Abstract
Understanding the organization of reaction fluxes in cellular metabolism from the stoichiometry and the topology of the underlying biochemical network is a central issue in systems biology. In this task, it is important to devise reasonable approximation schemes that rely on the stoichiometric data only, because full-scale kinetic approaches are computationally affordable only for small networks (e.g., red blood cells, ≈50 reactions). Methods commonly used are based on finding the stationary flux configurations that satisfy mass-balance conditions for metabolites, often coupling them to local optimization rules (e.g., maximization of biomass production) to reduce the size of the solution space to a single point. Such methods have been widely applied and have proven able to reproduce experimental findings for relatively simple organisms in specific conditions. Here, we define and study a constraint-based model of cellular metabolism where neither mass balance nor flux stationarity are postulated and where the relevant flux configurations optimize the global growth of the system. In the case of Escherichia coli, steady flux states are recovered as solutions, although mass-balance conditions are violated for some metabolites, implying a nonzero net production of the latter. Such solutions furthermore turn out to provide the correct statistics of fluxes for the bacterium E. coli in different environments and compare well with the available experimental evidence on individual fluxes. Conserved metabolic pools play a key role in determining growth rate and flux variability. Finally, we are able to connect phenomenological gene essentiality with “frozen” fluxes (i.e., fluxes with smaller allowed variability) in E. coli metabolism.
Keywords: fluxomics, growth, stoichiometry, conserved moieties, gene essentiality
Cellular metabolism involves a complex network of interactions and cross-regulations among metabolites, proteins, and genes. Although our knowledge of regulatory functions at the genetic level and at the level of protein–protein interactions is still in its infancy, the biochemical network of reactions that describes metabolism has been characterized in detail for many organisms (1–3). Most information about the metabolic network is contained in the matrices B = {biμ} and A = {aiμ} describing, respectively, the input and output stoichiometric coefficients of each metabolite μ (ranging from 1 to P) for all of the metabolic reactions i (1 to N). Their knowledge allows for the definition of constraint-based models from which a prediction of metabolic fluxes is possible (4–6). These models typically rely on a steady-state assumption that reflects the time scale separation between the (faster) equilibration of metabolic processes and the (slower) dynamics of genetic regulation. Under this condition, the concentration Xμ of metabolite μ and the flux νi ≥ 0 of reaction i are constant in time and globally linked by a set of mass-balance conditions:
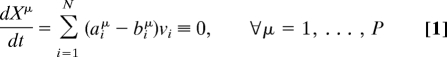 |
or, in matrix notation, (A − B)ν = 0, where ν = {ν1,…,νN} is a flux vector. The problem is that of finding a flux vector satisfying the set of P linear equations (1). For real metabolic networks, the above system is usually underdetermined because N > P [e.g., Escherichia coli has ≈1,100 reactions and <800 metabolites (1)], so that multiple solutions exist, and one has to specify which of these are the working, physical states of the network.
In organisms with high functional specificity or whose main objective is to produce certain specified metabolites, like, e.g., red blood cells, physical states are taken to be all those consistent with the mass-balance conditions. In such cases, it is important to be able to explore the solution space of Eq. 1 uniformly. Uniform sampling can be achieved in small networks with Monte Carlo methods (7), and, more recently, message-passing algorithms have been used (8, 9). However, computational considerations still prevent the application of such techniques to explore more general aspects of larger metabolic networks.
For more complex organisms, one normally complements Eq. 1 with the assumption that the physical state of the network obeys a specific optimization principle. In flux-balance analysis (FBA) (5), the choice usually falls on the maximization of biomass production, a useful proxy of the growth capabilities of an organism. FBA has been widely applied to different organisms (mostly bacteria) to investigate general aspects of metabolism, like flux distributions in different environments (10), evolutionary dispensability of enzymes (11), or the plasticity of the reaction network (12). Minimization of metabolic adjustment (MOMA) (6) is instead able to predict the reorganization of fluxes after a reaction knockout by minimizing the distance between the FBA solution and that obtained from Eq. 1 after the knockout.
