellii str. OSU 85-389 genomes, few gene positions are shared with R. bellii and R. canadensis or the derived taxa ( Figure 2 ), and switching the positions of the R. bellii strains in the alignment does not result in more conserved synteny between either strain and the derived taxa ( Figure S1-C, D ).
Figure 2. Alignment of 10 rickettsial genomes.
Taxa are in the same position as in estimated trees in Figure 3, with taxon abbreviations explained in the Figure 1 legend. Alignment created using Mauve [189] after reindexing the R. sibirica genome (see text for details).
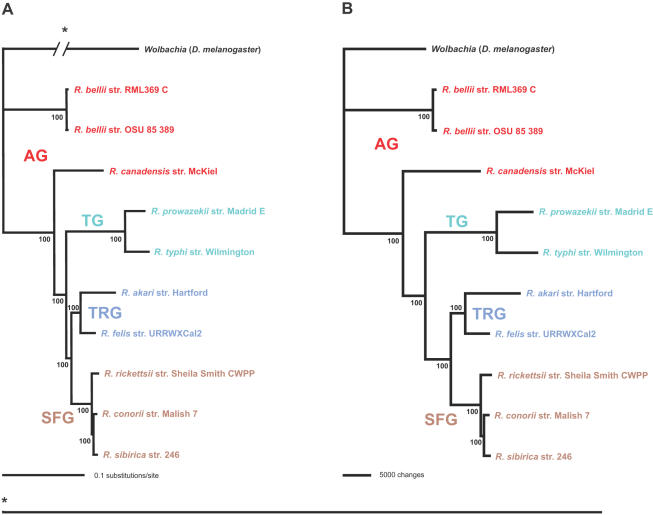
Phylogenetic analyses implementing both maximum likelihood and parsimony of the 731 representative core rickettsial proteins (discussed below) resulted in robust estimates for these 10 taxa ( Figure 3 ). The estimated tree topologies are identical in branching pattern and are congruent with the tree from our previous analysis of 716 fewer genes [28], suggesting that ten or more concatenated (and well-behaved, with high signal to noise ratio) genes are sufficient for obtaining a robust phylogenetic estimate for these rickettsial taxa. Thus, our recent classification scheme for Rickettsia consisting of 4 major groups (AG rickettsiae: R. bellii str. RML369-C, R. bellii str. OSU 85 389, R. canadensis str. McKiel; TG rickettsiae: R. prowazekii str. Madrid E, R. typhi str. Wilmington; TRG rickettsiae: R. akari str. Hartford, R. felis str. URRWXCal2; SFG rickettsiae: R. rickettsii str. Sheila Smith CWPP, R. conorii str. Malish 7, R. sibirica str. 246) is substantiated with a phylogenomic approach. In what follows, we use this evolutionary framework to analyze the distribution and relative conservation of all predicted genes for these ten rickettsial genomes.
Figure 3. Estimated phylogenies of ten rickettsial taxa based on 731 representative core proteins.
(A) Tree from Bayesian analysis. Three MCMC chains were primed with a neighbor-joining tree and run independently for 25000 generations in model-jumping mode. Burn-in was attained by 2500 generations for all chains, and a single tree topology with exclusive use of the Jones substitution model was observed in post burn-in data. The consensus tree shown here thus has 100% support for every branch. Branch support is from the distribution of posterior probabilities from all trees minus the burn-in. (B) Tree from exhaustive search using parsimony. Branch support is from one million bootstrap replicates.
Predicted OGs: Conservation and Representation
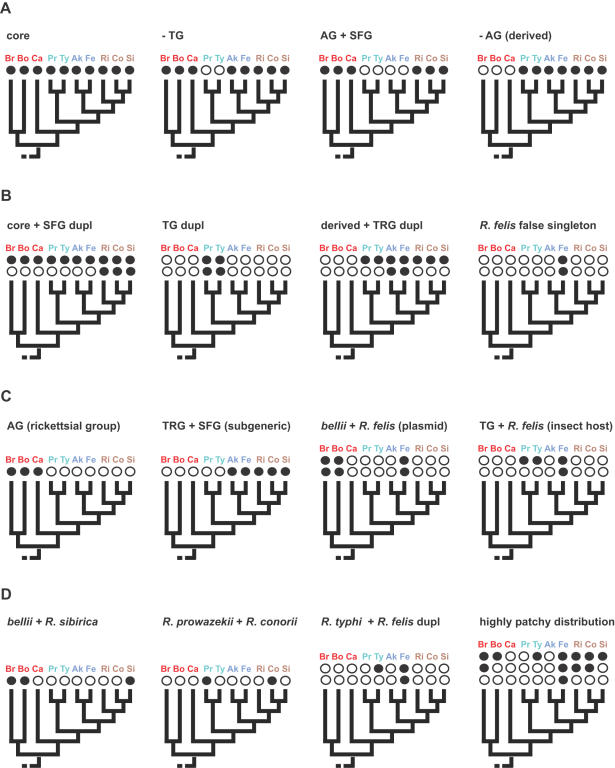
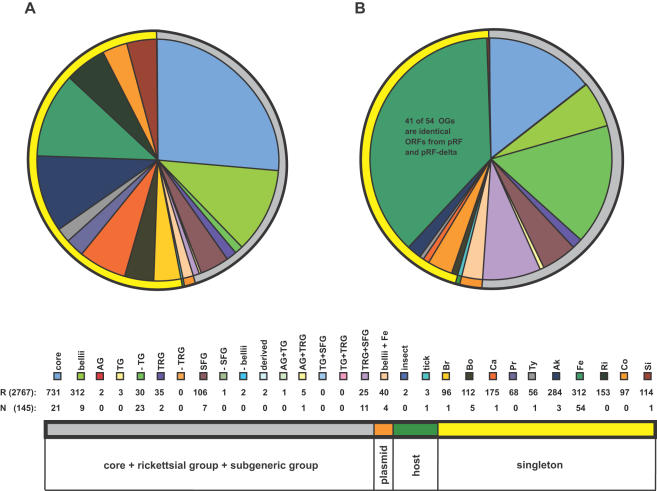
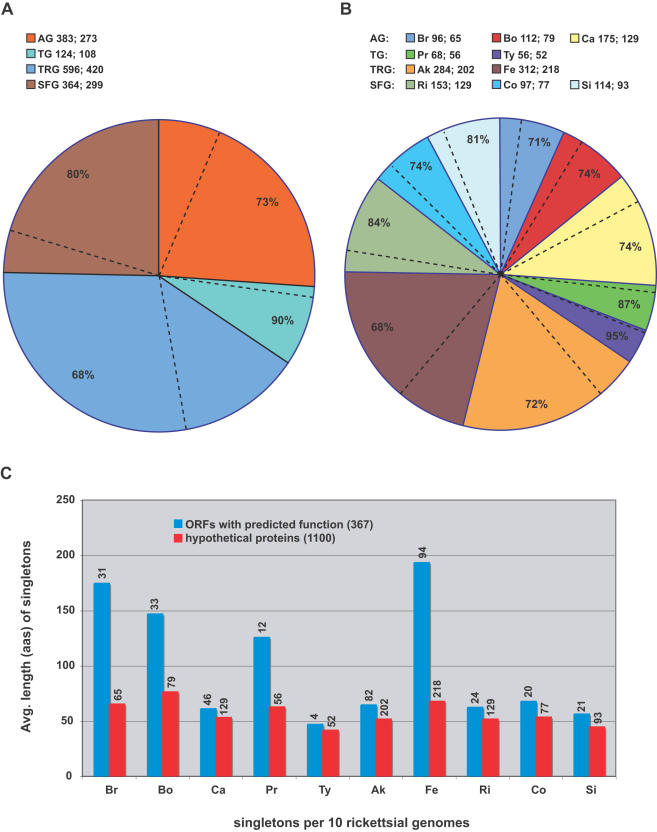
In the analysis of the rapidly growing list of rickettsial genomes we determined that OrthoMCL, a program that applies the Markov clustering algorithm of Van Dongen [33] to resolve the many-to-many orthologous relationships present within cross genome comparisons [34], outperformed more traditional approaches to establishing OGs, such as bidirectional best BLAST hits with and without cliques. Thus, we show here the results generated by OrthoMCL only, which grouped 12887 ORFs into 2082 total OGs ( Table 1 ). The bulk (88%) of these OGs are representative ( Figure 4A ), meaning they include only one CDS per strain, thus ranging in membership from 2–10 sequences. The remaining 12% of the OGs are non-representative ( Figure 4B ) and include multiple predicted ORFs from at least one member. Categorization of the OGs into two classes based on distribution across the rickettsial tree and other attributes, such as presence of plasmids and common arthropod hosts ( Figure 4C–D ), reveals that 69% of the OGs are comprised of single rickettsial groups (e.g., AG, TG, TRG, and SFG), shared rickettsial groups (subgeneric), plasmid-harboring genomes, and genomes with common arthropod hosts ( Table 1 ). These class 1 OGs (C1OGs) contain 76% of the predicted ORFs grouped into OGs by OrthoMCL, suggesting that our criteria for distinguishing biologically interesting protein families based empirically on robust phylogeny estimation, presence of extra-chromosomal DNA and shared arthropod hosts is valid. The remaining ORFs grouped into class 2 OGs (C2OGs) depict gene families drifting or sporadically lost from the core genetic repertoire of the rickettsial ancestor [32] or genes acquired laterally ( Figure S2 ). Interestingly, while the majority (71%) of representative OGs qualify as C1OGs, the non-representative OGs are distributed within C1OGs and C2OGs in near equal frequency ( Table 1 ), suggesting minimal conservation for gene duplications and laterally acquired genes in these rickettsial genomes.
Table 1. Distribution of representative and non-representative OGs predicted across 14354 ORFs from ten rickettsial genomes, and their categorization into Class 1 and Class 2 OGs.1 .
| Composition2 | All OGs | C1OGs3 | C2OGs4 | |||
| No. OGs | No. ORFs | No. OGs | No. ORFs | No. OGs | No. ORFs | |
| representative | 1823 (88%) | 11026 (86%) | 1300 (71%) | 8910 (81%) | 523 (29%) | 2116 (19%) |
| non-representative | 259 (12%) | 1861 (14%) | 145 (56%) | 930 (50%) | 114 (44%) | 931 (50%) |
| Tot. | 2082 | 12887 | 1445 (69%) | 9840 (76%) | 637 (31%) | 3047 (24%) |
Of 14354 total ORFs, 12887 were grouped by OrthoMCL, leaving 1467 singletons.
Containing either no duplications per each member within an OG (representative) or at least one member with a duplication within an OG (non-representative).
Figure 4. Illustration of representative and non-representative OGs and their categorization into Class 1 and Class 2 OGs.
Taxon abbreviations are explained in the Figure 1 legend. Dark circles depict gene presence, while open circles depict gene absence. (A) Representative OGs: orthologous groups with only one ORF per included genome. Our analysis includes ten rickettsial genomes, thus representative OGs only include from 2–10 ORFs. Four examples are shown. (B) Non-representative OGs: orthologous groups with multiple ORFs from at least one included genome, comprised of either recent (orthologs) or distant (paralogs) gene duplications (dupl). False singleton OGs are comprised of only one taxon, but with multiple ORFs from that taxon (example on right). Four examples are shown. (C) Class 1 OGs (C1OGs): orthologous groups comprising single rickettsial groups (e.g., AG, TG, TRG, and SFG), shared rickettsial groups (subgeneric), plasmid-harboring genomes, and genomes with common arthropod hosts. Two representative (left) and two non-representative (right) C1OGs are shown. (D) Class 2 OGs (C2OGs): orthologous groups with patchy distribution across the rickettsial tree, depicting gene losses and/or genes acquired laterally. Two representative and two non-representative C2OGs are shown.
The RiOGs range in membership from two to 31 ORFs, with few (<3%) OGs exceeding more than 10 ORFs ( Table 2 ). Representative C1OGs comprise a substantial portion (64%) of the OGs with membership of 10 or fewer ORFs. Regarding the OGs with more than 10 members, a range from 4% (R. prowazekii) to 32% (R. conorii) illustrates the frequencies at which a particular rickettsial genome contributes to non-representation. As expected due to their smaller genome sizes and few gene duplications [21], [25], TG rickettsiae make little contribution (avg. 5%) to larger non-representative OGs as compared to AG (avg. 19%), TRG (avg. 17%) and SFG (avg. 31%) rickettsiae ( Table 2 ). Thus, these three latter groups have genomes more tolerant of multicopy genes, particularly those resulting from transposases and other insertion sequences, which act to produce elevated levels of paralogous genes. For instance, analysis of the distribution of RiOGs containing genes associated with mobile DNA and/or horizontal gene transfer (HGT), such as genes coding for proteins with ankyrin (ANK) and tetratricopeptide repeat (TPR) motifs, proteins with rickettsial palindromic elements (RPE), proteins associated with transposable elements (TNP), proteins of toxin-antitoxin modules (TA), and phage related elements, revealed that they are nearly non-existent in TG rickettsial genomes ( Table 3 ). The remaining three lineages, all purportedly containing some species that harbor plasmids, have elevated levels of most of these gene groups compared to TG rickettsiae. Interestingly, nearly half (47%) of the C2OGs are comprised of these six gene groups, while only a small portion of the C1OGs (5%) and singletons (4%) contain them ( Table 3 ). Given the probable lateral inheritance of many of these genes, either as facilitators or products of HGT, it is evident that they are less conserved and of less importance to overall rickettsial fitness and survival. However, their contribution to species- and strain-specific pathogenicity cannot be overlooked. Interestingly, our observation that these more promiscuous gene families tend to occur predominantly within C2OGs is congruent with a recent study demonstrating that barriers to bacterial HGT are more stringent for single copy genes [35].
Table 2. Breakdown of membership (no. ORFs) across 2082 rickettsial OGs.
| OGs with 10 or fewer ORFs | |||
| No. ORFs | No. OGs | Representative C1OGs1 | Remaining OGs2 , 3 , 4 |
| 2 | 585 | 312 (bellii); 3 (TG); 35 (TRG); 40 (bellii+Fe) | 195 |
| 3 | 225 | 2 (AG); 106 (SFG); 2 (insect) | 115 |
| 4 | 128 | 0 (TG+TRG) | 128 |
| 5 | 90 | 25 (TRG+SFG); 1 (AG+TG); 5 (AG+TRG); 0 (TG+SFG) | 59 |
| 6 | 62 | 3 (tick) | 59 |
| 7 | 65 | 2 (derived); 1 (-SFG) | 62 |
| 8 | 65 | 2 (-bellii); 30 (-TG); 0 (-TRG) | 33 |
| 9 | 56 | 0 | 56 |
| 10 | 748 | 731 (core) | 17 |
| Tot | 2024 | 1300 | 724 (523 rep., 201 non-rep.) |
C1OGs (see Figure 4 for description and Figure 5 and Figure 7 for distribution of representative and non-representative
C1OGs across rickettsial phylogeny).
Comprising both representative and non-representative OGs.
Distributions of included C2OGs are shown over rickettsial phylogeny in Figure S2
First number is total no. ORFs within OGs; second number depicts no. of ORFs causing non-representation.
Taxon abbreviations are explained in the Figure 1 legend.
Table 3. Distribution across 10 rickettsial genomes of OGs and singletons containing proteins with ankyrin (ANK) and tetratricopeptide repeat (TPR) motifs, proteins with rickettsial palindromic elements (RPE), proteins associated with transposable elements (TPN), proteins of toxin-antitoxin modules (TA), and phage related proteins.
| C1OGs1 | Tot. OGs | Distribution2 | |||||||||||||||
| ANK | TPR | RPE | TNP | TA | PHAGE | Tot. | |||||||||||
| R | NR | R | NR | R | NR | R | NR | R | NR | R | NR | R | NR | R | NR | ALL | |
| core | 731 | 21 | 0 | 0 | 1 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 11 |
| AG | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| bellii | 312 | 9 | 10 | 0 | 3 | 0 | 0 | 0 | 3 | 5 | 1 | 0 | 1 | 0 | 18 | 5 | 23 |
| - bellii | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TG | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -TG | 30 | 23 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 3 | 3 | 6 |
| TRG | 35 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | 5 |
| SFG | 106 | 7 | 4 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 7 |
| -SFG | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| derived | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AG+TG | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AG+TRG | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| TRG+SFG | 25 | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | 4 |
| bellii +Fe | 40 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 1 | 0 | 7 | 0 | 7 |
| insect | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| tick | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tot. | 1300 | 79 | 16 | 0 | 4 | 2 | 13 | 0 | 6 | 5 | 14 | 0 | 2 | 2 | 55 | 9 | 64 |
| (5%) | |||||||||||||||||
C1OGs (see Figure 4 for description and Figure 5 and Figure 7 for distribution of representative and non-representative C1OGs across rickettsial phylogeny).
R = representative OGs, NR = non-representative OGs (see Figure 4 for description).
C2OGs (see Figure 4 for description and Figure S2 for distribution of representative and non-representative C2OGs across rickettsial phylogeny).
Percentage of 637 C2OGs present within each rickettsial genome. The 128 distributions of these OGs are illustrated in Figure S2 .
ORFs found in only one rickettsial genome. Does not include false singletons (see Figure 4 ).
A comparison of the distributions of both representative and non-representative C1OGs and their associated singletons uncovers the high occurrence of singleton genes (53%) per representative C1OGs ( Figure 5 ). While many singletons may be the product of gene overprediction (discussed below), some could possibly have important species- or strain-specific functions, such as host manipulation. “False singletons”, which depict non-representative OGs with all members from a single genome ( Figure 4C ), contribute less (17%) towards non-representation when identical genes from R. felis plasmids pRF and pRFδ are not considered (for speculation on the existence of pRFδ see Gillespie et al. [28]). Thus the biological causes of non-representation, such as HGT and gene duplication, tend to occur more within gene families common across multiple rickettsial genomes rather than in unique genes within individual genomes. This is congruent with our determination of the high occurrence within C2OGs of six gene families typically associated with mobile DNA and/or HGT (above).
Figure 5. Comparison of the distributions of 1300 representative and 145 non-representative class 1 OGs (C1OGs), 66 false singletons, and 1467 singleton ORFs.
Slices depict 16 generic and subgeneric groups, false singletons, singletons, plasmid associated groups, and two host-related groups, with outer circle colors depicted in schema. Taxon abbreviations, including subgeneric groups, are explained in the Figure 1 legend. (A) Distribution of 1300 representative C1OGs and 1467 singletons. (B) Distribution of 79 non-representative C1OGs and 66 false singletons.
