Abstract
A defining feature of basal-like breast cancer, a breast cancer subtype with poor clinical prognosis, is the high expression of “proliferation signature” genes. We identified B-Myb, a MYB family transcription factor that is often amplified and overexpressed in many tumor types, as being highly expressed in the proliferation signature. However, the roles of B-Myb in disease progression, and its mammary-specific transcriptional targets, are poorly understood. Here, we demonstrated that B-Myb expression is a significant predictor of survival and pathological complete response to neoadjuvant chemotherapy in breast cancer patients. We also identified a significant association between the G/G genotype of a nonsynonymous B-Myb germline variant (rs2070235, S427G) and an increased risk of basal-like breast cancer [OR 2.0, 95% CI (1.1-3.8)]. In immortalized, human mammary epithelial cell lines, but not basal-like tumor lines, cells ectopically expressing wild-type B-Myb or the S427G variant showed increased sensitivity to two DNA topoisomerase IIα inhibitors, but not to other chemotherapeutics. In addition, microarray analyses identified many G2/M genes as being induced in B-Myb overexpressing cells. These results confirm that B-Myb is involved in cell cycle control, and that dysregulation of B-Myb may contribute to increased sensitivity to a specific class of chemotherapeutic agents. These data provide insight into the influence of B-Myb in human breast cancer, which is of potential clinical importance for determining disease risk and for guiding treatment.
Keywords: B-Myb, MYBL2, breast cancer, basal-like
Introduction
Breast cancer is not one disease, but rather, represents at least six subtypes (Perou et al., 2000; Sorlie et al., 2003; Hu et al., 2006). These include luminal A, luminal B, normal-like, HER2-enriched, claudin-low and basal-like, each with unique gene expression profiles and distinct clinical outcomes. The majority of breast cancer cases (60-80 percent) comprise the luminal/estrogen receptor-α positive (ER+) tumors, while basal-like breast carcinoma accounts for 10-15 percent of all cases. The basal-like subtype, often clinically observed as “triple-negative” tumors (negative for ERα, PR, and HER2), is of particular interest because treatment options are limited to chemotherapy only and patients with this disease typically have poor outcomes. One contribution to the poor outcome of basal-like breast cancer patients may be their high cellular proliferation rates, which is manifested transcriptionally as the high expression of the “proliferation signature”; this is a dominant gene signature that is a marker of cell proliferation rates across multiple tumor types (Hu et al., 2006; Whitfield et al., 2006). B-Myb, a gene with known cell cycle control functions and implications in tumorigenesis (Sala, 2005), is one of approximately 100 genes that define the proliferation signature.
B-Myb is a member of the vertebrate MYB family of nuclear transcription factors. In humans this family is comprised of A-Myb (MYBL1), B-Myb (MYBL2), and c-Myb (MYB). Each family member is able to recognize and bind to the same DNA consensus sequence (PyAAC(G/T)G) to promote gene transcription; however, varying tissue-specific expression patterns, as well as protein-protein interactions with unique co-factors, suggests that distinct biological roles exist for each MYB family member (Rosinski & Atchley, 1998; Sala, 2005). Found in the genomes of both plants and animals, MYB proteins are conserved throughout evolution and control processes from flavonoid production to cellular proliferation (Rosinski & Atchley, 1998; Ito et al., 2001). In contrast to vertebrates, invertebrates contain only one MYB protein, which in Drosophila (dMYB) is phylogenetically and functionally complementary to vertebrate B-Myb, suggesting B-Myb to be the most ancient family member (Davidson et al., 2005). The expression of B-Myb, unlike c-Myb and A-Myb (Mucenski et al., 1991; Trauth et al., 1994; Toscani et al., 1997; Ness, 2003; Malaterre et al., 2007), is ubiquitously expressed in virtually all proliferating cells as a regulator of cell cycle progression and plays an essential role in vertebrate development; knocking out murine B-Myb causes early embryonic lethality (E4.5-6.5) resulting from unsuccessful inner cell mass formation (Tanaka et al., 1999).