Here, we consider a constraint-based model of metabolic activity with the aim of characterizing flux states corresponding to optimal net metabolic production. We neither assume constancy-in-time of fluxes nor do we impose mass-balance constraints on metabolites. Rather, we allow for these to be recovered as properties of the solutions. This assumption turns out to be able to reproduce the empirical statistics of fluxes for the bacterium E. coli in different environments with a remarkable accuracy, and the physical and biochemical origin of the robustness of the emerging picture can be completely understood. Further biological insight can be gained by comparing the variability of individual fluxes with data on phenomenological gene essentiality.
Model Definitions
The abstract setup we consider was originally introduced by J. Von Neumann (13) to model growth in production economies as an autocatalytic process. We consider a system of N chemical reactions between P reagents, with fluxes and concentrations evolving in discrete time t = 0,1,… (we leave the time scale unspecified). For now, the system is assumed to be purely autocatalytic, so that the total input of any given metabolite at a certain time step must come from (all of or part of) the output at the previous time step. Let Si(t) denote the scale at which reaction i operates at time t, so that the total input and output of metabolite μ are given by Iμ(t) = ΣiSi(t)biμ and Oμ(t) = ΣiSi(t)aiμ, respectively. Stability requires that Cμ(t) := Oμ(t) − Iμ(t + 1) ≥ 0 at all times, otherwise the system would need an outside metabolic source to survive. We focus our attention on the feasibility of dynamical paths with constant growth rate ρ > 0, where Iμ(t + 1) = ρIμ(t). It is easy to see that, in such paths, reaction rates must behave as Si(t) = siρt, with constants si ≥ 0 satisfying the P linear constraints
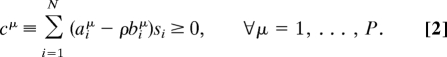 |
The main assumption now is that the network's physical state s* = {si*} corresponds to one of those for which the growth rate ρ attains its maximum possible value ρ* compatible with the constraints (Eq. 2). The rationale for this choice lies in the fact that, under general conditions, it can be proven that paths with the optimal use of resources coincide, apart from an initial transient, with those of maximal expansion (14). Note that the sis are essentially the discrete-time version of the continuous time fluxes νi of Eq. 1.
It is intuitive that the solution space of Eq. 2 shrinks as ρ is increased starting from ρ = 0, where any flux configuration is a viable state. Quite importantly, then, for a fixed set of input–output relations, the solutions of Eq. 2 are bound to coincide with those of Eq. 1 if ρ* = 1 and cμ = 0 ∀μ.
The typical behavior of Von Neumann's model can be fully appreciated analytically in the case of random graphs. Depending on their size and topology, autocatalytic networks with random stoichiometry give rise to very different optimal states. As in other constrained optimization problems (15), the key control parameter is the ratio N/P = n of variables-to-constraints. In random topologies (16, 17), as n increases, the system crosses over from a contracting phase with ρ* < 1 to an expanding one with ρ* > 1, passing through a stationary regime with ρ* = 1 in which reaction fluxes are constant in time as in FBA (although the values at which fluxes settle may be different).
Application to E. coli
A natural reaction network can be modeled as an autocatalytic system when the uptake reactions that provide resources and account for exchanges of metabolites with the surrounding environment are included. We have applied the Von Neumann scheme to the cellular metabolism of the bacterium E. coli, as reconstructed in ref. 1. To set the stage, 3 different operations have to be performed.
Environment Selection.
To fix the environment where the cell lives, we have defined a set of external metabolites on which we applied uptake fluxes. We have considered 3 distinct environmental conditions: (i) isolated cell, without uptakes; (ii) minimal environment, with uptakes on a restricted set of metabolites (10), namely CO2, K, NH4, PI, O2, SO4, and one of glu-L, succ, or glc; (iii) rich environment, with uptakes on all external metabolites.
“Leaf Removal.”