The Nature of Non-Representation
The degree of non-representation recovered by OrthoMCL is not a surprise as Rickettsia genomes are notorious for being highly reductive [e.g., 36]–[38], having a high occurrence of split genes and pseudogenes [e.g., 22], [23], [32], [39], [40] and limited conservation in important host-recognition proteins such as rickettsial outer membrane protein A (rOmpA) and other cell surface antigens (Scas) [e.g., 41]–[57]. Coupled with this, some of the more recently sequenced genomes (namely both R. bellii strains and R. felis) are riddled with gene rearrangements and elevated levels of repetitive elements and transposases [9], [27], and the staggering degree of repetitive sequences and gene duplications in the recently sequenced genome of Orientia tsutsugamushi [58] suggest the old paradigms for genome reduction and synteny in Rickettsiaceae need reevaluation. Furthermore, as we recently predicted [28], new evidence is mounting for the presence of plasmids in several members of AG, TRG and SFG rickettsiae (reviewed in Baldridge et al. [59]), with some proteins having high similarity to counterparts encoded on rickettsial chromosomes [e.g., 28], [60]. All of these factors confound the accurate assignment of gene orthology across genomes, and it is important to view our results as algorithm-dependent, which further required manual scrutiny and adjustment.
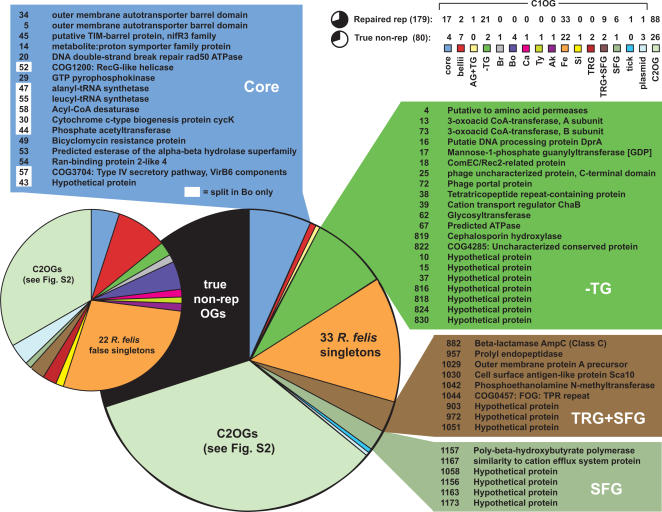
Manual inspection of the 259 non-representative OGs via multiple sequence alignment of each specific case revealed the high occurrence of split genes versus true gene duplications ( Table 4 ; Table S1 ). Including spurious duplications from the identical R. felis pRF and pRFδ plasmids, 387 problematic ORFs were eliminated or stitched together to create pseudogene ORFs, resulting in only 80 remaining non-representative OGs defined by true gene duplications. Notably, elimination of identical pRF and pRFδ plasmid genes created 33 additional R. felis singletons. After “repairing” OGs defined by split ORFs, four distributions contained the majority of C1OGs, illustrating the instances of gene decay from the core, -TG, TRG+SFG, and SFG distributions ( Figure 6 ). Regarding the repaired OGs with a core distribution, nearly half of the split genes were from the R. bellii str. OSU 85-389 genome and include critical genes such as those encoding alanyl- and leucyl-tRNA synthetases and one of the five virB6 components of the type IV secretion system. OGs containing split genes with a -TG distribution include two proteins possibly involved in DNA transformation: a ComEC/Rec2-related protein and a putative DNA processing protein DprA, plus two phage related proteins and a TPR motif-containing protein. This illustrates that genes deleted from the TG genomes involved in conjugation or other methods of foreign DNA uptake are in the process of decaying from the remaining rickettsial genomes. Through the comparison of the proportion of split genes to gene duplications per rickettsial genome ( Table 5 ), it is evident that split genes occur more frequently, particularly in SFG rickettsiae, and that both split genes and gene duplications are nearly nonexistent in TG rickettsiae. Interestingly, the genomes with plasmids and elevated levels of transposases and related elements, namely R. felis and R. bellii, also have elevated levels of gene duplications.
Table 4. Manual evaluation of 259 non-representative OGs across ten rickettsial genomes.
| Cause of non-representation1 | No. OGs | Tot. ORFs | Problem ORFs | Remaining non-rep. after manual curation |
| split genes only | 137 | 1217 | 280 split | 899 ORFs after concatenation; no non-rep. OGs |
| gene duplications only | 66 | 425 | 295 duplicated (207 duplications) | no change (all bona fide non-rep. OGs) |
| split genes+gene duplications | 6 | 78 | 9 split; 6 duplicated | 66 ORFs after concatenation; all non-rep. OGs |
| pRFδ only | 9 | 41 | 9 suspect duplications | 32 ORFs; no non-rep. OGs |
| pRFδ only (R. felis doublets) | 33 | 66 | 33 suspect duplications (pRFδ) | 33 R. felis singletons |
| pRFδ+gene duplications | 7 | 30 | 8 suspect duplications (pRFδ) | 22 remaining ORFs; all non-rep. OGs |
| pRFδ+split genes+gene duplications | 1 | 5 | 1 split; 2 suspect duplications (pRFδ) | 2 ORFs after concatenation; both non-rep. OGs |
| Tot. | 259 | 1862 | 387 split or spurious ORFs | 80 non-rep. OGs with 515 ORFs |
Split genes may be split multiple times, and multiple gene duplications may occur within single genomes (see Table S1 ).
Figure 6. Manual curation of 259 non-representative OGs predicted by OrthoMCL.
Schema depicts 179 OGs repaired to representative after stitching together split ORFs (larger pie chart) and remaining true non-representative OGs defined by in-paralogs.
Table 5. Characterization of 259 non-representative OGs per ten rickettsial genomes1.
| Group | Genome2 | Split genes3 | Gene duplications4 | Total5 | % Non-representation6 | |||
| AG | Br | 15 | 30 | 15 | 16 | 56 | 31 | 8% |
| Bo | 22 | 45 | 23 | 16 | 62 | 38 | 10% | |
| Ca | 20 | 41 | 21 | 2 | 11 | 22 | 5% | |
| Tot. | 57 | 116 | 59 | 34 | 129 | 91 | 23% | |
| TG | Pr | 3 | 6 | 3 | 0 | 0 | 3 | 0.70% |
| Ty | 1 | 2 | 1 | 2 | 4 | 3 | 0.70% | |
| Tot. | 4 | 8 | 4 | 2 | 4 | 6 | 1% | |
| TRG | Ak | 39 | 87 | 48 | 7 | 34 | 46 | 12% |
| Fe | 23 | 51 | 28 | 45 | 123 | 68 | 17% | |
| Tot. | 62 | 138 | 76 | 52 | 157 | 114 | 29% | |
| SFG | Ri | 59 | 128 | 69 | 7 | 14 | 66 | 17% |
| Co | 52 | 113 | 61 | 6 | 12 | 58 | 15% | |
| Si | 56 | 120 | 64 | 5 | 10 | 61 | 15% | |
| Tot. | 167 | 361 | 194 | 18 | 36 | 185 | 47% | |
| Tot. (all) | 290 | 623 | 333 | 106 | 326 | 396 | ||
Not including 52 instances where pRFδ ORFs cause or further contribute to non-representation.
Taxon abbreviations are explained in the Figure 1 legend.
Number of split genes, followed by number of ORFs resulting from splits, followed by overestimated ORFs. Note: split genes may be split more than once.
Number of gene duplications, followed by number of duplicated ORFs. Note: some genes are duplicated more than once, and pRF genes are considered duplications of R. felis chromosomal orthologs.
Total number of split ORFs and gene duplication events per genome.
Portion of each genome contributing to total non-representation.
Core and Group-Specific C1OGs
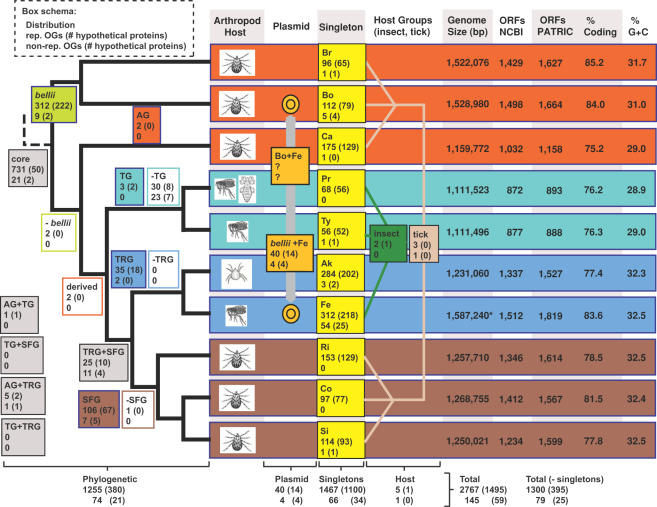
The distribution of representative (1300) and non-representative (79) C1OGs and singletons are shown over our estimated phylogeny ( Figure 7 ). Singletons (1467) are also shown but discussed in a separate section below. Of the 1379 C1OGs, 31% are annotated as hypothetical proteins (HPs), suggesting that a significant amount of even the conserved genes within these rickettsial genomes remain to be characterized. Not considering the bellii C1OG, which contains genes unique to the R. bellii genomes, the amount of HPs within the C1OGs decreases to 18%. The core and lineage specific C1OGs are discussed below.
Figure 7. Distribution of representative and non-representative class 1 OGs (C1OGs) and singleton ORFs over estimated rickettsial phylogeny.
Boxes depict the distribution of phylogenetic groups, singletons, plasmid associated groups, and host-related groups: Red = AG rickettsiae, aquamarine = TG rickettsiae, blue = TRG rickettsiae, brown = SFG rickettsiae, gray = higher-level groupings, light green = R. bellii strains only. Orange boxes depict genes found on the pRF plasmid of R. felis str. URRWXCal2 and chromosomes R. felis and both R. bellii strains (as of this publication the R. bellii plasmids remain unavailable). Genes specific to single rickettsial genomes (singletons) are in yellow boxes, with taxon abbreviations explained in the Figure 1 legend. Host specific groups are defined by green (insect) and tan (tick) boxes. Genome statistics were compiled from the PATRIC and NCBI databases. Cladogram is based on trees shown in Figure 3. Inset in dashed box describes general schema for each box. *Total R. felis genome size: 1,485,148 bp = chromosome; 62,829 bp = pRF and 39,263 bp = pRFδ.
Core rickettsial genes
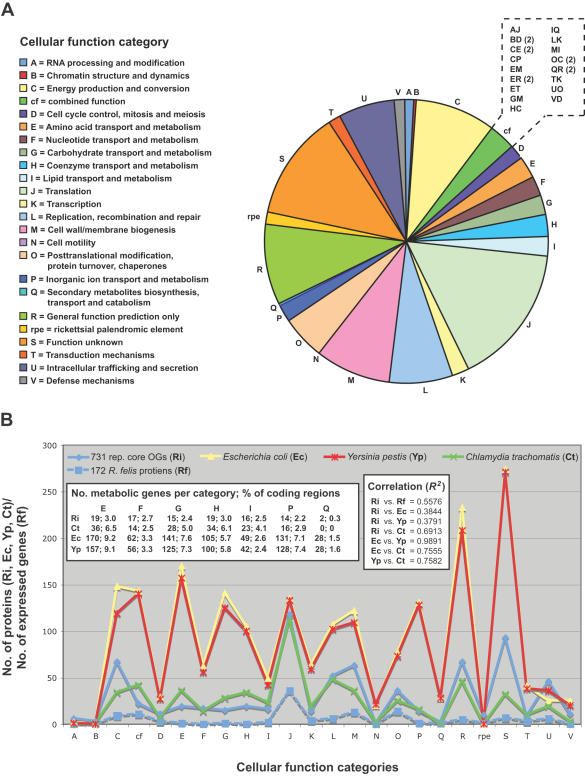
OrthoMCL grouped 731 representative and 21 non-representative protein families that are present in all ten analyzed rickettsial genomes ( Table S2 ). Thus, the genes encoding these proteins define the foundation of rickettsial biology, such as “house-keeping” functions, as well as rudimentary processes in host cell recognition, invasion and survival (but not necessarily virulence as not all Rickettsia spp. are known pathogens). The distribution of the assigned cellular functions of each of these core proteins provides insight on the conservation of cellular activities relative to other bacteria ( Figure 8A ). Not surprising, OGs involved in translation represent the largest functional category (16.14%), as other cellular functions such as amino acid (2.6%), carbohydrate (2.1%), nucleotide (2.3%), and lipid (2.2%) synthesis are less necessary when many of these resources can be obtained from host cells [61], [62]. Analyzing a crude depiction of the R. felis proteome, Ogawa et al. [40] reached a similar observation as their 172 identified proteins sorted into cellular function categories similar to those assigned for our core proteins, although with far fewer members per category ( Figure 8B ). The core rickettsial protein distribution across cellular function categories is also similar to another obligate intracellular pathogen, Chlamydia trachomatis, suggesting that this lifecycle is defined by reduction of many genes with conserved cellular functions (save translation) in facultative intracellular (Yersinia pestis) and extracellular (Escherichia coli) pathogenic bacteria. The percentage of ORFs coding for metabolic genes is lower in the obligate intracellular bacteria, with exception of the coenzyme transport/metabolism and lipid transport/metabolism genes of Chlamydia, which equal and exceed that of the two larger genomes, respectively.
Figure 8. Bioinformatic analysis of core representative OGs.
(A) Assignment of 731 core representative RiOGs to predicted cellular function categories. Format follows that established at the COG database (NCBI) except for cf = combined function and rpe = rickettsial palindromic element. (B) Comparison of the distribution of cellular function categories across 731 core rickettsial OGs (Ri), a recent protein expression profile for R. felis [40] (Rf), and COGs for three other bacteria: Escherichia coli (Ec), Yersinia pestis (Yp) and Chlamydia trachomatis (Ct). Inset at left shows the number of genes per genome for cellular function categories involved in organic and inorganic transport and metabolism (E, F, G, H, I, P, and Q) followed by the percentage these genes comprise of total protein-encoding genes. Results from a six-way regression analysis are shown in the right inset.
AG rickettsiae
Based on phylogeny estimation of over 30 proteins that placed R. canadensis basal to the TG, TRG and SFG rickettsiae, we categorized it with both R. bellii strains in the AG rickettsiae [28], a result recovered here and consistent with several previous studies [3; consensus tree of Vitorino et al. [63]]. Conversely, our analysis of OG distribution recovered only two proteins that are unique to AG rickettsiae: RiOG_1416 (Type I restriction-modification system, M subunit) and RiOG_1429 (F pilus assembly protein TraB). RiOG_1416 is truncated in R. bellii str. OSU 85-389 and extremely truncated in R. canadensis. Similarly, RiOG_1429 is truncated in R. canadensis; thus it is unlikely that either ORF is an important signature for AG rickettsiae. Furthermore, while both strains of R. bellii share 321 unique representative protein families ( Figure 7 , Table S3 ), R. canadensis only shares two unique proteins with the remaining derived rickettsiae: RiOG_925 (COG0419: ATPase involved in DNA repair) and RiOG_927 (methyltransferase family protein), with the latter likely part of a multigene family with other R. bellii homologs. Thus, OG distribution provides little evidence for placing R. canadensis either within AG rickettsiae or as derived. For instance, of the three derived rickettsial groups, R. canadensis shares more OGs with SFG (13; Figure S2-C8 ) than with either TG (3; Figure S2-B16 ) or TRG (5, Figure S2-B15 ) rickettsiae. However, the three OGs shared between R. canadensis and TG rickettsiae are all unique sugar transferases, and all three genomes share an unprecedented 52 lost OGs relative to the remaining seven rickettsial genomes ( Table 6 ; Figure S2-F3 ). Interestingly, R. canadensis shares zero lost genes with either TRG or SFG rickettsiae. It also shares with R. prowazekii a unique split gene, scaI, that is the most conserved member of the scas and is present in all analyzed Rickettsia spp. [57]. Thus, while phylogeny estimation places R. canadensis basal to the TG, TRG and SFG rickettsiae, and common OGs suggest an affinity to SFG and TRG rickettsiae over TG rickettsiae, the mode of gene loss across the lineages branching off after R. bellii suggests the position of R. canadensis within our generated phylogeny is well supported, but with possible affinities with TG rickettsiae, which were originally suggested based on serological cross reactivity studies [64]. Accordingly, phylogenetic analysis and signature proteins alone should not be solely used to characterize rickettsial groups, as shared absence of genes may reflect relatedness that is difficult to detect otherwise in these highly reductive genomes.
Table 6. OGs missing in the lineage spanning R. canadensis and TG rickettsiae.
| Missing from R. canadensis and TG rickettsiae (52)2 | Missing from TG rickettsiae (53)3 | ||
| RiOG1 | Annotation | RiOG | Annotation |
| 22 | COG1373: Predicted ATPase (AAA+ superfam) | 67 | Predicted ATPase |
| 973 | Acetylglutamate kinase | 62 | Glycosyltransferase |
| 958 | ADP-ribose pyrophosphatase MutT | 819 | Cephalosporin hydroxylase |
| 955 | Clavaminate synthase 1 | 879 | Acylamino-acid-releasing enzyme |
| 966 | DNA-damage-inducible protein J | 890 | AmpG protein |
| 964 | Optineurin | 886 | Blasticidin S-acetyltransferase |
| 982 | peptide deformylase | 872 | COG4912: Predicted DNA alkylation repair enzyme |
| 987 | Bacterioferritin comigratory protein | 915 | DNA repair protein radC homolog |
| 978 | Putative integral membrane protein | 916 | formamidopyrimidine-DNA glycosylase |
| 66 | Acetyltransferase | 913 | gabD |
| 40 | Beta-lactamase OXA-18 precursor | 888 | Magnesium and cobalt transport protein CorA |
| 893 | Dihydrofolate reductase type 9 | 889 | methylated-DNA-[protein]-cysteine S-methyltransferase |
| 898 | Flavodoxin | 875 | Periplasmic protein |
| 28 | Putative oxidoreductase protein | 884 | Phosphate regulon transcriptional regulatory protein phoB |
| 877 | Putative Zn-dependent hydrolase | 908 | Predicted metal-dependent hydrolase |
| 64 | Putative Zn-dependent hydrolase | 906 | ribose-phosphate pyrophosphokinase |
| 41 | Type I restriction enzyme EcoEI M protein | 904 | RNA methyltransferase, TrmH family, group 1 |
| 891 | Na+/H+ antiporter NhaA | 4 | Putative to amino acid permeases |
| 968 | ABC transporter ATP-binding protein | 13 | 3-oxoacid CoA-transferase, A subunit |
| 945 | RND efflux system, OM lipoprotein, NodT family | 73 | 3-oxoacid CoA-transferase, B subunit |
| 974 | Tellurite resistance protein-related protein | 16 | Putatie DNA processing protein DprA |
| 960 | Multidrug resistance protein mdtA precursor | 17 | Mannose-1-phosphate guanylyltransferase [GDP] |
| 943 | Multidrug resistance protein mdtB | 896 | Putative amino acid transporter yggA |
| 970 | COG0457: FOG: TPR repeat | 39 | Cation transport regulator ChaB |
| 65 | NT domain and HEPN domain | 18 | ComEC/Rec2-related protein |
| 11 | NT domain and HEPN domain | 25 | phage uncharacterized protein, C-terminal domain |
| 975 | addiction module toxin, Txe/YoeB family | 72 | Phage portal protein |
| 965 | prevent-host-death family protein | 38 | Tetratricopeptide repeat-containing protein |
| 977 | prevent-host-death family protein | 870 | Toxin of toxin-antitoxin system VapC |
| 70 | Prophage antirepressor | 876 | Arp2/3 complex activating protein rickA |
| 949 | COG5510: Predicted small secreted protein | 901 | Ecotin precursor |
| 961 | CHP TIGR02217 | 897 | Trichohyalin |
| 950 | COG1598: Uncharacterized conserved protein | 867 | Rickettsial palindromic element (RPE) domain |
| 979 | COG3755: Unchar. protein conserved in bacteria | 902 | Transposase |
| 985 | COG5449: Uncharacterized conserved protein | 894 | CHP TIGR00481 |
| 881 | COG4804: Uncharacterized conserved protein | 822 | COG4285: Uncharacterized conserved protein |
| 967 | UPF0246 protein FTH_1656 | ||
Underscored RiOGs depict non-representative OGs.