MYB family members have been implicated in tumorigenesis for several decades. The c-Myb proto-oncogene was first identified as the mammalian homolog of v-myb, which is the transforming gene transmitted by the avian myeloblastosis and E26 retroviruses causing acute leukemia in birds (Klempnauer et al., 1982; Leprince et al., 1983). A- and B-Myb were later discovered during low stringency screening of human cDNA libraries (Nomura et al., 1988). The B-Myb chromosomal locus, 20q13, is amplified and/or highly expressed in a variety of tumor types including breast, prostate, liver and ovarian carcinomas, and in most cases this high expression portends a poor prognosis (Sala, 2005). B-Myb is also an important marker of poor outcome in embryonal tumors of the central nervous system (CNS) (Pomeroy et al., 2002). Recently, a nonsynonymous B-Myb germline variant (rs2070235) causing a serine to glycine amino acid change (S427G) was linked to a decrease in overall cancer risk for neuroblastomas, chronic myelogenous leukemia, and colon cancers in a combined dataset of cases and controls (Schwab et al., 2007). However, the molecular roles of B-Myb in disease progression, as well as its transcriptional target genes in the mammary gland, are still poorly understood. To gain insight into B-Myb and its involvement in breast cancer, we analyzed the expression of B-Myb across the breast cancer subtypes, examined its relationship to survival and pathological complete response and the correlation of variant rs2070235 to disease risk. We also manipulated the expression of B-Myb and the S427G variant in normal and tumor derived mammary cell lines and observed alterations in drug sensitivity and cell cycle profiles.
RESULTS
High B-Myb Expression in Breast Tumors Predicts Poor Outcome
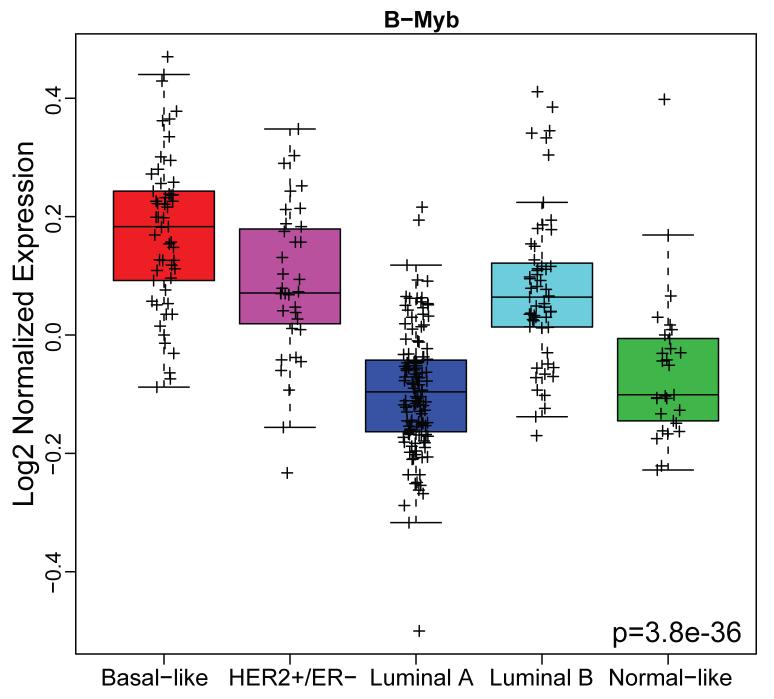
To asses the relevance of B-Myb gene expression across the breast cancer subtypes, breast tumor microarray data from the Netherlands Cancer Institute (NKI-295, n=295, (van de Vijver et al., 2002)) was analyzed. Tumor samples were classified into five breast cancer subtypes using a single sample centroid-based predictor as described (Hu et al., 2006). An ANOVA analysis performed on these stratified samples showed that B-Myb expression differs significantly across the subtypes and was highest in basal-like tumors (Figure 1).