Once the network is built, one has to remove from the internal metabolites those that are never produced, because the corresponding constraints are satisfied only by taking ρ = 0 or by applying a null flux to the corresponding reactions (as is actually done in FBA).
Accounting for Reversibility.
We have disposed of reversible reactions by introducing 2 separate fluxes and taking the absolute difference as the positive net flux.
The size of the resulting network, i.e., N and P, is ultimately different for different environments.
We calculate the maximum growth rate ρ* numerically via a generalized MinOver algorithm (18), detailed in ref. 17, in which solutions are found at fixed ρ, and then ρ is gradually increased. It has been shown rigorously (17) that this algorithm converges to a solution at a fixed ρ if at least 1 solution exists. Moreover, when multiple solutions occur (in which case they form a connected set), the algorithm provides a uniform sampling of the solution space [see supporting information (SI) for a test of this property at low dimensions]. Anticipating that indeed many flux vectors satisfy Eq. 2 at ρ* for E. coli, the latter is a particularly important advantage because a uniform sampling is required to characterize the solution space.
Characterization of the Solutions
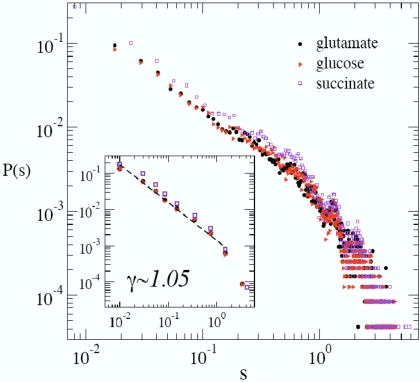
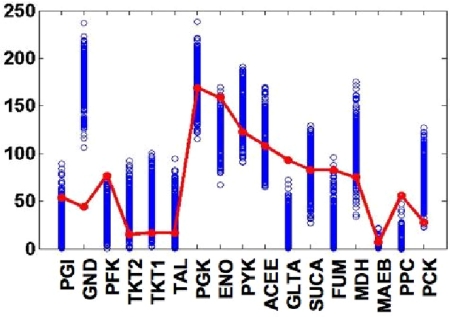
For the metabolic network of E. coli, we have found ρ* = 0.999 ± 0.001 independently of the environmental conditions we have tested, so that the state of optimal growth is compatible with one with constant fluxes. The distribution of reaction fluxes at ρ* (see Fig. 1) displays a regime (<2 decades) with a scaling form P(s) ≈ s−γ with exponent close to −1 [in reasonable agreement with the experimental evidence based on a limited set of measured fluxes (19, 10)], followed by a cutoff. Note that the flux histogram is remarkably stable over different solutions. This scenario has been partially reproduced by FBA (10), but the solution obtained optimizing the biomass production systematically overestimates γ. To compare individual fluxes with experiments, we aimed at studying the solution obtained in conditions similar to those described in ref. 19, focusing our attention on 17 fluxes from the central metabolism, as in ref. 6. Unfortunately, we are unable to reproduce in detail the M9 medium used in ref. 19, and can fix the uptakes of only 3 metabolites, namely glucose, CO2, and O2 identically to ref. 19. For the remaining part of the environment, we chose to simulate a minimal medium and fixed the 4 remaining external uptakes at arbitrary values (we did not observe a strong dependency of the results on these parameters). Results are shown in Fig. 2. We stress that the medium we consider is not strictly identical to that used in experiments. It is worth noting (see also ref. 20) that experiments employ 13C-labeled carbon sources as substrates for a growing bacterial culture kept at constant density, and that the relative abundance of metabolites can be captured by nuclear magnetic resonance or mass spectroscopy. Reaction fluxes of different metabolic pathways are then inferred, assuming a model of the reaction network.
Fig. 1.
Flux distributions at ρ* for E. coli in a minimal environment with 3 different carbon sources: glutamate (glu), succinate (suc), and glucose (glc). Here, N = 1,053 and P = 630 (glu); N = 1,054 and P = 630 (suc); N = 1,057 and P = 631 (glc). (Inset) The same distributions plotted after a binning of the abscissa.