Including six representative HPs and nine non-representative HPs.
Including eight representative HPs and seven non-representative HPs.
Interestingly, Vitorino et al. [63] recently demonstrated an affinity between R. canadensis and R. helvetica based on phylogeny estimation from eight genes, although they concluded that the phylogenetic position of R. canadensis was unstable, which is consistent with previous studies. For instance, like SFG rickettsiae, R. canadensis was isolated from ixodid ticks and is maintained transstadially and transovarially [65], [66], grows within the nuclei of its host [65], and contains both rOmpA and rOmpB genes [67], [68]. However, like TG rickettsiae, R. canadensis grows abundantly in yolk sac, lyses red blood cells, is susceptible to erythromycin, and forms smaller plaques as compared to SFG rickettsiae [69]. Genomic characteristics are just as anomalous, as despite sharing the same G+C% [26], [69] and only a slightly larger genome size than TG rickettsiae ( Figure 7 ), R. canadensis shares more common repetitive elements with SFG rickettsiae genomes than with any other group [26] and has many similar genes found within the tra cluster of R. massiliae [70]. Switching the position of R. canadensis in our genome alignment to reflect a derived relationship relative to TG rickettsiae did not improve synteny with the other rickettsial genomes, and despite a large central inversion, R. canadensis gene order is highly conserved with most of the derived taxa ( Figure S1-D ). In an effort to test a putative affinity between R. canadensis and R. helvetica (genome sequence unavailable), we selected 16 existing full or partial gene sequences for R. helvetica and estimated a phylogeny ( Figure 9 ). R. helvetica is supported as basal to the remaining SFG rickettsiae in an otherwise identical phylogeny estimated from the 731 core rickettsial genes ( Figure 3 ), thus refuting an affinity between R. canadensis and R. helvetica. The recent phylogenies estimated from 16S rDNA and groEL nucleotide sequences, the VirB4 protein and 14 concatenated proteins of the T4SS complex, and entire genome sequences placed R. canadensis between TG and TRG rickettsiae [26]; however, R. bellii was not sampled, likely affecting character polarity with the absence of an ancestral taxon. Thus, given our estimation of phylogeny from all available annotated rickettsial genomes, we are confident in the placement of R. canadensis as basal to the TG, TRG and SFG rickettsiae, although limited similarity is apparent to both R. bellii genomes as revealed by OG distribution and synteny. It is not unreasonable to predict that R. canadensis will ultimately group within a fifth distinct rickettsial group once more genomes are sequenced from lesser known rickettsiae, particularly species non-pathogenic to humans.
Figure 9. Phylogeny estimation of the ten analyzed rickettsial taxa plus R. helvetica and R. australis based on 16 proteins.
See Table S13 for gene names and sequence accession numbers. Tree estimated under parsimony (see text).
TG rickettsiae
Despite being distinct from the other rickettsial groups with its highly reductive genomes and strictly insect-specific lifestyles, TG rickettsiae were predicted to contain only three unique representative OGs: a putative GTP pyrophosphokinase (RiOG_2080) and two HPs (RiOG_2081 and RiOG_2082). RiOG_2080 is part of a probable multigene family that is duplicated in most rickettsial genomes. These enzymes catalyze the synthesis of guanosine 5′-triphosphate 3′-diphosphate (pppGpp) as well as guanosine 3′,5′-bispyrophosphate (ppGpp) by transferring pyrophosphoryl groups from ATP to GTP or GDP respectively [71], functioning as mediators of the stringent response that coordinate a wide range of cellular activities in reaction to changes in nutritional abundance [72]. While common in multiple variable copies across the sampled genomes, the role lineage specific GTP pyrophosphokinases play in accommodating the different modes of intracellular replication and intercellular spreading by different rickettsial groups is worth exploring. RiOG_2081 is an uncharacterized protein conserved in a limited number of other bacteria (COG3274) and unknown from non-TG rickettsiae. The distribution of this protein, a putative membrane associated acyltransferase, in many pathogenic bacterial species and one bacteriophage, PhiV10, is interesting ( Table 7 ). Finally, RiOG_2082 is a small putative ORF that BLASTs to no other organisms, with the start codon missing in R. typhi.
Table 7. Results of a BLASTP search for RiOG_2081 using RP338 (R. prowazekii) as a query1.
| Accession no. | Taxon/annotation | score (bits) | E value |
| NP_220721 | Rickettsia prowazekii str. Madrid E; HP RP338 | 546 | 7.00E-154 |
| YP_067290 | Rickettsia typhi str. Wilmington; HP RT0328 | 506 | 8.00E-142 |
| YP_157885 | Azoarcus sp. EbN1; conserved HP, predicted acyltransferase 3 family | 92.8 | 3.00E-17 |
| YP_039445 | Bacillus thuringiensis serovar konkukian str. 97-27; HP BT9727_5136 | 81.3 | 8.00E-14 |
| ZP_00239274 | Bacillus cereus G9241; membrane protein, putative | 79 | 4.00E-13 |
| NP_847850 | Bacillus anthracis str. Ames; HP BA5704 | 78.6 | 5.00E-13 |
| YP_897634 | Bacillus thuringiensis str. Al Hakam; possible membrane protein | 78.2 | 6.00E-13 |
| YP_086718 | Bacillus cereus E33L; probable membrane protein | 78.2 | 7.00E-13 |
| NP_932896 | Vibrio vulnificus YJ016; HP VV0103 | 77 | 2.00E-12 |
| EDK27457 | Unclassified Vibrionales; putative inner membrane protein | 76.6 | 2.00E-12 |
| ZP_01261849 | Vibrio alginolyticus 12G01; putative inner membrane protein | 76.3 | 3.00E-12 |
| NP_799345 | Vibrio parahaemolyticus RIMD 2210633; putative inner membrane protein | 74.7 | 7.00E-12 |
| NP_760091 | Vibrio vulnificus CMCP6; HP VV1_1144 | 74.7 | 7.00E-12 |
| ZP_01066487 | Vibrio sp. MED222; putative inner membrane protein | 70.5 | 1.00E-10 |
| ZP_01474781 | Vibrio sp. Ex25; HP VEx2w_02002647 | 69.3 | 3.00E-10 |
| ZP_00833544 | Yersinia intermedia ATCC 29909; COG3274 | 68.6 | 6.00E-10 |
| YP_001008263 | Yersinia enterocolitica subsp. enterocolitica 8081; HP YE4126 | 67.4 | 1.00E-09 |
| YP_206230 | Vibrio fischeri ES114; integral membrane protein | 67.4 | 1.00E-09 |
| ZP_00992296 | Vibrio splendidus 12B01; putative inner membrane protein | 67.4 | 1.00E-09 |
| YP_512280 | Phage phiV10; putative acetyltransferase | 65.9 | 4.00E-09 |
| ZP_00823633 | Yersinia bercovieri ATCC 43970; COG3274 | 64.7 | 7.00E-09 |
| ZP_00829271 | Yersinia frederiksenii ATCC 33641; COG3274 | 64.3 | 1.00E-08 |
| NP_521411 | Ralstonia solanacearum GMI1000; HP RSc3292 | 63.9 | 1.00E-08 |
| YP_100876 | Bacteroides fragilis YCH46; HP BF3599 | 62 | 6.00E-08 |
| YP_213008 | Bacteroides fragilis NCTC 9343; HP BF3402 | 61.6 | 6.00E-08 |
| ZP_01237231 | Vibrio angustum S14; HP VAS14_21937 | 61.2 | 9.00E-08 |
| ZP_00826782 | Yersinia mollaretii ATCC 43969; COG3274 | 60.8 | 1.00E-07 |
| ZP_01160312 | Photobacterium sp. SKA34; HP SKA34_16770 | 60.5 | 1.00E-07 |
Only sequences with a score greater than 60 bits are shown; of 88 subjects, no other rickettsiae were retrieved.
While a wealth of unique genes seemingly does not define TG rickettsiae, 53 unique gene loss events may offer insight into the streamlined manner of their evolution ( Table 6 ). The loss of the Arp2/3 complex activating protein, rickA, from TG rickettsiae has been well-documented, and distinguishes this group in its mode of host cell spreading [73], [74]. Interestingly, our comparative analysis has revealed two other curious proteins that are present and conserved in all other non-TG rickettsiae genomes. The first is RiOG_897, a putative trichohyalin, which are intermediate filament-associated proteins found predominantly in the hair follicle cells of mammals [75], [76] but also expressed in the hard palate, tongue, nail bed, and a suite of pathological epidermal tissues [77], [78]. We discuss more about trichohyalins below in regards to insect-associated rickettsiae containing a unique trichohyalin-like homolog that is different from the gene found in all other non-TG rickettsiae. The second interesting OG (RiOG_901) found exclusively in non-TG rickettsiae is an ecotin-like protein. Ecotin is a dimeric periplasmic protein described in Escherichia coli that belongs to the protease inhibitor I11 (ecotin) family (PF03974). Ecotin inhibits several pancreatic serine proteases, including chymotrypsin, trypsin, elastases, factor X, kallikrein, as well as a variety of other proteases [79]–[81]. Eggers et al. [82] have shown that ecotin protects E. coli from neutrophil elastase (NE), a mammalian serine protease demonstrated to be important for neutrophil killing of several gram-negative bacteria. Specifically, NE cleaves ompA causing increased permeability to the bacterial outer membrane [83]. Once NE translocates across the vulnerable outer membrane, it functions in inhibiting bacterial cell growth and repair, causing cell death. The presence of ecotin in the periplasm inhibits NE function, thus fostering recovery and growth of the invading bacterial cells [82]. Given the diversity of rickettsial outer membrane surface proteins, particularly the Scas [55], it is reasonable to suggest that one or several surface proteins present in all non-TG rickettsiae may be dependent upon the putative NE inhibitory function of RiOG_901.
TRG rickettsiae
Based on the monophyly of its sampled members (R. felis and R. akari), its strongly supported position in our estimated rickettsial phylogeny, an affinity with AG rickettsiae plasmid-associated genes, and the use of both acarines and insects as primary invertebrate hosts, we erected the TRG rickettsiae as a third derived lineage of Rickettsia [28]. OrthoMCL predicted 37 OGs unique to TRG rickettsiae ( Table 8 ). Of the three other rickettsial lineages, TRG shares more common OGs with SFG rickettsiae (36) than with TG rickettsiae (0) or AG rickettsiae (6) ( Figure 7 ), reflecting its shared common ancestry with the “true” spotted fever group taxa. However, exclusion of R. canadensis sheds light on our previously described affinities of TRG rickettsiae with AG rickettsiae ( Table 9 ). For instance, 26 OGs are shared between the R. bellii genomes and TRG rickettsiae ( Figure S2-C25 ), with six of these annotated as members of toxin-antitoxin (TA) modules, and another two annotated as bacteriophage-derived proteins. Additionally, the R. felis genome shares 44 OGs with the R. bellii genomes ( Figure 7 ), six of which are annotated as members of TA modules, with another one annotated as bacteriophage-derived protein. Furthermore, the R. akari genome shares 10 OGs with the R. bellii genomes ( Figure S2-B23 ), and two of these OGs are predicted members of TA modules. This high presence of TA system components, as well as bacteriophage-derived proteins, attests to our previous observations that AG (at least R. bellii) and TRG rickettsiae are linked via conjugative systems and have a pronounced presence of similar plasmid (and now phage) related ORFs, likely the end products of various lateral gene exchanges between these distantly related groups.
Table 8. OGs present only in TRG rickettsiae.
| RiOG1 | Annotation (37)2 |
| 2043 | COG1670: Acetyltransferases, incl. N-acetylases of ribosomal proteins |
| 2078 | Predicted acetyltransferase |
| 2062 | Predicted hydrolase or acyltransferase |
| 2038 | Putative cysteine protease yopT-like |
| 2047 | 5-Formyltetrahydrofolate cyclo-ligase |
| 1125 | alanine racemase |
| 2033 | Outer membrane protein A precursor |
| 2037 | Outer membrane protein A precursor |
| 2046 | Outer membrane protein A precursor |
| 2076 | Outer membrane protein A precursor |
| 2049 | ABC transporter, ATP-binding protein |
| 2075 | Cell surface antigen-like protein Sca7 |
| 2059 | Ankyrin repeat |
| 2066 | COG1487: Predicted nucleic acid-binding protein, contains PIN domain |
| 2056 | Probable antitoxin of toxin-antitoxin stability system |
| 2069 | addiction module toxin, Txe/YoeB family |
| 2050 | Virulence-associated protein B |
| 1483 | CHP |
| 2068 | CHP |
Underscored RiOGs depict non-representative OGs.
Including 18 representative HPs.
Table 9. OGs present only in R. bellii strains and TRG rickettsiae.
| Present in R. bellii strains and TRG rickettsiae (26)2 | |
| RiOG1 | Annotation |
| 1245 | HicB family |
| 1261 | Phage-related transcriptional regulator |
| 1215 | phage host specificity protein |
| 1128 | Transcriptional regulator |
| 1266 | PIN domain containing protein |
| 1256 | Antitoxin of toxin-antitoxin system StbD |
| 1262 | Cytotoxic translational repressor of toxin-antitoxin (TA) system RelE |
| 1251 | Cytotoxic translational repressor of toxin-antitoxin system RelE |
| 1243 | Growth inhibitor |
| 1240 | putative addiction module antidote protein, CC2985 family |
| 1269 | Transposase |
| 1 | Probable transposase for insertion sequence element |
| 1260 | CHP |
Underscored RiOGs depict non-representative OGs.
Including 12 representative HPs and 1 non-representative HP.
Including 14 representative HPs and 4 non-representative HPs.
Including 3 HPs.
Despite the abovementioned characteristics shared between AG and TRG rickettsiae, the TRG rickettsiae also share three TA components exclusively with SFG rickettsiae ( Table S4 ). Additionally, SFG rickettsiae and the R. bellii genomes have three TA components not found in the other analyzed genomes ( Figure S2-D7 ). This alludes to the likelihood that SFG rickettsiae and R. bellii have also had lateral exchange between plasmids at some point in their evolution, although not nearly to the degree that TRG and the R. bellii genomes have had. For instance, of the 27 OGs shared between R. felis and SFG rickettsiae ( Figure S2-C1 ), only three are components of TA modules ( Table S4 ). And of the 22 OGs shared between R. akari and SFG rickettsiae ( Figure S2-C2 ), none are predicted as components of TA modules. This distinction of the close relatedness of TRG to AG rickettsiae (at least the R. bellii genomes) relative to its sister clade, SFG rickettsiae, based on plasmid associated gene distribution is critical in understanding the mode of gene loss from the last common ancestor of Rickettsia, as well as the degree conjugative systems have contributed to the architecture of these genomes.
Based on phylogeny estimation of 16S rDNA sequences, the largest clade recovered to date for TRG rickettsiae included R. akari, R. felis, R. australis, and poorly characterized rickettsiae from booklouse (Liposcelis sp.) and parasitic wasp (Neochrysocharis sp.) hosts [16]. In addition, Reeves et al. [84] recently identified two novel rickettsial genotypes from the mite Ornithonyssus bacoti from Egypt that are closer to TRG rickettsiae than the other rickettsial groups based on partial sequence comparison of the 17 kD antigenic gene. Aside from R. australis, which has been found exclusively in tick hosts, none of these taxa purportedly parasitize ticks, with R. akari found in mites [85], R. felis found in fleas [51], [86]–[89], and the other unnamed Rickettsia spp. known only from their booklouse, wasp and mite hosts. Thus the group is interesting from an arthropod host perspective as well as from its apparent affinities to the R. bellii genomes. In light of this, we suggested that R. australis would continue to group within the TRG rickettsiae [28], as it has previously done in some cases wherein one or few genes were analyzed [e.g., 16], [63], [90]–[92]. Our dataset including 16 gene sequences from R. helvetica (discussed above) also contained eight sequences from R. australis and grouped this taxon with R. akari in a clade subtended by R. felis with strong bootstrap support ( Figure 9 ). However, while the TRG rickettsiae is still recovered when R. akari and R. australis are analyzed in the absence of R. felis [49], [92], [93], the exclusion of R. akari in the presence of R. australis and R. felis [51] failed to recover a monophyletic TRG rickettsiae. Furthermore, while four of the eight single gene phylogeny estimates by Vitorino et al. [63] recovered the TRG rickettsiae, the consensus tree did not, as the TG rickettsiae was placed within the TRG rickettsiae, splitting the R. akari/R. australis clade from R. felis. Thus, the TRG rickettsiae is not easily demonstrated as a distinct lineage of rickettsiae unless the taxon and character sampling is robust enough for this intriguing lineage to emerge ( Figure 9 ; [28]).
SFG rickettsiae
The majority of the described species of Rickettsia fall within the SFG rickettsiae. The analyzed spotted fever group genomes form a monophyletic cluster of taxa with little sequence divergence relative to the other rickettsial groups ( Figure 3 ). OrthoMCL predicted 113 OGs that are unique to SFG rickettsiae ( Table 10 ). Of note, in addition to the four core rickettsial proline/betaine transporters ( Table S2 ), SFG rickettsiae contain two variant copies (RiOG_1314 and RiOG_1332). Other transporters unique to SFG rickettsiae include three ATPase and permease components of an ABC-type multidrug transporter (RiOG_1347, RiOG_1364 and RiOG_1365), an ATP-binding protein similar to ABC transporter (RiOG_1376), an MSF-like sugar transporter (RiOG_1355), and an RND family efflux transporter (RiOG_1294). While high numbers of transporters are expected in Rickettsia to counterbalance depleted metabolic pathways and acquire host resources, it is unclear why the SFG rickettsiae have elevated levels of unique components of organic and inorganic transport systems relative to the other three rickettsial groups. As with TG rickettsiae, there are group-specific GTP pyrophosphokinases (RiOG_1350 and RiOG_1361) in SFG rickettsial genomes, and their role in a group-specific stringent response is worthy of attention. Like AG and TRG rickettsial genomes, SFG rickettsiae have group-specific ANK repeat containing proteins, with a particular one (RiOG_1344) similar to metazoan tankyrases, telomeric repeat binding factor-interacting ANK-related ADP-ribose polymerases. Aside from potentially playing key roles in the maintenance of telomere function [e.g., 94], tankyrases have been implicated in mitogen-activated protein kinase signaling [95], regulation of cell death [96], [97] and viral inhibition [98].