Figure 1. B-Myb expression across breast cancer subtypes.
The NKI breast tumor microarray dataset (n=295) was classified into the five intrinsic subtypes and box plots used to visualize B-Myb expression according to breast cancer subtypes. Statistical significance was calculated by ANOVA.
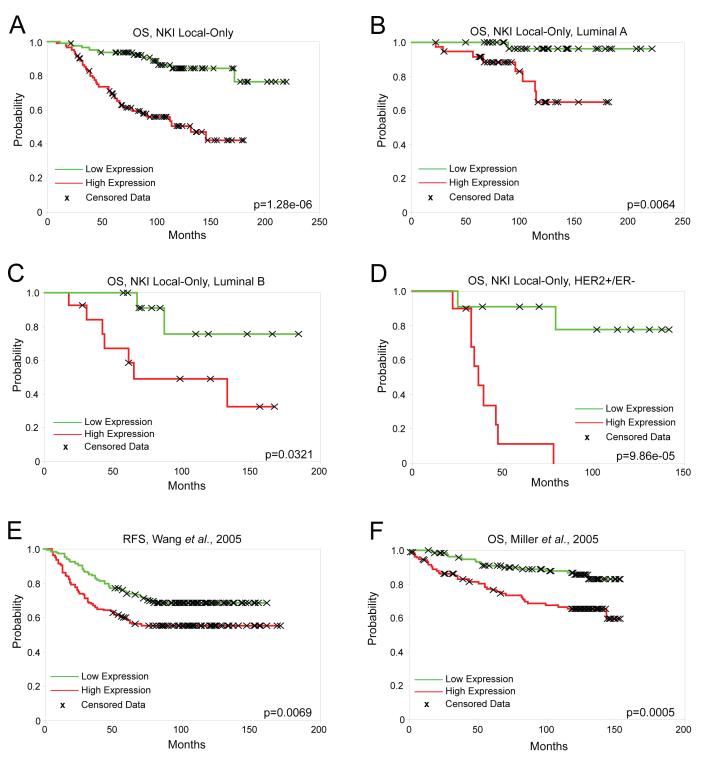
To test for correlations between B-Myb mRNA expression alone and patient outcome, we analyzed the NKI patients not receiving adjuvant systemic treatment (i.e. local treatment only; n=165). This allowed us to better identify the prognostic abilities of B-Myb without the confounding data of treatment response. The NKI “local-only” tumors were rank ordered into halves (low/high) based on their B-Myb expression levels and analyzed for overall survival (OS) and relapse free survival (RFS) by Kaplan-Meier analysis. Poor OS and RFS were highly correlated (p<0.001) with B-Myb high expression levels in these NKI samples (Figure 2A, and RFS data not shown). B-Myb expression alone was also able to significantly predict OS on local-only treated luminal A subtype tumors (n=72) (Figure 2B), luminal B (n=26) (Figure 2C), HER2+/ER− (n=21) (Figure 2D), but not basal-like tumors (n=30) (Supplementary Figure 1A). We then evaluated the prognostic ability of B-Myb using two other published breast tumor microarray datasets (Miller et al., 2005; Wang et al., 2005). Wang et al., 2005 (n=286) consisted of microarrays on untreated, lymph-node-negative primary ER+ and ER-breast cancers with relapse data, and B-Myb was capable of predicting RFS in these patients (Figure 2E). On this same dataset, B-Myb also predicted RFS in the ER+ patient subset (n=209), but not the ER− subset (n=77) (Supplementary Figures 1B, C). Another dataset consisting of primary invasive tumors (Miller et al., 2005) (n=234) was tested and similar results were found (Figure 2F).
Figure 2. High expression of B-Myb correlates with poor outcome.