Fig. 2.
Comparison of reaction fluxes predicted by the Von Neumann scheme with 17 fluxes measured in ref. 19 and analyzed in ref. 6. The different reactions (in no specific order) are reported on the horizontal axis (labeled as in ref. 6), their corresponding fluxes (relative to the glucose uptake) on the vertical axis. Red markers denote experimental values, blue ones theoretical predictions (see, however, Characterization of the Solutions for details on the environmental conditions considered). Note that many flux vectors satisfy Von Neumann's conditions at optimal growth. Three hundred different solutions are reported here.
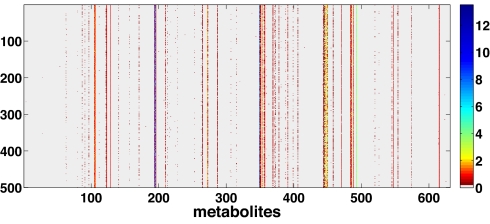
With stationarity recovered at ρ* = 1, as discussed above, the difference between the Von Neumann solution and that of FBA arises from the fact that not all cμs attain their lowest allowed value. This is clearly seen in Fig. 3. In each solution, some metabolites exist with a nonzero cμ, implying that in the steady state, a net production of such metabolites occurs. This may also signal an incompleteness of the stoichiometric data.
Fig. 3.
Values of cμ for different metabolites (horizontal axis) in 500 different solutions. The color code is reported on the right. The white background corresponds to cμ = 0. Colored marks indicate a net production of the corresponding metabolite in that solution or a mass balance violation. One sees that although mass balance holds for most metabolites, some are consistently unbalanced, whereas others may or may not be, depending on the solution.
To get further insight on the existence of multiple flux states compatible with Eq. 2 at ρ* and on the shape of the solution space, a rough but effective way consists in monitoring the mean overlap between different solutions. Given 2 solutions α and β at fixed ρ, we define their overlap qαβ as:
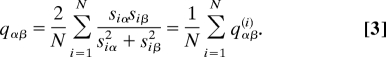 |
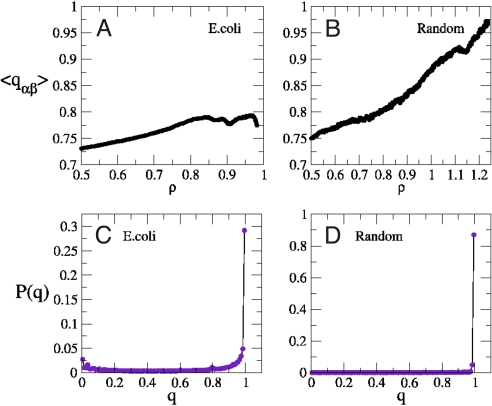
This quantity equals 1 when sα and sβ coincide and becomes smaller and smaller as the distance between sα and sβ in the space of flux vectors increases. In computing it, one should consider a flux to be zero whenever its value is below a threshold ε, to take into account the fact that there is a loss of information about relative fluctuations between different solutions in fluxes smaller than ε. We have chosen ε = 10−5 to ensure that overlaps are not overestimated. Results obtained with ε = 10−5, 10−6, and 10−7 are identical. We have furthermore taken into account the fact that the overlap between null fluxes must be defined by consistency to be equal to 1. In Fig. 4, we report the behavior of the mean overlap 〈qαβ〉 (the angular brackets representing an average over many pairs of solutions) for E. coli and for a network with the same topology and N/P but random Gaussian stoichiometry. Random networks provide an important benchmark against which the behavior of real networks can be tested, to highlight the extent to which observations are specific of the particular organism that is being studied. In particular, the role of topology and stoichiometry can be fully analyzed. For instance, in ref. 17, it is shown that the power-law behavior of the flux distribution cannot be ascribed to the specific network topology.
Fig. 4.