Table 10. OGs present only in SFG rickettsiae.
| RiOG1 | Annotation (113)2 |
| 1312 | COG0522: Ribosomal protein S4 and related proteins |
| 1378 | Acetate kinase |
| 1342 | Acetyltransferase |
| 1313 | COG1835: Predicted acyltransferases |
| 1317 | COG0840: Methyl-accepting chemotaxis protein |
| 1350 | GTP pyrophosphokinase |
| 1361 | GTP pyrophosphokinase |
| 1284 | Predicted NTPase |
| 1334 | Prolyl endopeptidase precursor |
| 1330 | Putative DNA processing protein DprA |
| 1398 | similarity to D-alanyl-D-alanine dipeptidase |
| 1344 | Tankyrase-1 |
| 1286 | Type I restriction enzyme EcoBI specificity protein |
| 1363 | P pilus assembly protein FimD |
| 1386 | P pilus assembly protein FimD |
| 1157 | Poly-beta-hydroxybutyrate polymerase |
| 1360 | Cell surface antigen Sca3 |
| 1349 | Cell surface antigen-like protein Sca8 |
| 1383 | Cell surface antigen-like protein Sca8 |
| 1347 | ABC-type multidrug transport syst., ATPase and permease components |
| 1364 | ABC-type multidrug transport syst., ATPase and permease components |
| 1365 | ABC-type multidrug transport syst., ATPase and permease components |
| 1376 | similarity to ABC transporter ATP-binding protein |
| 1314 | Proline/betaine transporter |
| 1332 | Proline/betaine transporter |
| 1294 | RND family efflux transporter |
| 1355 | MFS type sugar transporter |
| 1167 | similarity to cation efflux system protein |
| 1307 | Multidrug resistance protein mdtB |
| 1345 | Rickettsial palindromic element (RPE) domain |
| 1357 | Rickettsial palindromic element (RPE) domain |
| 1380 | Ankyrin repeat |
| 1388 | Ankyrin repeat |
| 1315 | Ankyrin repeat domain-containing protein 28 |
| 1392 | putative transposable insertion element |
| 1382 | COG4804: Uncharacterized conserved protein |
| 1299 | CHP |
| 1324 | CHP |
| 1341 | CHP |
| 1348 | CHP |
| 1354 | CHP |
Underscored RiOGs depict non-representative OGs.
Including 67 representative HPs and five non-representative OGs.
Using EasyGene [99], a program that ranks prokaryotic predicted ORFs based on statistical significance, Nielsen and Krogh [100] determined that the R. conorii str. Malish 7 genome was over-annotated by 16%, ranking 7th among most over-annotated replicons in a sample of 143 prokaryotic genomes. Specifically, EasyGene determined 225 RefSeq genes to be false, with 34 additional genes predicted by EasyGene that were not called in the original study [22], [23]. Aside from possible gross ORF over-prediction in all ten rickettsial genomes (discussed below), our analysis yielded many OGs with imperfect representation within the SFG group, as 54 OGs are found exclusively in the R. conorii and R. sibirica genomes ( Figure S2-A1 ), 52 are found exclusively in the R. rickettsii and R. sibirica genomes ( Figure S2-A2 ), and 36 are found exclusively in the R. rickettsii and R. conorii genomes ( Figure S2-A3 ). Given that the SFG rickettsial genomes have elevated split genes as compared to other rickettsial genomes ( Table 5 ; Table S1 ), our findings and those of Nielsen and Krogh [100] hint at a pronounced rate of pseudogenization in SFG rickettsiae depicted by a patchy distribution of split and truncated ORFs decaying from the ancestral SFG genome.
One hallmark occurrence of probable pseudogenization in SFG rickettsiae involves a Sec7-domain-containing protein known in prokaryotes only from Rickettsia and Legionella species [101]. The Legionella counterpart of this curious protein, named RalF, is a guanine nucleotide exchange factor that recruits ADP-ribosylation factor to occupied phagosomes, permitting Legionella to replicate free from the host immune system [102]. The rickettsial RalF xenolog (RiOG_19), including the N-terminal Sec7 domain and immediate flanking Sec7-capping-domain [103], is present in all rickettsial genomes except for SFG rickettsiae and R. canadensis, suggesting a biological mechanism that has been lost from the true spotted fever group and R. canadensis. Unlike Legionella RalF, which has a short (44 aa) C-terminal tail containing a type 4 secretion system (T4SS) signal sequence [104], the rickettsial genes encode an additional variable domain (97–315 aa) between the Sec7-capping-domain and the C-terminal tail. Within this third domain lies a region immediately flanking the predicted T4SS signal sequence that is extraordinarily rich in proline residues, much like the P-rich domain of rickA proteins [74]. Interestingly, the SFG genomes each contain small ORFs corresponding to the tails of the RalF-like sequences. A similar sequence within the R. canadensis genome (not annotated) also spans this region yet is riddled with frame-shift mutations. Given that Rickettsia, unlike Legionella, quickly lyse the phagosome upon host cell entry, the function of a RalF xenolog, particularly given its curious distribution in the rickettsial tree, is worthy of investigation. Finally, full intact RalF xenologs in both TRG rickettsial genomes further attest the distinction of this lineage from the SFG rickettsiae [28].
Arthropod Host-Specific OGs
Several studies have demonstrated the presence of certain rickettsial species outside of their natural arthropod hosts. For example, the louse (and less often flea) associated R. prowazekii has been found in ticks in Africa [105] and Mexico [106], and was also reported in acarids from flying squirrels in the United States [107]. However, it should be recognized that many blood-feeding arthropods have a wide range of vertebrate hosts and likely act as reservoirs for a variety of bacteria that incidentally fall outside of their natural arthropod vector. To this extent reports of pathogenic bacteria (i.e., R. prowazekii) in unusual vectors need to be substantiated beyond simple detection in these foreign hosts, and caution should be taken when immediately assigning novel host associations. Given the low frequency of resident bacteria in many natural arthropod populations [108], substantiation of novel arthropod hosts can be achieved in the field by robustly sampling other invertebrate and vertebrate animals from the same locality that may actually be the true host of the incidentally collected bacterium. Furthermore, laboratory studies would be needed to determine the pathogenicity, if any, that the bacterium causes in its novel host. However, laboratory inoculation of an animal may result in pathogenesis only because the number of bacteria far exceeded what occurs in nature, thus compromising an immune system that under natural circumstances is quite capable of killing the pathogen. Furthermore, demonstrating laboratory bacterial infection or vectorization in a foreign host, for example R. conorii in the body louse [109], may initially prove successful, but eventually will clear from the host as it would from natural populations. For instance, Rickettsia have been grown in mosquito cell lines, yet to our knowledge no wild caught mosquitoes to date have been shown to act as hosts to any Rickettsia. In fact, based on the analysis of the highly divergent sca genes in rickettsiae, which are suspected to directly interact with host cell proteins [47], [110], Blanc et al. [55] concluded that rapid evolution of such important host colonization genes likely keep Rickettsia host ranges quite narrow.
Given our conservative stance on definitive rickettsial arthropod hosts, we have chosen to present the predicted genes that are exclusive to insect associated Rickettsia and tick associated Rickettsia (as depicted in Figure 7 ). Because only one analyzed genome is from a mite-associated species (R. akari), we have no comparative analysis to describe potential mite-specific rickettsial genes. However, the list of singleton genes found in R. akari may provide a start to such an approach (see below).
Insect-associated rickettsiae
Three of the ten analyzed rickettsial genomes have definitive insect hosts, with R. typhi and R. felis reported from rodent, shrew and feline [51], [86]–[88] associated fleas, and R. prowazekii predominantly pathogenic in lice, as well as fleas in the sylvatic form. Thus these three rickettsial lineages share common arthropod hosts at least in fleas. Regarding R. typhi, It has become apparent that the ecology of murine typhus in both south Texas and southern California, where the classic cycle of R. typhi involving commensal rats and primarily the rat flea (Xensopsylla cheopis), has been replaced by the Virginia opossum (Didelphis virginiana)/cat flea (Ctenocephalides felis) cycle. For instance, Sorvillo et al. [111] demonstrated the association of 33 cases of locally acquired murine typhus in Los Angeles County with seropositive domestic cats and opossums. However, urban rat/flea populations are still the main reservoir of R. typhi worldwide and particularly in many cities where urban settings provide a constellation of factors for the perpetuation of murine typhus, including declining infrastructures, increased immunocompromised populations, homelessness, and high population density of rats and fleas. Thus, aside from the reported louse host of R. prowazekii and a laboratory demonstration that R. typhi infection is lethal for human body lice [112] despite R. typhi being unknown from wild lice, these three rickettsial taxa are all capable of infecting and causing pathogenicity in an overlapping range of flea species, prompting a genomic comparison to detect common genes possibly involved in flea cell invasion and pathogenicity.
Despite the vast evolutionary divergence between arachnids and hexapods, two lineages with a common ancestor estimated to have split over 500 million years ago [113], only two OGs (RiOG_1496 and RiOG_1497) specific to the R. prowazekii, R. typhi and R. felis genomes were predicted by OrthoMCL ( Figure 5 , Figure 7 , Table 11 ). However, these genes are exceptionally interesting from two perspectives. First, while the ORFs encoding both OGs are contiguous in all three genomes, they are present only on the pRF plasmid and not the chromosome of R. felis, suggesting a possible lateral exchange of these genes between TG rickettsiae and the R. felis genome. Second, these ORFs share little homology with genes from other organisms, and the taxonomic distribution of these organisms is quite intriguing. RiOG_1496 is annotated as myosin-11 and has close similarities to RiOG_1454, which is annotated as a HP found in the R. felis genome as well as both R. bellii genomes. Furthermore, RiOG_897 (discussed above), a predicted trichohyalin-like protein found in all analyzed rickettsial genomes but TG rickettsiae, has limited similarity with RiOG_1496. Aside from the more general functions described above, trichohyalin also acts as a cross-bridging protein that assists in the coordination of mechanical strength between the peripheral cell envelope barrier structures and cytoplasmic keratin filament networks [114]. The lysosomal cysteine protease, cathepsin L, which is critical for skin and hair follicle homeostasis, likely uses trichohyalin as a substrate [115]. Recently, Ou et al. [116] determined that a trichohyalin homolog, DYF-14, in the nematode Caenorhabditis elegans is essential for cilium biogenesis. Thus, this group of proteins seems to be critical for epithelial cell maintenance in a wide range of animals, and the presence of similar proteins in TG rickettsiae may hint at a molecular function involved with epithelial (invertebrate host) or endothelial (vertebrate host) cell entry and modification, as both R. typhi, R. prowazekii and R. felis enter their vertebrate hosts transdermally through inoculation or inhalation of insect feces.
Table 11. Results of BLASTP searches evaluating two OGs (1496 and 1497) predicted by OrthoMCL to contain only insect-associated rickettsiae.
| RiOG_1496 | |||||
| Accession No. | Annotation | Taxon | score (bits) | E value | OG |
| NP_220662 | HP RP278 | Rickettsia prowazekii str. Madrid E | 484 | 5.00E-135 | 1496 A |
| YP_067231 | CHP | Rickettsia typhi str. Wilmington | 431 | 3.00E-119 | 1496 A |
| YP_247443 | HP RF_p27 | Rickettsia felis URRWXCal2 | 102 | 3.00E-20 | 1496 A |
| YP_246459 | HP RF_0443 | Rickettsia felis URRWXCal2 | 67 | 2.00E-09 | 1454B |
| ZP_01379825 | HP RbelO_01000612 | Rickettsia bellii OSU 85-389 | 50.1 | 2.00E-04 | 1454B |
| YP_537715 | HP RBE_0545 | Rickettsia bellii RML369-C | 48.9 | 5.00E-04 | 1454B |
| NP_975020 | Ribose/Galactose ABC transporter, permease component | Mycoplasma mycoides subsp. mycoides SC str. PG1 | 43.9 | 0.014 | ------ |
| ZP_01380625 | HP RbelO_01001434 | Rickettsia bellii OSU 85-389 | 37.4 | 1.3 | 897C |
| YP_537282 | HP RBE_0112 | Rickettsia bellii RML369-C | 37.4 | 1.3 | 897C |
| XP_973544 | PREDICTED: similar to SMC6 protein | Tribolium castaneum | 35.4 | 5 | ------ |
| Q805A1 | SMC protein 5 | Xenopus laevis | 35 | 6.6 | ------ |
| XP_956017 | HP | Neurospora crassa OR74A | 35 | 7.7 | ------ |
| XP_783551 | PREDICTED: HP | Strongylocentrotus purpuratus | 34.7 | 7.9 | ------ |
| XP_001254413 | PREDICTED: similar to citron, partial | Bos taurus | 34.7 | 9.5 | ------ |
Myosin-11.
Consensus annotation = HP.
Trichohyalin. OG_897 also contains VBI2812RCa_1005 (ZP_01347956.1), VBI0166RF1_1469 (YP_247242.1), VBI0269RA_1318 (ZP_00340773.1), VBI0113RR_1403 (ZP_00154140.1), VBI2627RCo_1353 (NP_360825.1), and VBI0076RS_1050 (ZP_00142696.1) (all but TG rickettsiae).
Consensus annotation = HP. OG_1439 also contains VBI0166RF1_0910 (YP_246763.1).
Aside from sharing limited homology to these other OGs, RiOG_1496 is also similar to a predicted permease component of a ribose/galactose ABC transporter from the bacterium Mycoplasma mycoides (mollicutes: Spiroplasma group), the etiological agent of contagious bovine pleuropneumonia. Interestingly, a similar ORF is present in the cow genome, possible hinting at a horizontal exchange between M. mycoides and its bovine host. RiOG_1496 also Blasts to sequences from three other metazoans, the rust red flour beetle, Tribolium castaneum, the African clawed frog, Xenopus laevis, and the California purple sea urchin, Strongylocentrotus purpuratus. The beetle and frog ORFs are predicted as structural maintenance of chromosomes (SMC) proteins 6 and 5, respectively. SMC proteins are involved in such cellular processes as chromosome condensation, sister chromatid cohesion, chromosome partitioning, dosage compensation, DNA repair, and recombination [e.g.], [ 117]–[119]. In Bacillus subtilis, an SMC protein (BsSMC) plays a role in chromosome organization and partitioning, and has been shown to affect supercoiling in vivo, most likely by constraining positive supercoils, an activity contributing to the compaction and organization of chromosomes [120]. The ORF from the sea urchin, as well as one final BLASTP hit to a sequence from Neurospora crassa, a type of red bread mold of the phylum Ascomycota, are annotated as HPs.
Like RiOG_1496, RiOG_1497 had only a few BLASTP hits with significant alignments, yet they cover a range of diverse organisms. RiOG_1497 shares limited similarity with RiOG_1439, which is annotated as a HP and found only in the R. bellii genomes and the chromosomal genome of R. felis. Regarding eukaryotes, RiOG_1497 shares limited similarity with HPs from the green spotted pufferfish, Tetraodon nigroviridis, and the rice blast fungus, Magnaporthe grisea. RiOG_1497 also Blasts to a HP from another α-proteobacterium, Stappia aggregata (Rhodobacterales). Interestingly, there is also limited similarity between RiOG_1497 and a serine/threonine protein kinase from the marine filamentous cyanobacterium, Trichodesmium erythraeum.
OrthoMCL predicted zero non-representative OGs for the insect-associated Rickettsia ( Figure 5 , Figure 7 ), and only two representative and two non-representative OGs are present in all other genomes except the insect-associated rickettsiae (depicting shared lost genes in the insect-associated genomes) ( Figure S2-F6 ). Both representative OGs (RiOG_948 and RiOG_951) are HPs, while the two non-representative OGs, RiOG_814 and RiOG_817, are annotated as a conserved uncharacterized bacterial protein (COG4374) and a HP, respectively. Thus, only the poorly characterized tandem gene group of RiOG_1496 and RiOG_1497 exists for attempting to distinguish the insect-associated Rickettsia from the other lineages with non-insect hosts.
Although the similarity of both RiOG_1496 and RiOG_1497 to the sequences described above is limited, it is nonetheless interesting that their distribution as contiguous ORFs in the TG rickettsiae and the R. felis pRF plasmid is unique amongst the analyzed rickettsial genomes. It is also interesting that at least one of the ORFs (RiOG_1496) has homology to vertebrate smooth muscle protein myosin-11, which is known to be expressed in the esophagus and trachea of humans, as well as trichohyalin, a protein associated with various healthy and pathological epithelial cell types. Both of these proteins are present at the infection interface between insect associated Rickettsia and vertebrate hosts and, at the very least, provide our best guess for a means to distinguish, at the genomic level, insect-associated vertebrate cell invasion from that of acarine. This result of a few examples from the comparative analysis of ten genomes is surprising, and perhaps can be improved upon by the sequencing of more insect-associated rickettsial genomes.
While much of the genome sequencing of rickettsiae has focused on medically important species, it is imperative to consider the species non-pathogenic to humans for comparative biological reasons, in particular for determining the mode of insect-cell invasion and pathogenicity. Studies demonstrating pathogenicity exclusively in insect hosts are limited to 1. male killing in two ladybird beetles (Coleoptera: Coccinelidae), Adalia bipunctata [10] and A. decempuncata [121], and the buprestid beetle Brachys tessellatus (Coleoptera: Buprestidae) [122], 2. thelytoky (female parthenogenesis) induction in the serpentine leafminer endoparasitoid, Neochrysocharis formosa (Hymenoptera: Eulophidae), [123], 3. reduced weight and fecundity in the pea aphid, Acrythosiphon pisum (Hemiptera: Aphididae), [11], [124], and 4. oogenesis induction in the booklouse Liposcelis bostrychophila (Psocoptera: Liposcelididae) [125] and in the date stone beetle, Coccotrypes dactyliperda (Coleoptera: Scolytidae), [126]. Other organisms beneficial to humans that are affected by insect-associated Rickettsia will also be of interest in evaluating insect cell invasion and pathogenicity. For instance, the leafhopper Empoasca papayae (Hemiptera: Cicadellidae) is seemingly unaffected by a resident species of Rickettsia (PBT) that devastates commercial papaya production (papaya bunchy top disease) [7]. However, the effects on insects by some poorly characterized resident Rickettsia species are currently unknown, including those from the springtail Onychiurus sinensis (Collembola: Onychiuridae), the bluetongue virus vector Culicoides sonorensis (Diptera: Ceratopogonidae), the sweet potato whitefly Bemisia tabaci (Hemiptera: Aleyrodidae), the bruchine beetle Kytorhinus sharpianus (Coleoptera: Chrysomelidae), and the crane fly Limonia chorea (Diptera: Limoniidae) [7], [13], [126]–[128]. Nevertheless, all of these less-understood insect-associated Rickettsia spp. are good candidates for comparative genomic analysis with R. prowazekii, R. typhi and R. felis for improving the current knowledge of the mechanisms underlying insect cell invasion and pathogenicity.