Kaplan-Meier survival analyses based on B-Myb expression values rank ordered into halves (low/high). (A-D) Overall survival (OS) of locally treated NKI tumor samples: (A) All subtypes combined (n=165), (B) Luminal A (n=72), (C) Luminal B (n=26), (D) HER2+/ER− (n=21). (E) RFS, Wang et al., 2005 (n=286), a locally treated, lymph-node-negative tumor microarray dataset. (F) Miller et al., 2005 breast tumor microarray dataset (n=234).
To determine if B-Myb expression was involved with pathologic complete response (pCR), we used the data of Hess et al., 2006, where microarrays were performed on pre-treatment breast tumors from patients receiving neoadjuvant paclitaxel, followed by 5FU-Adriamycin-Cyclophosphamide (T/FAC; n=133). Again, samples were split into two groups based on B-Myb expression (low/high). B-Myb high expression was again associated subtype (data not shown) and with pCR, as calculated by chi-square test (p=0.008; Supplementary Table 1).
B-Myb Germline Variant (rs2070235) Increases Risk of Basal-Like Breast Cancer
A nonsysnonymous germline B-Myb variant exists that causes a serine to glycine substitution (rs2070235, S427G). This non-conservative change prompted us to look for correlations between this variant and baseline susceptibility risk in the population-based Carolina Breast Cancer Study/CBCS (Newman et al., 1995). Odds ratios for the B-Myb genotype and all breast cancer cases, luminal A, and basal-like cases, versus controls are presented in Table 1. There was no association between B-Myb genotype and all breast cancer (p=0.71); however a statistically significant association was observed for basal-like breast cancer (p=0.047), but not luminal A (p=0.14). No association was observed for B-Myb genotype and the other breast cancer subtypes (luminal B, HER2+/ER− and unclassified: data not shown). Odds ratios were similar in African-Americans and Caucasians (likelihood-ratio tests/LRTs for interaction with race were not statistically significant). Among controls in the CBCS, allele frequencies for the B-Myb G allele were higher in African-Americans (0.27) than Caucasians (0.08).
Table 1. B-Myb variant genotype (G/G) increases the risk of basal-like breast cancer.
Odds ratios for the B-Myb genotype and basal-like, luminal A, and all breast cancer cases. Patient samples were genotyped for the B-Myb polymorphism at codon 427, rs2070235 (A: wild-type; G: variant).
| B-Myb Genotype |
Controls (N=1814) |
Basal-like cases (N=206) |
OR (95% Cl)a | Luminal A cases (N=698) |
OR (95% Cl)a | All cases (N=1256) |
OR (95% Cl)a |
|---|---|---|---|---|---|---|---|
| A/A | 1319 | 131 | Referent | 531 | Referent | 912 | Referent |
| A/G | 436 | 60 | 1.2 (0.8-1.7) | 149 | 0.8 (0.7-1.1) | 301 | 1.O (0.8-1.2) |
| G/G | 59 | 15 | 2.0 (1.1-3.8) | 18 | 0.8 (0.5-1.4) | 43 | 1.O (0.6-1.5) |
| Trend test | p=0.047 | p=0.14 | p=0.71 |
Adjusted for offsets, age, and race
Ectopic Expression of B-Myb Increases Sensitivity to TOP2A Inhibitors
Given B-Myb’s expression within the proliferation signature, evidence suggesting that rapidly growing tumors may be more chemotherapy sensitive, and the correlation between B-Myb expression and pathological complete response, we sought to determine if ectopic expression of B-Myb in vitro had an effect on sensitivity to chemotherapeutics. B-Myb, and the B-Myb S427G variant, were overexpressed in two hTERT-immortalized human mammary epithelial cell lines (HME-CC, ME16C) and two basal-like tumor derived lines (SUM102, SUM149) (Supplementary Figure 2). Low endogenous B-Myb levels were detectable by western blot in the tumor lines, and in all lines by microarray analysis for mRNA levels (Supplementary Figure 2 and data not shown). All cell lines were also genotyped for rs2070235 and identified as homozygous for the major allele. It is important to note that the two normal tissue derived cell lines have a basal-like phenotype when assessed by gene expression analysis (Troester et al., 2004), thus, these lines represent appropriate counterparts to the two basal-like tumor lines.