Overlap-based comparison between the solution spaces of E. coli and of its random counterpart. (A and B) Mean overlap among 500 different solutions as function of ρ in E. coli and in a random metabolic network; the last point on the abscissa corresponds to the value of ρ* estimated by simulation. (C and D) Overlap histogram P(q) at ρ* in E. coli and in the random network. Note the different scales of the y axes in the lower images.
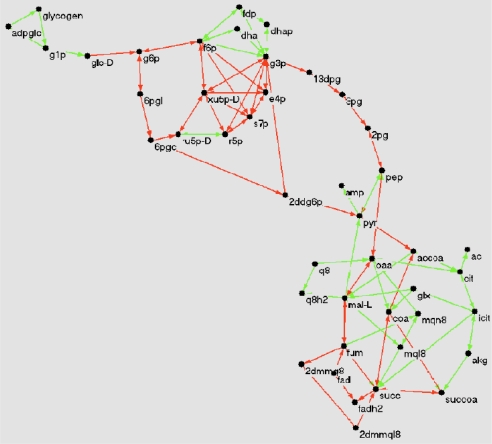
One sees that for the artificial metabolic network, 〈qαβ〉 is an increasing function of ρ that approaches 1 as ρ→ ρ* (Fig. 4B). Moreover, the overlap histogram has a marked δ-peak at q = 1 whose mass increases as ρ → ρ*. These results indicate that the optimal solution is unique. On the other hand, the behavior of 〈qαβ〉 for E. coli suggests that the volume of solutions stops contracting when ρ reaches ≈0.8 from below. In particular, at ρ*, multiple solutions survive. From the corresponding histogram, Fig. 4C, we see that only ≈30% of reactions have an overlap close to 1. In a different jargon, one may say that ≈30% of the variables are frozen (i.e., assume the same value on all solutions of the constrained optimization problem), whereas the remaining are free. The existence of frozen variables characterizes many random constraint satisfaction problems (21), but it is normally hard to identify topological motifs where variables are more likely to be frozen. In the present case, it is reasonable to expect that for purely structural reasons, reaction chains are entirely frozen when the first reaction of the chain is frozen. This is indeed confirmed by the map of frozen/free fluxes in E. coli's central metabolism, displayed in Fig. 5. The chain-like part of glycolysis appears indeed to be frozen.
Fig. 5.
E. coli's central metabolism: Nodes represent metabolites; an arrow joining 2 nodes is present when a reaction exists converting one into the other. Red (green) links denote frozen (free) reactions, with overlap larger (smaller) than 0.9.
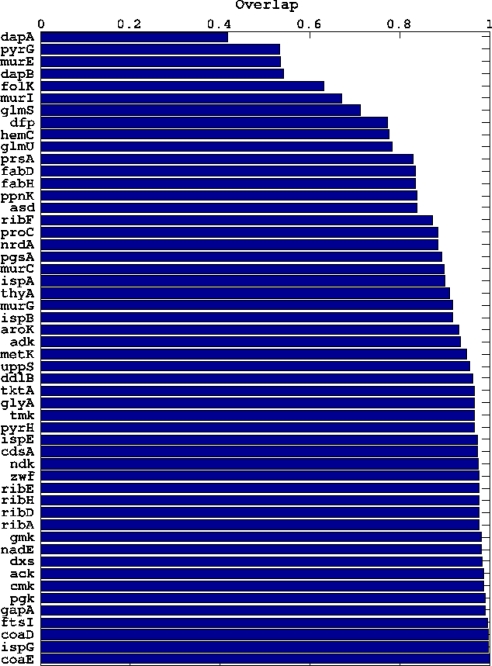
The existence of frozen fluxes raises the obvious question of their biological significance. To address this point, we have correlated reaction overlaps qαβ(i) with the essentiality of the corresponding genes according to the notion of universal essentiality used in ref. 22, which combines phenomenological relevance (a gene is essential if knocking it out causes the cell to die) with evolutionary retention (the presence of the gene in different species). In ref. 22, 55 essential genes of E. coli involved in metabolism have been identified that are also present in 80% of 32 different bacterial genomes. We have been able to link 52 of such genes to reactions in the reconstructed network. It turns out, see Fig. 6, that 43 of such genes correspond to reactions with overlap >0.8 and that only 7 genes relate to reactions with an overlap significantly <80%. Whether a gene is “essential” or not (according to the definition of ref. 22) depends on the choice of the environment. In particular, ref. 22 considers a rich medium as the environment. For comparison, we have considered theoretical values of the overlaps for fluxes calculated assuming a rich environment. Note that, in principle, the same gene can be linked to a more or less variable flux in different environments. However, the results presented are qualitatively preserved in the minimal environment with different carbon sources. This suggests that frozen fluxes, which in Von Neumann's framework are allowed a very limited variability if a state of optimal growth is to be kept, may carry an evolutionary significance.