Tick-associated rickettsiae
Six of the ten analyzed rickettsial genomes have definitive tick hosts, including both R. bellii genomes, R. canadensis, R. rickettsii, R. conorii, and R. sibirica. In general, little is known about the definitive host ranges of members of the AG and SFG rickettsiae, partly because few host-specific characteristics have been described for any rickettsial/acarine relationship, but also because multiple arthropod or vertebrate (or other eukaryote) hosts are seldom sampled from a given locality to distinguish between true rickettsial hosts and incidental vectors (discussed above). R. bellii seems to parasitize the widest range of tick genera [17], while of the pathogenic taxa, only R. conorii seems to be limited to one vector species [129]. OrthoMCL predicted one non-representative (RiOG_866) and three representative (RiOG_1005, RiOG_1012 and RiOG_1021) OGs specific to the tick-associated rickettsial genomes ( Figure 5 , Figure 7 ). RiOG_866 is an alpha-(1,3)-fucosyltransferase that is highly truncated in all but the R. bellii genomes and further split in R. conorii and R. sibirica ( Table S1 ), depicting a gene undergoing decay. Similarly, RiOG_1021, annotated as a poly-beta-hydroxybutyrate polymerase, is also experiencing pseudogenization, as it depicts an artifact of the clustering process. RiOG_1021 is related to RiOG_834 (core distribution), which has full-length (∼583 aa) proteins in TG and TRG rickettsiae, but parts of split genes from the tick-associated taxa. The corresponding halves of these split genes constitute RiOG_1021. Thus, if only the full sized ORFs are functional, alpha-(1,3)-fucosyltransferase is the lone signature protein found exclusively in TG and TRG genomes (the converse of the tick-associated rickettsiae).
RiOG_1005 has mild similarity to fic (filamentation induced by cAMP) proteins ( Table 12 ), which are involved in cell division and folate metabolism (IPR003812). Aside from R. canadensis, which is highly truncated, the rickettsial sequences contain the central conserved HPFXXGNG motif characteristic of this protein family. Critical for the production and maintenance of new cells [130], folate is especially important during periods of rapid cell division and growth. While the exact molecular function of fic proteins is unknown, it is possible RiOG_1005 is involved in some aspect of folate synthesis, an incomplete pathway in Rickettsia likely requiring energy-coupled transporters to uptake host stores of the vitamin and/or its derivatives [61], [62]. However, the absence of this gene in insect- and mite-associated rickettsial genomes and the loss of the majority of the protein in R. canadensis hint more toward the decaying of this gene family. The identification of a core rickettsial transporter involved in folate/folate derivative uptake would support this hypothesis.
Table 12. Results of BLASTP searches evaluating two OGs (RiOG_1005 and RiOG_1012) predicted by OrthoMCL to contain only tick-associated rickettsiae.
| RiOG_1005 | |||
| Accession No. | Taxon/annotationA | score (bits) | E value |
| YP_538109 | Rickettsia bellii RML369-C; Cell filamentation protein Fic | 636 | 0 |
| ZP_01379987 | Rickettsia bellii OSU 85-389; HP RbelO_01000779 | 634 | 2.00E-180 |
| NP_360166 | Rickettsia conorii str. Malish 7; similarity to cell filamentation proteins (fic) | 518 | 2.00E-145 |
| ZP_00142033 | Rickettsia sibirica 246; hypothetical cell filamentation proteins (fic) | 517 | 4.00E-145 |
| ZP_00153572 | Rickettsia rickettsii ; COG3177: Uncharacterized conserved protein | 514 | 2.00E-144 |
| ZP_01254701 | Psychroflexus torquis ATCC 700755; HP P700755_13960 | 286 | 8.00E-76 |
| ZP_01048880 | Cellulophaga sp. MED134cell filamentation protein-like (fic) | 286 | 1.00E-75 |
| ZP_01202287 | Flavobacteria bacterium BBFL7putative cell filamentation protein Fic | 285 | 2.00E-75 |
| YP_860923 | Gramella forsetii KT0803; filamentation induced by cAMP (Fic) family protein | 275 | 4.00E-72 |
| NP_973239 | Treponema denticola ATCC 35405; Fic family protein | 242 | 2.00E-62 |
| YP_378503 | Chlorobium chlorochromatii CaD3; Fic family protein | 241 | 4.00E-62 |
| YP_790458 | Pseudomonas aeruginosa UCBPP-PA14; HP PA14_28800 | 236 | 2.00E-60 |
| ABQ20395 | Vibrio cholerae O395; Fic family protein | 234 | 6.00E-60 |
| YP_388986 | Desulfovibrio desulfuricans G20; HP Dde_2494 | 232 | 2.00E-59 |
| YP_901526 | Pelobacter propionicus DSM 2379; transcriptional regulator, Fis family | 228 | 4.00E-58 |
| ZP_01673548 | Candidatus Desulfococcus oleovorans Hxd3; conserved HP | 226 | 2.00E-57 |
| YP_064910 | Desulfotalea psychrophila LSv54; HP DP1174 | 224 | 5.00E-57 |
| NP_603868 | Fusobacterium n. nucleatum ATCC 25586; Huntington interacting Protein HYPE | 221 | 5.00E-56 |
| EDK50213 | Shewanella baltica OS223; filamentation induced by cAMP protein Fic | 218 | 3.00E-55 |
| YP_064922 | Desulfotalea psychrophila LSv54; HP DP1186 | 218 | 3.00E-55 |
| YP_750639 | Shewanella frigidimarina NCIMB 400; filamentation induced by cAMP protein Fic | 218 | 3.00E-55 |
| ZP_01704525 | Shewanella putrefaciens 200; filamentation induced by cAMP protein Fic | 218 | 5.00E-55 |
| ABA87022 | Vibrio cholerae; HP | 216 | 2.00E-54 |
| YP_847967 | Syntrophobacter fumaroxidans MPOB; filamentation induced by cAMP protein Fic | 212 | 2.00E-53 |
| ZP_00143967 | Fusobacterium nucleatum subsp. vincentii ATCC 49256; hypothetical cytosolic protein | 206 | 1.00E-51 |
| NP_931130 | Photorhabdus luminescens subsp. laumondii TTO1; HP plu3930 | 202 | 2.00E-50 |
| YP_516911 | Desulfitobacterium hafniense Y51; HP DSY0678 | 196 | 2.00E-48 |
| NP_634630 | Methanosarcina mazei Go1; HP MM2606 | 118 | 5.00E-25 |
| ZP_00121191 | Bifidobacterium longum DJO10A; COG3177: uncharacterized conserved protein | 92.4 | 3.00E-17 |
| ZP_01347715 | Rickettsia canadensis str. McKiel; HP RcanM_01000664 | 92 | 4.00E-17 |
| NP_695410 | Bifidobacterium longum NCC2705; narrowly conserved HP | 89 | 3.00E-16 |
| YP_064096 | Desulfotalea psychrophila LSv54; HP DP0360 | 88.6 | 4.00E-16 |
| YP_001112298 | Desulfotomaculum reducens MI-1; filamentation induced by cAMP protein Fic | 88.2 | 5.00E-16 |
| YP_001048169 | Methanoculleus marisnigri JR1; filamentation induced by cAMP protein Fic | 85.9 | 3.00E-15 |
| YP_155075 | Idiomarina loihiensis L2TR; Uncharacterized protein containing Fic domain | 85.5 | 4.00E-15 |
| YP_001213658 | Dehalococcoides sp. BAV1; filamentation induced by cAMP protein Fic | 82.4 | 3.00E-14 |
| XP_972015 | Tribolium castaneum; PREDICTED: similar to CG9523-PA | 82 | 4.00E-14 |
| NP_396283 | Agrobacterium tumefaciens str. C58; HP AGR_pAT_503 | 80.9 | 9.00E-14 |
| NP_268827 | Streptococcus pyogenes M1 GAS; HP SPy0558 | 80.5 | 1.00E-13 |
| YP_281824 | Streptococcus pyogenes MGAS5005; hypothetical cytosolic protein | 80.1 | 1.00E-13 |
| ZP_01199959 | Xanthobacter autotrophicus Py2; conserved HP PA0574 | 80.1 | 2.00E-13 |
| NP_714132 | Leptospira interrogans serovar Lai str. 56601;Huntingtin interacting protein E-like protein | 80.1 | 2.00E-13 |
HP.
Truncated ORF from R. canadensis (ZP_01347690.1) annotated as HP with bit score = 45.8.
RiOG_1012 is highly similar to macrolide, virginiamycin A, chloramphenicol, and streptogramin A acetyltransferases, acetyltransferases of the isoleucine patch superfamily and transferases with hexapeptide repeats from many different bacterial species, several of which are highly pathogenic ( Table 12 ). In particular, streptogramin A and virginiamycin A acetyltransferases confer gram-positive bacteria resistance to A-type compounds of virginiamycin-like (Vml) antibiotics [e.g.], [ 131]–[134]. Because gram-negative bacteria typically have an innate resistance to Vml antibiotics [e.g.], [135,136], the presence of Vml acetyltransferases in certain gram-negative bacterial genomes went unnoticed until their discovery early this decade in Yersinia enterocolitica [137]. With the rapid accumulation of bacterial genome sequences it became apparent that many gram-negative bacterial genomes harbor Vml acetyltransferases (e.g., Table 12 ). Interestingly, the predominant presence of Vml acetyltransferases on plasmids in gram-positive bacteria versus their typical chromosomal location in gram-negative bacteria suggests that the genes encoding these variable proteins likely spread via conjugation and possibly equip gram-positive bacteria with resistance to Vml antibiotics [137]. While all six sequences within RiOG_1012 are highly similar in the C-terminal region, the N-terminal halves of the proteins are highly divergent between SFG rickettsiae, the R. belii sequences, and R. canadensis ( Table 12 ). This is consistent with the initial studies that concluded streptogramin, chloramphenicol and related acetytransferases belong to a vast family of enzymes with varying substrates [131], [138]. The presence of a Vml acetyltransferase only in tick-associated rickettsiae is interesting and implores further laboratory investigation.
As with insect-associated rickettsiae, OrthoMCL predicted few signatures for tick-associated rickettsiae. Despite the diversity between insects and ticks, all of the analyzed rickettsial species are capable of infecting vertebrates; thus the identified host-specific OGs likely do not contain proteins involved in vertebrate host cell invasion and pathogenicity. The likelihood that these signatures are involved in arthropod cell entry is also low, given the incidental collection of rickettsial species outside the range of their expected hosts (discussed above). However, these signature genes may be involved in mechanisms specific to arthropod host lifestyle, aiding long-term infection and the ability to persist in tick (via transstadial and transovarial transmission) and insect (via fecal inoculation and inhalation) populations despite the rapid generation times of these arthropods.
Plasmid Associated OGs
We recently analyzed the genetic composition of the pRF plasmid of R. felis and determined that the replicon is composed of genes with likely origins to AG rickettsiae and other plasmid-containing bacteria [28]. This suggests that the last common ancestor of all rickettsiae likely harbored plasmids, with R. bellii [139], R. felis [27], likely R. akari [140] and other members of TRG rickettsiae, and some members of SFG rickettsiae either maintaining plasmids despite the constraints of shrinking genomes, or acquiring plasmids later in their evolution. Given the plasticity of plasmid presence/absence in other obligate intracellular bacteria [[e.g., 141]–[145]], as well as other medically-important pathogenic bacteria [e.g., 146]–[150], it is probable that the presence of plasmids may be variable at the strain level in Rickettsia, particularly when only one of the two sequenced R. bellii genomes harbors a plasmid [9], [139]. Past reports of pulsed-field gel electrophoresis (PGE) on rickettsial species that do not correlate with the sizes of recently sequenced genomes [151], [152] may also allude to plasmid plasticity in populations of species and strains of Rickettsia.
Our previous suspicion that plasmids are likely to be found in some lineages of SFG rickettsiae [28] has recently been confirmed, as the plasmid pRM from R. monacensis was identified by transposon insertion and further characterization and sequencing [60]. Subsequently, the same research group used PGE and southern blotting to identify plasmids of variable size and composition in R. helvetica, R. peacockii, R. amblyommii, and R. massiliae [59]. The entire plasmid sequence of R. massiliae was later reported [70]. Furthermore, the duplication of several ORFs associated with the type IV secretion system (T4SS) in rickettsiae (VirB4, VirB6, VirB8, and VirB9), coupled with phylogenetic evidence for an ancestral plasmid origin of all T4SSs [153], suggests plasmid systems and related chromosomal genes are a major constituent of rickettsial genomes, possibly contributing to pathogenicity in many lineages. The recent discovery of extraordinarily duplicated conjugative operons, as well as extremely elevated levels of transposons, TPR and ANK motif-containing proteins, integrases, and potential T4SS effector proteins in the Orientia tsutsugamushi genome further attests to the phenomena of plasmid plasticity and HGT amongst the Rickettsiales [58], implying that the rickettsiae progenitor was larger and less stream-lined than its modern descendants [32] and likely equipped with a suite of conjugative machineries [28].
Plasmids
OrthoMCL grouped 58 predicted pRF ORFs into 49 OGs, with 11 pRF ORFs left as singletons ( Table 13 ). Of these 49 OGs, six contained two pRF ORFs (RiOG_920, RiOG_1057, RiOG_1279, RiOG_1282, RiOG_1283, and RiOG_1596), and one contained three pRF ORFs (RiOG_928), depicting the presence of duplicated genes on the plasmid, including the chromosomal replication initiator protein DnaA (pRF04 and pRF19), a probable transposase of the mutator family (pRF01, pRF30 and pRF55), an epsilon subunit-like protein of DNA polymerase III (pRF34 and pRF53), two TPR motif-containing proteins: (pRF12 and pRF15) and (pRF16 and pRF18), a site specific recombinase similar to DNA invertase Pin homologs and TnpR resolvase (pRF32 and pRF66), and a predicted transcription regulatory protein (pRF02 and pRF29). The remaining representative OGs containing single pRF ORFs generally reflect the distribution reported by Gillespie et al. [28] based on BLASTP results, except for a few instances (italicized OGs in Table 13 ). In comparison to the recently discovered SFG rickettsial plasmids, it is apparent that at least three proteins, namely a DnaA-like replication initiation protein, a Sca12-like protein and a small heat shock protein, are common to all rickettsial plasmids [59]. Thus, despite the growing number of plasmids in Rickettsia, their unknown origin in the rickettsial tree and lack of conserved genes involved in conjugation keep their exact function and essentiality elusive.
Table 13. Distribution of the 68 R. felis pRF plasmid ORFs within the OGs predicted by OrthoMCLA.
| ORFs present exclusively on the pRF plasmid | ||||||
| (pRF+pRFδ) | ||||||
| ORF | Name | AnnotationB | OGC , D , E | R/NF | No. taxa | No. ORFs |
| pRF04 | -------- | R. felis specific proteinG | 1604 | R* | 1 | 2 |
| pRF05 | -------- | chromosomal replication initiator protein DnaA-like protein G | 920 | N* | 6 | 8 |
| pRF07 | HsdR | type I restriction-modification system methyltransferase subunit H | 1223 | R* | 3 | 4 |
| pRF09 | -------- | R. felis specific protein (not found in other life) G | 1605 | R* | 1 | 2 |
| pRF12 | tpr | tetratricopeptide repeat domain (TPR) G , I | 1279 | N* | 1 | 2 |
| pRF14 | ank | ankyrin-repeat containing gene (ANK) J | 1597 | R* | 1 | 1 |
| pRF39 | -------- | MobA_MobL (plasmid transfer)/RecD (exonuclease V) hybrid I , K | 1086 | R* | 4 | 4 |
| pRF40 | -------- | R. felis specific protein I | 1603 | R* | 1 | 1 |
| pRF44 | traDF | putative conjugative transfer protein TraD (E. coli F plasmid) G | 1073 | R* | 4 | 4 |
| pRF45 | -------- | R. felis specific protein G | 1593 | R* | 1 | 1 |
| pRF46 | traGF | putative conjugative transfer protein TraG (E. coli F plasmid) G | 1085 | R* | 4 | 4 |
| pRF47 | traGF | putative conjugative transfer protein TraG (E. coli F plasmid) | 1588 | R* | 1 | 1 |
| pRF48 | rve | integrase (integration of viral DNA into the host chromosome) L | 1402 | R* | 2 | 2 |
| pRF49 | -------- | similar to integrase L | 1578 | R* | 1 | 1 |
| pRF50 | -------- | HP conserved in a few other bacteria | 1582 | R* | 1 | 1 |
| pRF53 | -------- | DNA polymerase III, epsilon subunit-like protein I , M | 1057 | N* | 1 | 3 |
| pRF56 | -------- | hyaluronidase (increases tissue permeability/antigenic disguise) I | 1583 | R* | 1 | 1 |
| pRF57 | trp_20 | transposase 20: IS116/IS110/IS902 family [pfam02371] N | 1591 | R* | 1 | 1 |
| pRF58 | trp | COG3547: transposase and inactivated derivatives N | 1585 | R* | 1 | 1 |
| pRF59 | -------- | R. felis specific protein (not found in other life) G | 1580 | R* | 1 | 1 |
| pRF60 | -------- | similar to IS element transposase (E. coli) G | 1592 | R* | 1 | 1 |
| pRF62 | -------- | R. felis specific protein; possible tldD/PmbA protein I | 1589 | R* | 1 | 1 |
| pRF63 | -------- | R. felis specific protein; similar to Wolbachia repA G | 1590 | R* | 1 | 1 |
| pRF66 | -------- | site-specific recombinases (DNA invertase Pin homologs) O | 1282 | N* | 1 | 2 |
| pRF67 | -------- | similar to transposase ISSag8 (Streptococcus agalactiae A909) P | 1602 | R* | 1 | 1 |
| pRF68 | -------- | rickettsial HP | 1054 | R* | 5 | 5 |
Results include 44 predicted ORFs from the putative smaller R. felis plasmid, pRFδ (see Gillespie et al., 2007).
Follows Gillespie et al. (2007). Additional annotation is listed below in footnotes G-Z.
Blank values depict R. felis singletons.
Bold OGs depict singletons upon the removal of doubtful orthologs from pRFδ. Italicized OGs Blast to chromosomal proteins on the R. felis chromosome.
Underlined OGs contain more than one pRF ORF.
Representative (R) or non-representative (N) family. Groups containing ORFs from pRFδ are noted with an asterisk.
HP.
Type I restriction enzyme EcoEI M protein.
CHP.
The pRFδ protein in PATRIC, VBI0166RF3_0019, is slightly different than the pRF protein.
Conjugal transfer protein TraA.
Putative transposase.
DNA polymerase III polC-type
Transposase for insertion sequence element IS1328.
R46 site-specific recombinase, Transposon Tn917 resolvase.
ISBma2, transposase.
Transposon Tn917 resolvase, R46 site-specific recombinase.
The pRFδ protein in PATRIC, VBI0166RF3_0020, is slightly different than the pRF protein.
Probable transposase for transposon Tn903.
COG1396: Predicted transcriptional regulators.
COG3706: Response regulator containing a CheY-like receiver domain and a GGDEF domain.
Protein virD4.
Spore protein SP21.