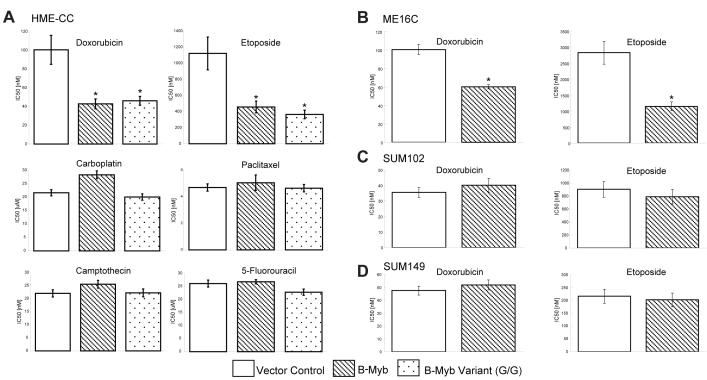
Cells ectopically expressing B-Myb, or S427G variant, were treated with a panel of chemotherapy agents including two DNA topoisomerase IIα (TOP2A) inhibitors (doxorubicin, etoposide), a DNA topoisomerase I inhibitor (camptothecin), a microtubule stabilizer (paclitaxel), a DNA alkylating agent (carboplatin), and an antimetabolite (5-flurouracil), most of which are commonly used in breast cancer treatment. HME-CC cells overexpressing B-Myb, or variant, were approximately twice as sensitive to the two DNA topoisomerase IIα inhibitors, based on IC50 assays, compared to the parental cell lines, but showed no significant change in sensitivity to the other drugs (Figure 3A). To further test B-Myb effects on chemotherapy sensitivity, another immortalized HMEC line (ME16C) and two basal-like tumor derived cell lines (SUM102, SUM149) were tested. The ME16C cell line was also sensitized to TOP2A inhibitors by B-Myb expression, but not to treatment with other chemotherapeutics (Figure 3B and data not shown); however, this sensitivity profile was not observed in either of the two basal-like tumor derived lines (Figures 3C and D), where B-Myb expression had no apparent effect. The B-Myb S427G variant was also tested in each cell line for chemosensitivity and behaved similarly to the cells overexpressing wild-type B-Myb (Figure 3A and data not shown).
Figure 3. Drug sensitivities in B-Myb overexpressing cell lines.
IC50 doses (72h) of chemotherapy on cell lines stably expressing vector control, B-Myb, or B-Myb S427G variant. Each MTT experiment was performed in triplicate and error bars represent 95% confidence intervals (*p<0.001 relative to vector control). (A) hTERT-immortalized HMEC line HME-CC. (B) hTERT-immortalized HMEC line ME16C. (C) Basal-like tumor derived cell line SUM102 and (D) SUM149.
Gene Expression Analysis of Cell Lines Ectopically Expressing B-Myb
To look for gene expression changes in cells overexpressing B-Myb, and to further assess the chemotherapy phenotype, microarrays were performed on the HME-CC cell lines. Under normal, non-confluent conditions, the only statistically significant expression difference (Significance Analysis of Microarrays/SAM analysis (Tusher et al., 2001) with a 3% false discovery rate/FDR) was B-Myb itself; also, no gene expression differences were observed between the B-Myb and B-Myb S427G-expressing cell lines. Since a chemotherapy-related phenotype was observed with TOP2A inhibitors, we tested the cell lines after treatment with the 72 hour IC50 dose of doxorubicin. In a two-class SAM analysis (i.e. doxorubicin-treated HME-CC control vs. doxorubicin-treated HME-CC+B-Myb), 217 genes were identified (FDR <3%; Supplementary Table 2).