Fig. 6.
Essential genes (vertical axis) versus overlap of the corresponding reactions in the reconstructed metabolic network of E. coli (horizontal axis).
Role of Conserved Moieties
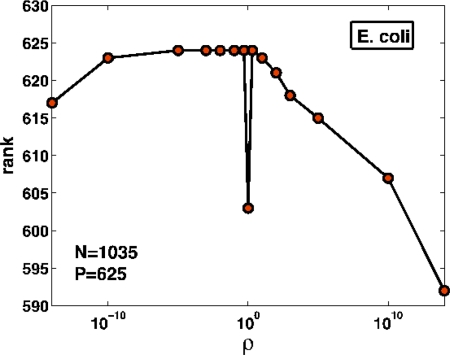
To trace back the physical origin of the results presented above, we have studied the rank of the matrix A − ρB associated to the P linear constraints (Eq. 2) as a function of the parameter ρ, see Fig. 7.
Fig. 7.
The rank of the matrix associated to the system of inequalities (2) for E. coli (isolated cell) as a function of ρ. Around the singularity, the rank equals the number of metabolites; conserved pools of metabolites are present only at exactly ρ = 1.
The singularity occurring at ρ = 1 is related to the presence of conserved pools of metabolites, groups of reagents whose total concentration is constant in time (23). Their existence is due to the fact that the concentrations of metabolites of a certain pool are always coupled with a common functional group. For example the 3 metabolites ATP, AMP, and ADP belong to a pool of cofactors that preserve the adenylate moiety (24). This implies that a change in the concentration of a given metabolite cannot be accomplished without considering the entire pool to which that metabolite belongs.
A conserved pool g is formally defined by a P-dimensional Boolean vector of elements zgμ such that zgμ = 1 if μ ∈ g and zgμ = 0 otherwise satisfying
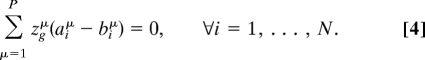 |
Such conservation laws manifest themselves precisely at ρ = 1, By virtue of Eq. 2, the relation Σμ zgμcμ ≥ 0 must hold for any g. It follows that either ρ* ≤ 1, or all fluxes connected to a metabolite belonging to a conserved pool must be equal to zero. Because the null solution s = 0 must be discarded on obvious physical grounds, if all fluxes are connected to metabolites in a conserved pool then necessarily ρ* ≤ 1. This is consistent with the results obtained in different environmental conditions and implies that this scenario must be stable against small perturbations of the network topology.
Moreover, from Eqs. 4 and 2, it is easy to see that, for ρ = 1, μ ∈ g implies cμ = 0, i.e., for a metabolite belonging to a conserved pool the mass-balance condition must be strictly valid. We have shown instead (Fig. 3) that the values of the constraints are not always zero: For some metabolites, the mass balance condition is not reached, and we can assert that they do not belong to any conserved pool (within the stoichiometric description used).
We conclude that in the state of optimal growth with ρ* = 1, in addition to the stationarity of reaction rates, a spontaneous condition of mass balance holds for most, not all, metabolites.