Probable transposase for insertion sequence element.
Cell division cycle protein 27 homolog.
Myosin-11 (R. prowazekii), M protein, serotype 2.1 precursor (R. typhi).
Toxin-antitoxin modules
Many plasmid-containing bacteria have associated toxin-antitoxin (TA) systems encoded on plasmids, typically as two-component operons, for the control of plasmid partitioning and stable inheritance [154]–[157]. Antitoxins, usually highly labile in their mature form, are constitutively expressed and neutralize the accumulation of their counterpart toxins, which are more stable. Upon imperfect segregation of plasmids after cell division, plasmidless daughter cells are destroyed by elevated toxin levels due to the rapid breakdown of the unstable antitoxin and lack of its further synthesis [158], [159]. Although originally described as mediators of bacterial programmed cell death, studies now suggest that TA modules also act as regulators of the stringent response (reviewed in [159]) and are widely present on chromosomes of diverse bacteria [160]. While TA systems are found in many free-living bacteria, they are typically uncommon among obligate intracellular pathogens [160], [161]. However, the genome sequences for both R. bellii strains and R. felis contain elevated levels of chromosomally encoded TA loci, the majority of which seem to be degraded [9], [27]. Moreover, these genomes typically retain only one component of the TA modules, possibly alluding to a neofunctionalization [153] of the remaining genes for adaptation to eukaryotic hosts, as has been suggested for at least R. felis toxin and antitoxin genes [27]. However, given that the reductive nature of rickettsial genomes may result in high levels of constitutively expressed loci and reduced operons, and that many antitoxins contain motifs common to two, three and even four different DNA-binding-proteins [162], incomplete and noncontiguous rickettsial TA modules may still interact with one another to coordinate a response to stress within host cells. Alternatively, the presence of incomplete TA modules may reflect vertically acquired plasmid-associated genes that are in the process of pseudogenization. In support of this, of the numerous TA components in the R. felis genome, only one VapB antitoxin (RiOG_941) was recovered in a proteome screen [40].
Our bioinformatic analysis reveals that components of 5 TA systems (relBE, phd/doc, vapBC/vag, mazEF, and parDE) are recurrent in all rickettsial genomes save the TG rickettsiae and R. canadensis ( Table S4 ). Of the predicted 56 toxin and 86 antitoxin ORFs, zero occur in the TG rickettsiae and only two are found in R. canadensis. The majority of these ORFs occur in the R. bellii genomes and TRG rickettsiae (avg. of 22 and 26 TA ORFs per genome, respectively), although slightly lower levels also occur in SFG rickettsiae (avg. of 14.3 TA ORFs per genome). However, there are more occurrences of similar TA module components shared between the R. bellii genomes and TRG rickettsiae (12) than between SFG rickettsiae and either the R. bellii genomes (1) or TRG rickettsiae (5) ( Table S4 ). Thus, the presence and distribution of these TA ORFs correlates to the lineages of sampled rickettsiae that contain plasmids, and further supports the TRG rickettsiae having affinities with AG rickettsiae [28]. Furthermore, the R. belli and TRG rickettsiae genomes have elevated levels of predicted PIN-domain proteins (homologs of the pilT N-terminal domain), which in eukaryotes function as ribonucleases [163], [164] involved in RNAi and nonsense-mediated RNA degradation [162], [163]. Most of the described prokaryotic PIN-domain proteins are toxins of chromosomally-encoded TA operons [159]–[161] that are present in a diverse array of unrelated bacteria, likely having arisen due to the advantages they bestow on competing mobile elements [165], [166]. Indeed, the PilT protein of the pathogenic Neisseria meningitidis has been hypothesized to interact with the T4SS due to its limited homology to the DotB protein of the Legionella T4SS [167].
While the exact manner of their origin and current functional significance is debatable, it is apparent that TA systems have arisen via HGT in a wide range of bacteria [168]. Given the distribution of plasmids and associated TA systems in the analyzed rickettsial genomes, it is likely that conjugation via plasmids befits some rickettsial lineages with genes important for survival in stressful environments, allowing for dormancy and slow growth. However, it remains to be determined if those rickettsial species that harbor plasmids use TA modules for mediating the partitioning and stable inheritance of said plasmids.
Singleton ORFs
OrthoMCL failed to group 1467 ORFs (10.2% of total predicted ORFs) from the ten analyzed genomes into any OG ( Figure 5 , Figure 7 ; Table S5 , S6 , S7 , S8 , S9 , S10 , S11 , S12 , S13 , S14 ). The range across rickettsial groups shows TG genomes contribute the least (8.5%) and TRG genomes contribute the most (41%) to the total count of singletons ( Figure 10A ). The individual genome contributions to the overall singleton count range from 4% (R. typhi) to 21% (R. felis), with the rank of all genomes matching the group ranking (TG<SFG<AG<TRG) ( Figure 10A ). However, an inherent bias of these comparisons is difficult to avoid, as OrthoMCL grouped 321 ORFs present only from both R. bellii genomes ( Figure 5 , Figure 7 , Table S3 ). Accounting for these R. bellii doubletons, the rank and proportion of singletons per rickettsial group is modified: TG (7%)<SFG (20%)<TRG (34%)<AG (39%), and illustrates that TRG and AG genomes are more similar in their number of singleton ORFs relative to TG and SFG rickettsiae. This brings up a practical concern with phylogenomic analysis in that sampling one genome per species (or strain) may not suffice for capturing the true composition of genes within the bacterial population. This is consistent with a recent study that cautioned on the very same idea in relation to vaccine design for Streptococcus agalactiae, an organism that has a core genome of approximately 80% across various strains, with the accessory genome quite plastic [169]. Using mathematics, a rather daunting conclusion was reached suggesting even after sampling hundreds of additional S. agalactiae genome sequences, novel genes would still be added to the accessory genome [169]. Nonetheless, inclusion of the R. bellii doubletons illustrated the similar composition of singletons in AG and TRG genomes and further adds to the similarities these genomes share as a result of related conjugation systems.
Figure 10. Analysis of the distribution of 1467 singleton ORFs omitted from OG prediction across 10 rickettsial genomes.
(A) Singleton ORFs across four rickettsial groups. (B) Singleton ORFs across 10 rickettsial genomes. First number is total number of singleton ORFs per taxon, with second number the total singleton ORFs annotated as HPs. Dashed lines in pie charts separate characterized proteins from HPs, with percentages given only for HPs. (C) Average lengths of singleton ORFs with predicted functions versus singleton ORFs annotated as HPs for all ten analyzed rickettsial genomes.
Unsurprisingly, the majority of singleton ORFs are annotated as HPs, ranging from 68% (R. felis) to 95% (R. typhi) across the analyzed genomes ( Figure 10A, B ). In an effort to identify the degree of over-prediction of ORFs, we plotted the average lengths of singleton ORFs with predicted functions versus singleton ORFs annotated as HPs for all ten rickettsial genomes ( Figure 10C ). The rationale for this is that the majority of singletons under 100 amino acids in length should be HPs, with many having arisen by chance [100]. Aside from R. felis, R. prowazekii and the R. bellii genomes, there is minimal difference between the average lengths of singletons with predicted functions and singletons annotated as HPs. The much larger average lengths of singletons with predicted functions versus singleton HPs are expected in the R. bellii and R. felis genomes, as many of the larger singletons in these genomes are probable products of HGT events (e.g., larger transposases, ANK- and TPR-motif containing proteins). This same pattern in the R. prowazekii singletons, however, is unexpected, yet is skewed in part due to the presence of several large split ORFs that did not cluster into their respective OGs. While the shorter singleton HPs may have arisen by chance, it is likely that some of them are functional genes that are difficult to homologize with other closely related sequences, given the problems with assessing percent conservation across short sequences with even minimal differences. For instance, small ORFs are found in a variety of protein classes, including ribosomal proteins, transcriptional regulators, chaperonins, thioredoxins, metal ion chelators, proteolipids, stress proteins, nucleases, and mating pheromones [170]. Of the original 299 Saccharomyces cerevisiae small ORFs annotated as HPs, 170 have since been assigned cellular functions, with the majority of information coming from laboratory evidence [171]. Given the probable plasticity of the accessory genomes of rickettsial strains (discussed above) and the growing importance small ORFs have garnered in the literature [e.g.], [ 171]–[174], the high number of small singleton HPs in Rickettsia should not be ignored. Experimental evidence has confirmed the translation of several small HPs in R. felis [40] and future microarray data will help lend resolution to this poorly understood characteristic of rickettsial genomes.
Conclusion
This study analyzed 14354 predicted ORFs from ten rickettsial genomes and generated OGs ranging from two to 31 sequences for 90 percent of the total ORFs. A conserved core rickettsial genome consisting of 731 OGs (51% of total predicted ORFs) was identified, and a phylogeny was estimated from this core genome to allow for subsequent phylogenomic comparison of the remaining accessory genome. This robust phylogeny estimate is congruent with our recent reclassification of rickettsial lineages into four groups [28] and OGs specific to each group provide the first signature genes possibly involved in the phenotypic characteristics defining each group. The unstable phylogenetic position of R. canadensis, coupled with it only sharing three OGs with the R. bellii genomes, reflects that the base of the rickettsial tree is poorly defined. However, an unprecedented mode of gene loss was discovered in the lineage spanning R. canadensis and TG rickettsiae, illustrating that gene signatures alone may not well-characterize specific rickettsial groups, but instead the modes of gene loss (and stricter reliance on host resources) may be the defining features [175]. Given the emerging diversity of Rickettsia [16], particularly species associated with medically non-important metazoans and ancestrally related to the pathogenic species analyzed here, the origins of pathogenicity from primitive rickettsial symbionts may not be elucidated without a broader genomic comparison reflective of the overall diversity within the genus.
As a consequence of distinguishing OGs comprising single rickettsial groups (e.g., AG, TG, TRG, and SFG), shared rickettsial groups (subgeneric), plasmid-harboring genomes, and genomes with common arthropod hosts (C1OGs) from OGs with a patchy distribution across the rickettsial tree (C2OGs), two interesting results were obtained. First, C2OGs comprise 31% of all generated OGs, implying a significant portion of the rickettsial accessory genome is comprised of gene decay and laterally acquired genes. Supporting this is the presence of the majority of split ORFs within C2OGs ( Table S1 ) and the high proportion of gene families typically associated with the bacterial mobile gene pool in C2OGs (47%) versus the low proportions in C1OGs (5%) and singleton ORFs (4%). Second, the ratio of representative OGs to non-representative OGs is skewed within C1OG distributions (71–29%) but nearly equal in C2OG distributions (56–44%), suggesting that gene duplications (paralogs) and HGT events (xenologs) are more prevalent in C2OGs. Taken collectively, these observations yield the manner in which the rickettsial genomes have acquired their variation: a conserved core genome is supplemented with a highly variable accessory genome that is comprised of gene decay and many horizontally acquired genes. However, the nature of the horizontally acquired genes remains unknown: for example, did the products of HGT arise ancestrally in the analyzed taxa, becoming shuffled over time through recombination and high rates of decay, or are HGT products continually sculpting the variation within the accessory genome overtop of a highly reductive nature of all genes within the genome? The recent explosion of reported cases of plasmids in all rickettsial groups except TG rickettsiae argues for the latter scenario, and is congruent with our findings of nearly zero instances of plasmid associated genes, genes typical of HGT events and gene duplications within TG rickettsial genomes. Thus, while many Rickettsia seem to be able to accept and pass genes of the mobile gene pool, the contribution of HGT products to pathogenicity is unknown and seemingly nonessential to all known rickettsial pathogens. The role lineage specific virulence factors play in pathogenic strains is thus an important aspect of future laboratory work. While HGT was traditionally considered rare in Rickettsia, we recently suggested, based on a detailed analysis of the R. felis pRF genes, that it is more common, particularly among species in which conjugation systems had yet been discovered [28]. Our suspicions have recently been verified [70] and the exact degree HGT contributes to rickettsial diversification will only be elicited with the accumulation of more rickettsial genome sequences. Such endeavors will challenge our existing classification scheme; however, a preliminary analysis of two recently published SFG rickettsiae genomes (R. massiliae str. MTU5 and R. africae str. ESF 5) using genome alignment ( Figure S1-E ) and phylogeny estimation ( Figure S3 ) does not overturn our results, and we predict that OGs generated with the inclusion of these new genomes will not alter the conclusions reached herein.
Finally, we present two concerns regarding phylogenomic analysis of Rickettsia. First, the high degree of pseudogenization in rickettsial genomes means that OG prediction programs and related methods alone are insufficient for grouping related genes. Manual inspection of algorithm output is imperative, as the high occurrence of split genes will lead to overestimation of non-representative OGs as well as inaccuracies in ORF clustering (see Table S1 ). Second, and perhaps more pressing, is the revelation that rickettsial species may be comprised of highly variable genomes, particularly across exceedingly divergent strains. Attesting to this, our analysis of predicted OGs included two strains of R. bellii that shared 321 species-specific genes but contained 97 (str. RML369-C) and 117 (str. OSU 85 389) strain-specific genes. Similarly, a recent genomic comparison of R. rickettsii str. Sheila Smith CWPP with the avirulent R. rickettsii str. Iowa revealed 143 deletions and 492 SNPs between the two genomes [176]. Altogether, these issues challenge future genomic studies on Rickettsia, particularly regarding which species/strains to select for genome sequencing, but also for justifying approaches for vaccine design with little understanding of what exactly are rickettsial virulence factors. The complexity Rickettsia has posed on laboratory work has plagued researchers for decades, and it is apparent from our study that genomic comparison is not immune from these associated difficulties.
Materials and Methods
Gene and protein prediction
Complete protocols for manual and automated curation and annotation of predicted rickettsial ORFs are listed at the PATRIC website (http://patric.vbi.vt.edu/about/standard_procedures.php). The number of ORFs per rickettsial genome differ from the previously published studies ( Figure 7 ).
Generation of orthologous groups
Complete lists (in FASTA format) of all predicted proteins encoded by each of the ten analyzed rickettsial genomes were used as templates for evaluating the performance of a suite of OG prediction methods. All methods began with all-vs-all BLASTP [177], [178] of the complete protein set. The OrthoMCL program [34], a graph-based clustering method centered on the Markov clustering algorithm of Van Dongen [33], was compared with other clustering methods. A reciprocal-best-hit clustering was performed, in which the blast results were first filtered for reciprocal best hits. In the resulting OGs, each member was the reciprocal-best-hit of each other member. Another method used these reciprocal-best-hit clusters as seed groups, which were augmented using Hidden Markov Model (HMM) searches of the complete protein set. A comparison of the resulting OG sets indicated superior performance by OrthoMCL, using the criteria of least number of ungrouped singleton ORFs and most number of OGs with perfect representation (10 ORFs from 10 genomes). Files containing all results from OrthoMCL are posted on PATRIC (http://patric.vbi.vt.edu/about/publications.php).
Phylogeny estimation
Rickettsial protein sequences comprising the 731 core representative OGs (dataset 1) were exported from the PATRIC database and aligned locally using default parameters in the command-line version of the program MUSCLE [179], [180]. Related sequences from Wolbachia (Drosophila melanogaster symbiont) were included when possible. Alignments were analyzed under maximum likelihood using Bayesian inference in the program MrBayes v3.1.2 [181]. A starting tree was generated with BIONJ using the WAG amino acid substitution matrix [182] and estimating all parameters with four substitution rate categories [183]. This tree was used to prime the Bayesian analysis, which was run in model-jumping mode with a single chain implemented, assessing burn-in (arrival at a likelihood plateau) as described previously [184]. We also analyzed the data under parsimony in an exhaustive search in the program PAUP* version 4.10 (Altivec) [185]. Branch support was assessed using the bootstrap [186] with default settings in PAUP*. We performed one million bootstrap replications. Tree files from both Bayesian and parsimony analyses were used to draw trees in PAUP*.
The second phylogenetic analysis (dataset 2) incorporating additional rickettsial taxa for which a genome sequence is not available (R. helvetica, R. australis) was initiated by performing BLASTP searches against the NCBI protein database using the following 16 R. helvetica amino acid sequences as queries: citrate synthase I (Q59741; RiOG_175), ATP synthase F1 alpha subunit (AAM93518; RiOG_208), type IV secretion/conjugal transfer ATPase, VirB4 family (ABG74480; RiOG_225), DNA polymerase III alpha subunit (CAB56077; RiOG_230), DNA polymerase I (Q9RLB6; RiOG_231), signal recognition particle-docking protein FtsY (CAB56072; RiOG_232), recombinase A (ABG74458; RiOG_245), translation elongation factor Tu (Q8KT99; RiOG_305), 10 kDa chaperonin 5 (GroES) (ABD93985; RiOG_335), chaperonin GroEL (ABD93984; RiOG_336), chromosomal replication initiator protein DnaA (ABG74394; RiOG_356), antigenic heat-stable 120 kDa protein Sca4 (AAL23857; RiOG_432), chaperone protein DnaK (ABG74418; RiOG_667), DNA-directed RNA polymerase, beta subunit (AAM93506; RiOG_701), translation elongation factor G (Q8KTB4; RiOG_708), and outer membrane autotransporter barrel domain (190 KD antigen precursor sca1) (AAU06440; RiOG_797). The nr (All GenBank+RefSeq Nucleotides+EMBL+DDBJ+PDB) database was used, coupled with a search against the Conserved Domains Database. Searches were performed across ‘all organisms’ with composition-based statistics. No filter was used. Default matrix parameters (BLOSUM62) and gap costs (Existence: 11 Extension: 1) were implemented, with an inclusion threshold of 0.005. Subjects from the ten genomic sequences were retrieved from BLAST results with the R. helvetica query sequences. When available (8 out of 16) sequences for R. australis were also retrieved ( Table S15 ). Fasta-formatted sequence files were aligned using MUSCLE, with aligned datasets converted to Nexus format using the program seqConverter.pl, version 1.1 [187]. Each Nexus file was concatenated manually into a combined executable Nexus file and analyzed under parsimony in a heuristic search implementing 500 random sequence additions saving 100 trees per replicate. Branch support was assessed from 1000 bootstrap replications.
The third phylogenetic analysis (dataset 3) used the same query sequences as the second analysis but performed tBLASTN [188] searches against the NCBI whole-genome shotgun reads (wgs) database to retrieve homologous sequences from the unannotated R. massiliae and R. africae genomes. Parameters were the same as used in the BLASTP searches, and the data were aligned and analyzed in the same manner as the second phylogenetic analysis.
Genome alignment
Six genome sequence alignments were performed using Mauve v.2.0.0 [189]. Unmodified Fasta files for each rickettsial genome were used as input, except that the R. sibirica genome sequence was reindexed using the reverse-complement of its circular permutation from the original position 668301.
Supporting Information
Analysis of synteny across aligned rickettsial genomes. Taxon abbreviations are explained in the Figure 1 legend. Five alignments are shown that are all permutations of the alignment presented in Figure 2. (A) Removal of R. felis. (B) Swapping of R. felis and R. akari. (C) Swapping of the R. bellii genomes. (D) Swapping of the R. bellii genomes plus the repositioning of the R. canadensis genome between TG and TRG rickettsiae. (E) Inclusion of the recently sequenced genomes of R. massiliae str. MTU5 and R. africae str. ESF 5, both SFG rickettsiae. Alignments performed using Mauve (Darling et al., 2004) (see text for details).