An EASE (Expression Analysis Systematic Explorer) analysis (Hosack et al., 2003) was performed and many cell cycle related gene ontology categories were identified as being significantly enriched (Table 2). Therefore, in doxorubicin treated cells overexpressing B-Myb there was significantly higher expression of many cell cycle genes compared to the treated control.
Table 2. Top 15 significant gene ontology categories determined by EASE analysis for B-Myb associated genes.
Significant genes, as determined by SAM, for doxorubicin-treated B-Myb overexpressing HME-CC cells versus doxorubicin-treated controls were input to EASE and analyzed for enriched gene ontology categories.
| Doxorubicin-induced genes: | |||||
|---|---|---|---|---|---|
|
| |||||
| Gene Ontology Category | List Hits | List Total | Population Hits | Population Total | Bonferroni p-value |
| Mitotic cell cycle | 58 | 138 | 352 | 13248 | 2.59E-51 |
| Cell cycle | 65 | 138 | 745 | 13248 | 8.60E-41 |
| M phase | 37 | 138 | 174 | 13248 | 1.22E-34 |
| Nuclear division | 35 | 138 | 167 | 13248 | 2.84E-32 |
| Mitosis | 32 | 138 | 131 | 13248 | 2.31 E-31 |
| M phase of mitotic cell cycle | 32 | 138 | 133 | 13248 | 3.87E-31 |
| Cell proliferation | 65 | 138 | 1116 | 13248 | 4.33E-30 |
| DNA replication and chromosome cycle | 29 | 138 | 199 | 13248 | 2.21 E-21 |
| DNA metabolism | 36 | 138 | 579 | 13248 | 4.60E-15 |
| DNA replication | 20 | 138 | 156 | 13248 | 1.22E-12 |
| S phase of mitotic cell cycle | 20 | 138 | 158 | 13248 | 1.54E-12 |
| Spindle | 17 | 135 | 97 | 12954 | 1.66E-12 |
| Regulation of cell cycle | 28 | 138 | 406 | 13248 | 4.68E-12 |
| Cytokinesis | 16 | 138 | 112 | 13248 | 3.14E-10 |
| Cell growth and/or maintenance | 81 | 138 | 3996 | 13248 | 3.44E-09 |
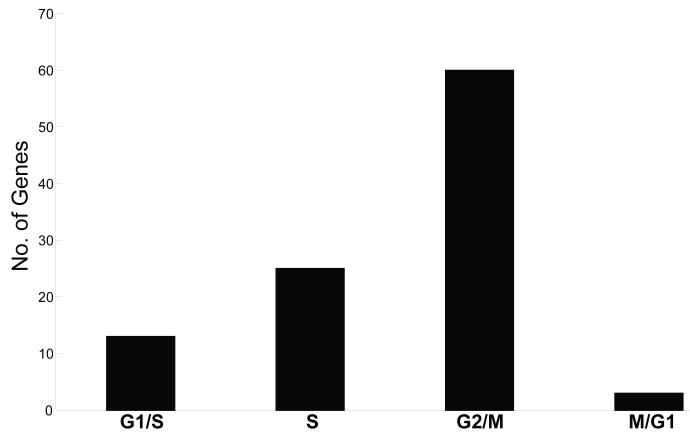
B-Myb is thought to be a transcriptional regulator of G2/M genes (Zhu et al., 2004). To determine if the 217-gene SAM list was enriched for genes within a particular phase of the cell cycle, each gene was assigned to a specific phase by comparing them to a known list derived from a precise cell cycle microarray time course experiment (Whitfield et al., 2002), or by literature search. Out of 217 significant genes, 101 genes were identified as being specifically induced during the cell cycle and 60/101 genes were assigned as G2/M-specific genes (Figure 4). In addition, previous known B-Myb target genes were present on the SAM list including CDC2, Cyclin B1, BIRC5, and the B-Myb binding partner, LIN-9.
Figure 4. Enrichment of G2/M phase genes in doxorubicin-treated B-Myb overexpressing HME-CC cells.