In summary, the Von Neumann model relies on the assumption that the arrangement of metabolic fluxes follows a principle of growth maximization. It does not imply a priori either the mass balance or the stationarity of fluxes, but these 2 conditions are essentially recovered at the maximum growth rate. This is due to the presence of conserved metabolic pools. The singularity in the rank of A − ρB at ρ = 1 implies that the physically relevant solutions are those to which the system tends as ρ → 1. In this limit, mass balance is recovered (i.e., cμ = 0) for most, but not all, metabolites. This provides a nontrivial correction to the picture extracted by FBA and reproduces the limited experimental evidence that is available.
Supplementary Material
Acknowledgments.
We gratefully acknowledge important discussions with G. Bianconi, R. Monasson, A. Pagnani, and F. Ricci Tersenghi and a number of useful suggestions. M.M. was supported by Information Society Technologies STREP GENNETEC contract no. 034952.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813229106/DCSupplemental.
References
- 1.Reed JL, Vo TD, Schilling CH, Palsson BØ. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4:R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiback SJ, Palsson BØ. Extreme pathway analysis of human red blood cell metabolism. Biophys J. 2002;83:808–818. doi: 10.1016/S0006-3495(02)75210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duarte NC, Herrgård MJ, Palsson BØ. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14:1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards JS, Ibarra RU, Palsson BØ. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotech. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 5.Kauffman KJ, Prakash P, Edwards JS. Advances in flux balance analysis. Curr Opin Biotech. 2003;14:491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiback SJ, Famili I, Greenberg HJ, Palsson BØ. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. J Theor Biol. 2004;228:437–447. doi: 10.1016/j.jtbi.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Bianconi G, Zecchina R. Viable flux distribution in metabolic networks. Networks Heterogeneous Media. 2008;3:361–369. [Google Scholar]
- 9.Braunstein A, Mulet R, Pagnani A. Estimating the size of the solution space of metabolic networks. BMC Bioinformatics. 2008;9:240. doi: 10.1186/1471-2105-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabási AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 11.Pal C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 12.Almaas E, Oltvai ZN, Barabási AL. The activity reaction core and plasticity in metabolic networks. PLoS Comput Biol. 2005;1:11e68. doi: 10.1371/journal.pcbi.0010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Neumann J. Über ein ökonomisches Gleichungssystem und eine Verallgemeinerung des Brouwerschen Fixpunktsatzes. Ergebn Math Kolloq. 1937;8:73–83. English translation in Rev Econ Studies 13:1 (1945) [Google Scholar]
- 14.McKenzie LW. Optimal economic growth, turnpike theorems and comparative dynamics. In: Arrow KJ, Intriligator MD, editors. Handbook of Mathematical Economics. Vol 3. Amsterdam: North Holland; 1986. pp. 1281–1355. [Google Scholar]
- 15.Monasson R. Complex Systems LXXXV, Lecture Notes of the Les Houches Summer School 2006. Amsterdam: Elsevier; 2007. Optimization problems and statistical physics; pp. 1–65. [Google Scholar]
- 16.De Martino A, Marsili M. Typical properties of optimal growth in the Von Neumann expanding model for large random economies. J Stat. 2005:L09003. [Google Scholar]
- 17.De Martino A, Martelli C, Monasson R, Perez Castillo I. Von Neumann's expanding model on random graphs. J Stat. 2007:P05012. [Google Scholar]
- 18.Krauth W, Mézard M. Learning algorithms with optimal stability in neural networks. J Phys A. 1987;20:L745–L752. [Google Scholar]
- 19.Emmerling M, et al. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol. 2002;184:152–164. doi: 10.1128/JB.184.1.152-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semerjian G. On the freezing of variables in random constraint satisfaction problems. J Stat Phys. 2008;130:251–293. [Google Scholar]
- 22.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Famili I, Palsson BØ. The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools. Biophys J. 2003;85:16–26. doi: 10.1016/S0006-3495(03)74450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolaev EV, Burgard AP, Maranas CD. Elucidation and structural analysis of conserved pools for genome-scale metabolic reconstructions. Biophys J. 2005;88:37–49. doi: 10.1529/biophysj.104.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.