(1.99 MB PDF)
Distribution of 637 representative and non-representative class 2 OGs (C2OGs) over estimated rickettsial phylogeny. These OGs likely include pseudogenes, genes with less conserved functions in rickettsiae, and laterally acquired genes. Black = strictly representative OGs, blue = strictly non-representative OGs, red = both representative and non-representative OGs. Top numbers depict total number of OGs and bottom numbers show proportion of hypothetical proteins. Numbers in parentheses depict the proportion of non-representative OGs made representative via concatenation of split ORFs (see Table S1). Asterisks denote distributions that are made entirely representative after split ORF concatenation (27 of 47 non-representative distributions; see Table 4 and Table S1).
(2.99 MB PDF)
Phylogenetic analysis of 14 rickettsial taxa. Tree estimated using the same 16 proteins as the analysis in Figure 9, with the addition of orthologous sequences from the recently completed genomes of R. massiliae str. MTU5 and R. africae str. ESF 5 (sequences obtained from WGS reads using tBlastn). Tree estimated under parsimony (see text for details).
(0.34 MB PDF)
Phylogenetic analysis of 14 rickettsial taxa. Tree estimated using the same 16 proteins as the analysis in Figure 9, with the addition of orthologous sequences from the recently completed genomes of R. massiliae str. MTU5 and R. africae str. ESF 5 (sequences obtained from WGS reads using tBlastn). Tree estimated under parsimony (see text for details).
(0.34 MB PDF)
Distribution and characterization of predicted ORFs within 259 non-representative OGs across ten rickettsial genomes, and the results after manual curation.
(0.18 MB PDF)
Seven hundred-fifty two core rickettsial OGs predicted across ten analyzed genomes.
(0.19 MB PDF)
OGs present only in the R. bellii genomes.
(0.06 MB PDF)
Distribution of putative toxin-antitoxin (TA) systems within the rickettsial OGs predicted by OrthoMCL.
(0.07 MB PDF)
Singletons present in the R. bellii str. RML369-C genome.
(0.05 MB PDF)
Singletons and false singletons present in the R. bellii str. OSU 85 389 genome.
(0.06 MB PDF)
Singletons and false singletons present in the R. canadensis str. McKiel genome.
(0.06 MB PDF)
Singletons present in the R. prowazekii str. Madrid E genome.
(0.05 MB PDF)
Singletons present in the R. typhi str. Wilmington genome.
(0.05 MB PDF)
Singletons and false singletons present in the R. akari str. Hartford genome.
(0.07 MB PDF)
Singletons and false singletons present only in the R. felis genome.
(0.08 MB PDF)
(0.06 MB PDF)
(0.05 MB PDF)
(0.05 MB PDF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is funded through NIAID contract HHSN266200400035C to BSS and NIH grants AI59118 and AI17828 to AFA.
References
- 1.Weisburg WG, Dobson ME, Samuel JE, Dasch GA, Mallavia LP, et al. Phylogenetic diversity of the Rickettsiae. J Bacteriol. 1989;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stothard DR, Fuerst PA. Evolutionary analysis of the spotted fever and typhus groups of Rickettsia using 16S rRNA gene sequences. Syst Appl Microbiol. 1995;18:52–61. [Google Scholar]
- 4.Boone DR, Castenholz RW, Garrity GM. Bergey's manual of systematic bacteriology. New York, NY: Springer; 2001. [Google Scholar]
- 5.Tamura A, Ohashi N, Urakami H, Miyamura S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995;45:589–591. doi: 10.1099/00207713-45-3-589. [DOI] [PubMed] [Google Scholar]
- 6.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis MJ, Ying ZT, Brunner BR, Pantoja A, Ferwerda FH. Rickettsial relative associated with papaya bunchy top disease. Curr Microbiol. 1998;36:80–84. doi: 10.1007/s002849900283. [DOI] [PubMed] [Google Scholar]
- 8.Dykova I, Veverkova M, Fiala I, Machackova B, Peckova H. Nuclearia pattersoni sp n. (Filosea), a new species of amphizoic amoeba isolated from gills of roach (Rutilus rutilus), and its rickettsial endosymbiont. Folia Parasitol. 2003;50:161–170. [PubMed] [Google Scholar]
- 9.Ogata H, LaScola B, Audic S, Renesto P, Blanc G, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werren JH, Hurst GDD, Zhang W, Breeuwer JAJ, Stouthamer R, et al. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata). J Bacteriol. 1994;176:388–394. doi: 10.1128/jb.176.2.388-394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen DQ, Campbell BC, Purcell AH. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr Microbiol. 1996;33:123–128. doi: 10.1007/s002849900086. [DOI] [PubMed] [Google Scholar]
- 12.Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997;63:3926–3932. doi: 10.1128/aem.63.10.3926-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukatsu T, Shimada M. Molecular characterization of Rickettsia sp. in a bruchid beetle, Kytorhinus sharpianus (Coleoptera: Bruchidae). Appl Entomol Zool. 1999;34:391–397. [Google Scholar]
- 14.Kikuchi Y, Sameshima S, Kitade O, Kojima J, Fukatsu T. Novel clade of Rickettsia spp. from leeches. Appl Environ Microbiol. 2002;68:999–1004. doi: 10.1128/AEM.68.2.999-1004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss E, Moulder JW. The rickettsias and chlamydias. Order l. Rickettsiales. In: Krieg NR, Holt JG, editors. Bergey's manual of systematic bacteriology, Vol. 1. Baltimore: Williams & Wilkins; 1984. pp. 687–729. [Google Scholar]
- 16.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–2106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azad AF, Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4:179–186. doi: 10.3201/eid0402.980205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azad AF, Radulovic S. Pathogenic rickettsiae as bioterrorism agents. Ann NY Acad Sci. 2003;990:734–738. doi: 10.1111/j.1749-6632.2003.tb07452.x. [DOI] [PubMed] [Google Scholar]
- 20.Azad AF. Pathogenic rickettsiae as bioterrorism agents. Clin Infect Dis. 2007;45(S1):S52–S55. doi: 10.1086/518147. [DOI] [PubMed] [Google Scholar]
- 21.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 22.Ogata H, Audic S, Barbe V, Artiguenave F, Fournier PE, et al. Selfish DNA in protein-coding genes of Rickettsia. Science. 2000;290:347–350. doi: 10.1126/science.290.5490.347. [DOI] [PubMed] [Google Scholar]
- 23.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 24.Malek JA, Wierzbowski JM, Tao W, Bosak SA, Saranga DJ, et al. Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica. Nucleic Acids Res. 2004;32:1059–1064. doi: 10.1093/nar/gkh254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, et al. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eremeeva ME, Madan A, Shaw CD, Tang K, Dasch GA. New perspectives on rickettsial evolution from new genome sequences of rickettsia, particularly R. canadensis, and Orientia tsutsugamushi. Ann NY Acad Sci. 2005;1063:47–63. doi: 10.1196/annals.1355.006. [DOI] [PubMed] [Google Scholar]
- 27.Ogata H, Renesto P, Audic S, Robert C, Blanc G, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM, et al. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE. 2007;2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 30.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder EE, Kampanya N, Lu J, Nordberg E, Rajasimha H, et al. The VBI PathoSystems Resource Integration Center (PATRIC). Nucleic Acids Res. 2007;35:D401–406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanc G, Ogata H, Robert C, Audic S, Suhre K, et al. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 2007;3:e14. doi: 10.1371/journal.pgen.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dongen S. Graph clustering by flow simulation. 2000. Ph.D thesis, University of Utrecht, The Netherlands.
- 34.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 36.Andersson SG, Eriksson AS, Naslund AK, Andersen MS, Kurland CG. The Rickettsia prowazekii genome: A random sequence analysis. Microb Comp Genomics. 1996;1:293–315. [PubMed] [Google Scholar]
- 37.Andersson SGE, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 38.Andersson JO, Andersson SGE. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev. 1999;9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 39.Ogata H, Audic S, Abergel C, Fournier PE, Claverie JM. Protein coding palindromes are a unique but recurrent feature in Rickettsia. Genome Res. 2002;12:808–816. doi: 10.1101/gr.227602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa M, Renesto P, Azza S, Moinier D, Fourquet P, et al. Proteome analysis of Rickettsia felis highlights the expression profile of intracellular bacteria. Proteomics. 2007;7:1232–1248. doi: 10.1002/pmic.200600721. [DOI] [PubMed] [Google Scholar]
- 41.Gilmore RD, Jr, Joste N, McDonald GA. Cloning, expression and sequence analysis of the gene encoding the 120 kDa surface-exposed protein of Rickettsia rickettsii. Mol Microbiol. 1991;5:3089. [PubMed] [Google Scholar]
- 42.Gilmore RD., Jr Comparison of the rompA gene repeat regions of Rickettsiae reveals species-specific arrangements of individual repeating units. Gene. 1993;125:97–102. doi: 10.1016/0378-1119(93)90752-o. [DOI] [PubMed] [Google Scholar]
- 43.Roux V, Fournier PE, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 1996;34:2058–2065. doi: 10.1128/jcm.34.9.2058-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Raoult D. Distribution of immunogenic epitopes on the two major immunodominant proteins (rOmpA and rOmpB) of Rickettsia conorii among the other rickettsiae of the spotted fever group. Clin Diagn Lab Immunol. 1997;4:753–763. doi: 10.1128/cdli.4.6.753-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M, Fan MY, Bi DZ, Zhang JZ, Chen XR. Sequence analysis of a fragment of rOmpA gene of several isolates of spotted fever group rickettsiae from China. Acta Virol. 1998;42:91–93. Erratum in: Acta Virol 42:196. [PubMed] [Google Scholar]
- 46.Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48 Pt 3:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Walker DH. RompA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microbial Pathogenesis. 1998;24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 48.Moron CG, Bouyer DH, Yu XJ, Foil LD, Crocquet-Valdes P, et al. Phylogenetic analysis of the rompB genes of Rickettsia felis and Rickettsia prowazekii European-human and North American flying-squirrel strains. Am J Trop Med Hyg. 2000;62:598–603. doi: 10.4269/ajtmh.2000.62.598. [DOI] [PubMed] [Google Scholar]
- 49.Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- 50.Stenos J, Walker DH. The rickettsial outer-membrane protein A and B genes of Rickettsia australis, the most divergent rickettsia of the spotted fever group. Int J Syst Evol Microbiol. 2000;50 Pt 5:1775–1779. doi: 10.1099/00207713-50-5-1775. [DOI] [PubMed] [Google Scholar]
- 51.Bouyer DH, Stenos J, Crocquet-Valdes P, Moron CG, Popov VL, et al. Rickettsia felis: Molecular characterization of a new member of the spotted fever group. Int J Syst Evol Microbiol. 2001;51:339–347. doi: 10.1099/00207713-51-2-339. [DOI] [PubMed] [Google Scholar]
- 52.Croquet-Valdes PA, Diaz-Montero CM, Feng HM, Li H, Barrett ADT, et al. Immunization with a portion of rickettsial outer membrane protein A stimulates protective immunity against spotted fever rickettsiosis. Vaccine. 2001;20:979–988. doi: 10.1016/s0264-410x(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 53.Diaz-Montero CM, Feng HM, Crocquet-Valdes PA, Walker DH. Identification of protective components of two major outer membrane proteins of spotted fever group Rickettsiae. Am J Trop Med Hyg. 2001;65:371–378. doi: 10.4269/ajtmh.2001.65.371. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama T. Adherence to and invasion of Vero cells by recombinant Escherichia coli expressing the outer membrane protein rOmpB of Rickettsia japonica. Ann NY Acad Sci. 2003;990:585–590. doi: 10.1111/j.1749-6632.2003.tb07431.x. [DOI] [PubMed] [Google Scholar]
- 55.Blanc G, Ngwamidiba M, Ogata H, Fournier PE, Claverie JM, et al. Molecular evolution of rickettsia surface antigens: Evidence of positive selection. Mol Biol Evol. 2005;22:2073–2083. doi: 10.1093/molbev/msi199. [DOI] [PubMed] [Google Scholar]
- 56.Jiggins FM. Adaptive evolution and recombination of Rickettsia antigens. J Mol Evol. 2006;62:99–110. doi: 10.1007/s00239-005-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngwamidiba M, Blanc G, Raoult D, Fournier PE. Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 2006;6:12. doi: 10.1186/1471-2180-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. PNAS USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldridge GD, Burkhardt NY, Felsheim RF, Kurttu TJ, Munderloh UG. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl Envirom Microbiol. 2007;74:645–652. doi: 10.1128/AEM.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldridge GD, Burkhardt NY, Felsheim RF, Kurttu TJ, Munderloh UG. Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis. Appl Envirom Microbiol. 2007;73:4984–4995. doi: 10.1128/AEM.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Renesto, Ogata H, Audic S, Claverie JM, Raoult D. Some lessons from Rickettsia genomics. FEMS Microbiol Rev. 2005;29:99–117. doi: 10.1016/j.femsre.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Fuxelius HH, Darby A, Min CK, Cho NH, Andersson SG. The genomic and metabolic diversity of Rickettsia. Res Microbiol. 2007;158:745–753. doi: 10.1016/j.resmic.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Vitorino L, Chelo IM, Bacellar F, Zé-Zé L. Rickettsiae phylogeny: A multigenic approach. Microbiology. 2007;153:160–168. doi: 10.1099/mic.0.2006/001149-0. [DOI] [PubMed] [Google Scholar]
- 64.McKiel JA, Bell EJ, Lackman DB. Rickettsia canada: a new member of the typhus group of rickettsiae isolated from Haemaphysalis leporispalustris ticks in Canada. Can J Microbiol. 1967;13:503–510. doi: 10.1139/m67-065. [DOI] [PubMed] [Google Scholar]
- 65.Burgdorfer W. Observations on Rickettsia canada, a recently described member of the typhus group rickettsiae. J Hyg Epid Microbiol Immunol. 1968;12:26–31. [PubMed] [Google Scholar]
- 66.Brinton LP, Burgdorfer W. Fine structure of Rickettsia canada in tissues of Dermacentor andersoni Stiles. J Bacteriol. 1971;105:1149–1159. doi: 10.1128/jb.105.3.1149-1159.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dasch GA, Bourgeois AL. Antigens of the typhus group of rickettsiae: importance of the species-specific surface protein antigens in eliciting immunity. In: Burgdorfer W, Anacker RL, editors. Rickettsiae and Rickettsial Diseases. New York: Academic Press; 1981. pp. 61–70. [Google Scholar]
- 68.Ching WM, Dasch GA, Carl M, Dobson ME. Structural analyses of the 120-kDa serotype protein antigens of typhus group rickettsiae: comparison with other S-layer proteins. Ann NY Acad Sci. 1990;590:334–351. doi: 10.1111/j.1749-6632.1990.tb42241.x. [DOI] [PubMed] [Google Scholar]
- 69.Myers WF, Wisseman CL., JR . The taxonomic relationship of Rickettsia canada to the typhus and spotted fever groups of the genus Rickettsia. In: Burgdorfer W, Anacker RL, editors. Rickettsiae and Rickettsial Diseases. New York: Academic Press; 1981. pp. 313–325. [Google Scholar]
- 70.Blanc G, Ogata H, Robert C, Audic S, Claverie JM, et al. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 2007;17:1657–1664. doi: 10.1101/gr.6742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metzger S, Dror IB, Aizenman E, Schreiber G, Toone M, et al. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 72.Metzger S, Sarubbi E, Glaser G, Cashel M. Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J Biol Chem. 1989;264:9122–9125. [PubMed] [Google Scholar]
- 73.Gouin E, Egile C, Dehoux P, Villiers V, Adams J, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 74.Jeng RL, Goley ED, D'Alessio JA, Chaga OY, Svitkina TM, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 75.Rothnagel JA, Rogers GE. Trichohyalin, an intermediate filament-associated protein of the hair follicle. J Cell Biol. 1986;102:1419–1429. doi: 10.1083/jcb.102.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fietz MJ, McLaughlan CJ, Campbell MT, Rogers GE. Analysis of the sheep trichohyalin gene: potential structural and calcium-binding roles of trichohyalin in the hair follicle. J Cell Biol. 1993;121:855–865. doi: 10.1083/jcb.121.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamilton EH, Payne RE, Jr, O'Keefe EJ. Trichohyalin: presence in the granular layer and stratum corneum of normal human epidermis. J Invest Dermatol. 1991;96:666–672. doi: 10.1111/1523-1747.ep12470590. [DOI] [PubMed] [Google Scholar]
- 78.O'Guin WM, Manabe M. The role of trichohyalin in hair follicle differentiation and its expression in nonfollicular epithelia. The molecular and structural biology of hair. Ann NY Acad Sci. 1991;642:51–62. doi: 10.1111/j.1749-6632.1991.tb24380.x. [DOI] [PubMed] [Google Scholar]
- 79.Chung CH, Ives HE, Almeda S, Goldberg AL. Purification from Escherichia coli of a periplasmic protein that is a potent inhibitor of pancreatic proteases. J Biol Chem. 1983;258:11032–11038. [PubMed] [Google Scholar]
- 80.Seymour JL, Lindquist RN, Dennis MS, Moffat B, Yansura D, et al. Ecotin is a potent anticoagulant and reversible tight-binding inhibitor of factor Xa. Biochemistry. 1994;33:3949–3958. doi: 10.1021/bi00179a022. [DOI] [PubMed] [Google Scholar]
- 81.Ulmer JS, Lindquist RN, Dennis MS, Lazarus RA. Ecotin is a potent inhibitor of the contact system proteases factor XIIa and plasma kallikrein. FEBS Lett. 1995;365:159–163. doi: 10.1016/0014-5793(95)00466-m. [DOI] [PubMed] [Google Scholar]
- 82.Eggers CT, Murray LA, Delmar VA, Day AG, Craik CS. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochemical Journal. 2004;379:107–118. doi: 10.1042/BJ20031790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289:1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 84.Reeves WK, Loftis AD, Szumlas DE, Abbassy MM, Helmy IM, et al. Rickettsial pathogens in the tropical mite Orithonyssus bacoti (Acari: Macronyssidae) from Egyptian rats (Rattus spp.). Exp Appl Acarol. 2007;41:101–107. doi: 10.1007/s10493-006-9040-3. [DOI] [PubMed] [Google Scholar]
- 85.Huebner RJ, Jellison WL, Pmerantz C. Rickettsialpox-a newly recognized rickettsial disease. IV. Isolation of a rickettsia apparently identical with the causative agent of rickettsialpox from Allodermanyssus sanguineus, a rodent mite. Public Health Rep. 1948;61:1677–1682. [PubMed] [Google Scholar]
- 86.Adams JR, Schmidtmann ET, Azad AF. Infection of colonized cat fleas, Ctenocephalides felis (Bouche), with a rickettsia-like microorganism. Am J Trop Med Hyg. 1990;43:400–409. doi: 10.4269/ajtmh.1990.43.400. [DOI] [PubMed] [Google Scholar]
- 87.Azad AF, Sacci JB, Jr, Nelson WM, Dasch GA, Schmidtmann ET, et al. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. PNAS USA. 1992;89:43–46. doi: 10.1073/pnas.89.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zavala-Velazquez JE, Zavala-Castro JE, Vado-Solis I, Ruiz-Sosa JA, Moron CG, et al. Identification of Ctenocephalides felis fleas as a host of Rickettsia felis, the agent of a spotted fever rickettsiosis in Yucatan, Mexico. Vector Borne Zoonotic Dis. 2002;2:69–75. doi: 10.1089/153036602321131869. [DOI] [PubMed] [Google Scholar]
- 89.Ogata H, Robert C, Audic S, Robineau G, Blanc G, et al. Rickettsia felis, from culture to genome sequencing. Ann NY Acad Sci. 2005;1063:26–34. doi: 10.1196/annals.1355.004. [DOI] [PubMed] [Google Scholar]
- 90.Sekeyova Z, Roux V, Raoult D. Phylogeny of Rickettsia spp. inferred by comparing sequences of 'gene D', which encodes an intracytoplasmic protein. Int J Syst Evol Microbiol. 2001;51:1353–1560. doi: 10.1099/00207713-51-4-1353. [DOI] [PubMed] [Google Scholar]
- 91.Ngwamidiba M, Blanc G, Ogata H, Raoult D, Fournier PE. Phylogenetic study of Rickettsia species using sequences of the autotransporter protein-encoding gene sca2. Ann NY Acad Sci. 2005;1063:94–99. doi: 10.1196/annals.1355.015. [DOI] [PubMed] [Google Scholar]
- 92.Jado I, Oteo JA, Aldámiz M, Gil H, Escudero R, et al. Rickettsia monacensis and human disease, Spain. Emerg Infect Dis. 2007;13:1405–1407. doi: 10.3201/eid1309.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 94.Donigian JR, de Lange T. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem. 2007;282:22662–22667. doi: 10.1074/jbc.M702620200. [DOI] [PubMed] [Google Scholar]
- 95.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 96.Kaminker PG, Kim S-H, Taylor RD, Zebarjadian Y, Funk WD, et al. TANK2, a new TRF1-associated Poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J Biol Chem. 2001;276:35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- 97.Bae J, Donigian JR, Hsueh AJW. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem. 2003;278:5195–5204. doi: 10.1074/jbc.M201988200. [DOI] [PubMed] [Google Scholar]
- 98.Deng Z, Atanasiu C, Zhao K, Marmorstein R, Sbodio JI, et al. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated Poly-ADP ribose polymerase that binds and modifies EBNA1. J Virol. 2005;79:4640–4650. doi: 10.1128/JVI.79.8.4640-4650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Larsen TS, Krogh A. EasyGene - a prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics. 2003;4:21. doi: 10.1186/1471-2105-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nielsen P, Krogh A. Large-scale prokaryotic gene prediction and comparison to genome annotation. Bioinformatics. 2005;21:4322–4329. doi: 10.1093/bioinformatics/bti701. [DOI] [PubMed] [Google Scholar]
- 101.Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 103.Amor JC, Swails J, Zhu X, Roy CR, Nagai H, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280:1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 104.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, et al. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. PNAS. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reiss-Gutfreund RJ. The isolation of Rickettsia prowazeki and mooseri from unusual sources. Am J Trop Med Hyg. 1966;15:943–949. doi: 10.4269/ajtmh.1966.15.943. [DOI] [PubMed] [Google Scholar]
- 106.Medina-Sanchez A, Bouyer DH, Cantara-Rodriguez V, Mafra C, Zavala-Castro J, et al. Detection of a typhus group Rickettsia in Amblyomma ticks in the state of Nuevo Leon, Mexico. Ann NY Acad Sci. 2005;1063:327–332. doi: 10.1196/annals.1355.052. [DOI] [PubMed] [Google Scholar]
- 107.Bozeman FM, Masiello SA, Williams MS, Elisberg BL. Epidemic typhus rickettsiae isolated from flying squirrels. Nature. 1975;255:545–547. doi: 10.1038/255545a0. [DOI] [PubMed] [Google Scholar]
- 108.Weinert LA, Tinsley MC, Temperley M, Jiggins FM. Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biol Lett. 2007;3:678–681. doi: 10.1098/rsbl.2007.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Houhamdi L, Raoult D. Experimentally infected human body lice (Pediculus humanus humanus) as vectors of Rickettsia rickettsii and Rickettsia conorii in a rabbit model. Am J Trop Med Hyg. 2006;74:521–525. [PubMed] [Google Scholar]
- 110.Uchiyama T. Role of major surface antigens of Rickettsia japonica in the attachment to host cell. In: Kazar J, Raoult D, editors. Rickettsiae and rickettsial diseases. Bratislava: Publishing house of the Slovak Academy of Sciences; 1999. pp. 182–188. [Google Scholar]
- 111.Sorvillo FJ, Gondo B, Emmons R, Ryan P, Waterman SH, et al. A suburban focus of endemic typhus in Los Angeles County: association with seropositive domestic cats and opossums. Am J Trop Med Hyg. 1993;48:269–273. doi: 10.4269/ajtmh.1993.48.269. [DOI] [PubMed] [Google Scholar]
- 112.Houhamdi L, Fournier PE, Fang R, Lepidi H, Raoult D. An experimental model of human body louse infection with Rickettsia prowazekii. J Infect Dis. 2002;186:1639–1646. doi: 10.1086/345373. [DOI] [PubMed] [Google Scholar]
- 113.Grimaldi D, Engel MS. Evolution of the Insects. New York: Cambridge University Press; 2005. p. 772. ISBN-13: 9780521821490. [Google Scholar]
- 114.Steinert PM, Parry DAD, Marekov LN. Trichohyalin mechanically strengthens the hair follicle: Multiple cross-bridging roles in the inner root sheath. J Biol Chem. 2003;278:41409–41419. doi: 10.1074/jbc.M302037200. [DOI] [PubMed] [Google Scholar]
- 115.Tobin DJ, Foitzik K, Reinheckel T, Mecklenburg L, Botchkarev VA, et al. The lysosomal protease cathepsin L is an important regulator of keratinocyte and melanocyte differentiation during hair follicle morphogenesis and cycling. Am J Pathol. 2002;160:1807–1821. doi: 10.1016/S0002-9440(10)61127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ou G, Koga M, Blacque OE, Murayama T, Ohshima Y, et al. Sensory ciliogenesis in Caenorhabditis elegans: Assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cobbe N, Heck MM. Review: SMCs in the world of chromosome biology-from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- 118.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. PNAS USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- 120.Lindow JC, Britton RA, Grossman AD. Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J Bacteriol. 2002;184:5317–5322. doi: 10.1128/JB.184.19.5317-5322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.von der Schulenburg JHG, Habig M, Sloggett JJ, Webberley KM, Bertrand D, et al. Incidence of male-killing Rickettsia spp. (alphaproteobacteria) in the ten-spot ladybird beetle Adalia decempunctata L. (Coleoptera: Coccinellidae). Appl Environ Microbiol. 2001;67:270–277. doi: 10.1128/AEM.67.1.270-277.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lawson ET, Mousseau TA, Klaper R, Hunter MD, Werren JH. Rickettsia associated with male-killing in a buprestid beetle. Heredity. 2001;86:497–505. doi: 10.1046/j.1365-2540.2001.00848.x. [DOI] [PubMed] [Google Scholar]
- 123.Hagimori T, Abe Y, Date S, Miura K. The first finding of a Rickettsia bacterium associated with parthenogenesis induction among insects. Curr Microbiol. 2006;52:97–101. doi: 10.1007/s00284-005-0092-0. [DOI] [PubMed] [Google Scholar]
- 124.Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl Environ Microbiol. 2005;71:4069–4075. doi: 10.1128/AEM.71.7.4069-4075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yusuf M, Turner B. Characterisation of Wolbachia-like bacteria isolated from the parthenogenetic stored-product pest psocid Liposcelis bostrychophila (Badonnel) (Psocoptera). J Stored Prod Res. 2004;40:207–225. [Google Scholar]
- 126.Zchori-Fein E, Borad C, Harari AR. Oogenesis in the date stone beetle, Coccotrypes dactyliperda, depends on symbiotic bacteria. Physiol Entomol. 2006;31:164–169. [Google Scholar]
- 127.Campbell CL, Mummey DL, Schmidtmann ET, Wilson WC. Culture-independent analysis of midgut microbiota in the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae). J Med Entomol. 2004;41:340–348. doi: 10.1603/0022-2585-41.3.340. [DOI] [PubMed] [Google Scholar]
- 128.Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, et al. Identification and localization of Rickettsia in Bemisia tabaci (Homoptera: Aleyrodidae). Appl Environ Microbiol. 2006;72:3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weiss E, Coolbaugh JC, Williams JC. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl Microbiol. 1975;30:456–463. doi: 10.1128/am.30.3.456-463.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kamen B. “Folate and antifolate pharmacology”. Seminars in oncology. 1997;24(5 Suppl 18):S18-30–S18-39. [PubMed] [Google Scholar]
- 131.Allignet J, Loncle V, Simenel C, Delepierre M, el Solh N. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91–98. doi: 10.1016/0378-1119(93)90350-c. [DOI] [PubMed] [Google Scholar]
- 132.Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother. 1993;37:2119–2125. doi: 10.1128/aac.37.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Allignet J, el Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2036. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Allignet J, Liassine N, el Solh N. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother. 1998;42:1794–1798. doi: 10.1128/aac.42.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Leclercq R, Courvalin P. Intrinsic and unusual resistance to macrolide, lincosamide, and streptogramin antibiotics in bacteria. Antimicrob Agents Chemother. 1992;35:1273–1276. doi: 10.1128/aac.35.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Verbist L, Verhaegen J. Comparative in-vitro activity of RP 59500. J Antimicrob Chemother. 1992;30(Suppl. A):39–44. doi: 10.1093/jac/30.suppl_a.39. [DOI] [PubMed] [Google Scholar]
- 137.Seoane A, García-Lobo JM. Identification of a streptogramin A acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother. 2000;44:905–909. doi: 10.1128/aac.44.4.905-909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Parent R, Roy PH. The chloramphenicol acetyltransferase gene of Tn2424: a new breed of cat. J Bacteriol. 1992;174:2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Eremeeva ME, Madan A, Dasch GA. Genome sequence of Rickettsia bellii OSU 85-389. 2006. 20th Meeting of The American Society for Rickettsiology in conjunction with the 5th International Conference on Bartonella as Emerging Pathogens. September 2–7, 2006. Asilomar Conference Grounds, Pacific Grove, California, USA. Abstract #11.
- 140.Eremeeva ME, Madan A, Halsell T, Dasch GA. Sequencing and characterization of Prak1, a 24.4 Kb plasmid from Rickettsia akari. 2007. 21st Meeting of The American Society for Rickettsiology. September 8–11, 2007. Colorado Springs, Colorado, USA. Abstract #81.
- 141.Lusher M, Storey CC, Richmond SJ. Plasmid diversity within the genus Chlamydia. J Gen Microbiol. 1989;135:1145–1151. doi: 10.1099/00221287-135-5-1145. [DOI] [PubMed] [Google Scholar]
- 142.Savinelli EA, Mallavia LP. Comparison of Coxiella burnetii plasmids to homologous chromosomal sequences present in a plasmidless endocarditis-causing isolate. Ann NY Acad Sci. 1990;590:523–533. doi: 10.1111/j.1749-6632.1990.tb42262.x. [DOI] [PubMed] [Google Scholar]
- 143.Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology. 1997;143:1847–1854. doi: 10.1099/00221287-143-6-1847. [DOI] [PubMed] [Google Scholar]
- 144.Willems H, Ritter M, Jager C, Thiele D. Plasmid-homologous sequences in the chromosome of plasmidless Coxiella burnetii Scurry Q217. J Bacteriol. 1997;179:3293–3297. doi: 10.1128/jb.179.10.3293-3297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McClenaghan M, Honeycombe JR, Bevan BJ, Herring AJ. Distribution of plasmid sequences in avian and mammalian strains of Chlamydia psittaci. J Gen Microbiol. 1988;134:559–565. doi: 10.1099/00221287-134-3-559. [DOI] [PubMed] [Google Scholar]
- 146.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. PNAS. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 148.Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. PNAS USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.van Passel MWJ, van der Ende A, Bart A. Plasmid diversity in Neisseriae. Infect Immun. 2006;74:4892–4899. doi: 10.1128/IAI.02087-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roux V, Raoult D. Genotypic identification and phylogenetic analysis of the spotted fever group rickettsiae by pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4895–4904. doi: 10.1128/jb.175.15.4895-4904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Eremeeva M, Balayeva N, Ignatovich V, Raoult D. Genomic study of Rickettsia akari by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:3022–3024. doi: 10.1128/jcm.33.11.3022-3024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Frank AC, Alsmark CM, Thollesson M, Andersson SG. Functional divergence and horizontal transfer of type IV secretion systems. Mol Biol Evol. 2005;22:1325–1336. doi: 10.1093/molbev/msi124. [DOI] [PubMed] [Google Scholar]
- 154.Jensen RB, Gerdes K. Programmed cell death in bacteria: Proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 155.Yarmolinsky MB. Programmed cell death in bacterial population. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 156.Couturier M, Bahassi EM, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 157.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and anti-death in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 158.Hayes F. Toxins–antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 159.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nature Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 160.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang YX, Guo XK, Wu C, Bi B, Ren SX, et al. Characterization of a novel toxin-antitoxin module, VapBC, encoded by Leptospira interrogans chromosome. Cell Res. 2004;14:208–216. doi: 10.1038/sj.cr.7290221. [DOI] [PubMed] [Google Scholar]
- 162.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Clissold P, Ponting C. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 164.Fatica A, Tollervey D, Dlakic M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cooper TF, Heinemann JA. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. PNAS USA. 2000;97:12643–12648. doi: 10.1073/pnas.220077897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cooper TF, Heinemann JA. Selection for plasmid post-segregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc R Soc Lond B Biol Sci. 2005;272:403–410. doi: 10.1098/rspb.2004.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pujol C, Eugene E, Marceau M, Nassif X. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. PNAS USA. 1999;96:4017–4022. doi: 10.1073/pnas.96.7.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arcus VL, Rainey PB, Turner SJ. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005;13:360–365. doi: 10.1016/j.tim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 169.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. PNAS. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 171.Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16:365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wilson GA, Bertrand N, Patel Y, Hughes JB, Feil EJ, et al. Orphans as taxonomically restricted and ecologically important genes. Microbiology. 2005;151:2499–2501. doi: 10.1099/mic.0.28146-0. [DOI] [PubMed] [Google Scholar]
- 173.Sopko R, Andrews B. Small open reading frames: not so small anymore. Genome Res. 2006;16:314–315. doi: 10.1101/gr.4976706. [DOI] [PubMed] [Google Scholar]
- 174.Wilson GA, Feil EJ, Lilley AK, Field D. Large-scale comparative genomic ranking of taxonomically restricted genes (TRGs) in bacterial and archaeal genomes. PLoS ONE. 2007;2:e324. doi: 10.1371/journal.pone.0000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Darby AC, Nam-Huyk C, Fuxelius HH, Westberg J, Andersson SGE. Intracellular pathogens go extreme. Trends Gen. 2007;23:511–520. doi: 10.1016/j.tig.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 176.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, et al. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun. 2007;76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 178.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 182.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 183.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 184.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. PNAS USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Swofford D. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4 ed. Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 186.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 187.Bininda-Emonds O. seqConverter. pl, version 1.1. ed, Institut fur Spezeille Zoologie und Evolutionsbiologie mit Phyletischem Museum, Friedrich-Schiller-Universitat Jena 2006.
- 188.Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of synteny across aligned rickettsial genomes. Taxon abbreviations are explained in the Figure 1 legend. Five alignments are shown that are all permutations of the alignment presented in Figure 2. (A) Removal of R. felis. (B) Swapping of R. felis and R. akari. (C) Swapping of the R. bellii genomes. (D) Swapping of the R. bellii genomes plus the repositioning of the R. canadensis genome between TG and TRG rickettsiae. (E) Inclusion of the recently sequenced genomes of R. massiliae str. MTU5 and R. africae str. ESF 5, both SFG rickettsiae. Alignments performed using Mauve (Darling et al., 2004) (see text for details).
(1.99 MB PDF)
Distribution of 637 representative and non-representative class 2 OGs (C2OGs) over estimated rickettsial phylogeny. These OGs likely include pseudogenes, genes with less conserved functions in rickettsiae, and laterally acquired genes. Black = strictly representative OGs, blue = strictly non-representative OGs, red = both representative and non-representative OGs. Top numbers depict total number of OGs and bottom numbers show proportion of hypothetical proteins. Numbers in parentheses depict the proportion of non-representative OGs made representative via concatenation of split ORFs (see Table S1). Asterisks denote distributions that are made entirely representative after split ORF concatenation (27 of 47 non-representative distributions; see Table 4 and Table S1).
(2.99 MB PDF)
Phylogenetic analysis of 14 rickettsial taxa. Tree estimated using the same 16 proteins as the analysis in Figure 9, with the addition of orthologous sequences from the recently completed genomes of R. massiliae str. MTU5 and R. africae str. ESF 5 (sequences obtained from WGS reads using tBlastn). Tree estimated under parsimony (see text for details).
(0.34 MB PDF)
Phylogenetic analysis of 14 rickettsial taxa. Tree estimated using the same 16 proteins as the analysis in Figure 9, with the addition of orthologous sequences from the recently completed genomes of R. massiliae str. MTU5 and R. africae str. ESF 5 (sequences obtained from WGS reads using tBlastn). Tree estimated under parsimony (see text for details).
(0.34 MB PDF)
Distribution and characterization of predicted ORFs within 259 non-representative OGs across ten rickettsial genomes, and the results after manual curation.
(0.18 MB PDF)
Seven hundred-fifty two core rickettsial OGs predicted across ten analyzed genomes.
(0.19 MB PDF)
OGs present only in the R. bellii genomes.
(0.06 MB PDF)
Distribution of putative toxin-antitoxin (TA) systems within the rickettsial OGs predicted by OrthoMCL.
(0.07 MB PDF)
Singletons present in the R. bellii str. RML369-C genome.
(0.05 MB PDF)
Singletons and false singletons present in the R. bellii str. OSU 85 389 genome.
(0.06 MB PDF)
Singletons and false singletons present in the R. canadensis str. McKiel genome.
(0.06 MB PDF)
Singletons present in the R. prowazekii str. Madrid E genome.
(0.05 MB PDF)
Singletons present in the R. typhi str. Wilmington genome.
(0.05 MB PDF)
Singletons and false singletons present in the R. akari str. Hartford genome.
(0.07 MB PDF)
Singletons and false singletons present only in the R. felis genome.
(0.08 MB PDF)
(0.06 MB PDF)
(0.05 MB PDF)
(0.05 MB PDF)