Significance Analysis of Microarray was used to identify 217 significant genes whose high expression was present in B-Myb overexpressing cells. These genes were then assigned to a specific phase of the cell cycle by comparing them to Whitfield et al., 2002, which identified 101/217 genes as being specifically induced during the cell cycle. The graph shows to which phase of the cell cycle these 101 genes mapped.
Cell Cycle Profiles of B-Myb Overexpressing Cells
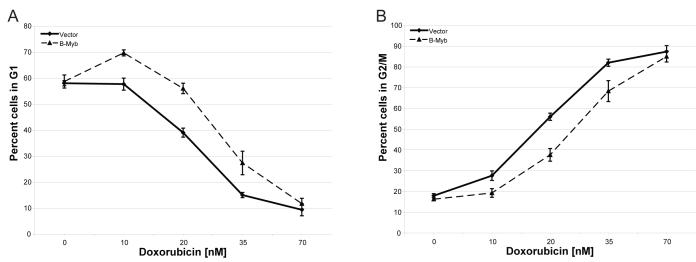
Since B-Myb expressing, doxorubicin treated cells produced a significant G2/M-enriched gene list, we hypothesized that the cell cycle profiles of these cells may be different than treated controls. B-Myb expressing HME-CC and empty vector controls were both treated with a range of doxorubicin doses for 48 hours and their cell cycle profiles analyzed for DNA content using flow cytometry. At zero dose or high dose of doxorubicin, the cell cycle profiles for both B-Myb overexpressing cells and controls were identical in terms of the percentage of cells in G1 and G2/M phase (Figure 5). However, at low and intermediate concentrations (10-35 nM) of doxorubicin there was a significant difference in the number of cells in G1 or G2/M with a larger percentage of B-Myb overexpressing cells in G1 versus controls, and a lower percentage of B-Myb overexpressing cells in G2/M (Figure 5A and 5B, respectively); very similar results were obtained with etoposide treatment (Supplementary Figure 3). At high doses of doxorubicin or etoposide, regardless of B-Myb expression, the majority of cells arrested in G2/M.
Figure 5. Cell cycle profile of HME-CC cells stably expressing B-Myb and treated with doxorubicin.
Cell cultures were treated with a range of doses of doxorubicin for 48 hours followed by propidium iodide DNA content analysis. Percentage of cells in (A) G1 phase and (B) G2/M were calculated by gating based on DNA content. Error bars indicate standard deviations between three independent experiments.
DISCUSSION
Clinically defined as ER-, PR-, and HER2 not amplified, the basal-like subtype of breast cancer portends a poor prognosis. Basal-like breast cancer has an inherently high proliferation rate, which by microarray is identified by high expression of the “proliferation signature” genes; this signature has been identified in many publications, in many tumor types, and is highly enriched for cell cycle regulated genes (Whitfield et al., 2006). Here, we find that B-Myb, a gene highly expressed within the proliferation cluster, plays an important role in regulating cell cycle progression, which likely has effects on patient prognosis and response to chemotherapy.
We showed that B-Myb high expression was significantly associated with the poor outcome, basal-like breast cancer subtype and that B-Myb gene expression levels alone predicted poor outcomes in the absence of therapy (i.e. prognosis, Figure 2) and are correlated with achieving a pCR (i.e. prediction, Supplementary Table 1). Stratification of B-Myb expression, even within the luminal A, B, and HER2+/ER− subtypes, or ER+ tumors, was significantly associated with survival. Stratification of B-Myb expression within basal-like tumors did not predict outcomes, nor was outcome predicted in the ER− subset of the Wang et al., 2005 dataset (which includes basal-like tumors); however, these are inherently poor outcome tumors that significantly trend towards high B-Myb expression (Figure 1).
The B-Myb chromosomal locus, 20q13, is found amplified in a variety of cancers, including breast (Chin et al., 2006), and this amplification is linked to poor prognosis (Bergamaschi et al., 2006). Gene copy number analysis across breast tumor subtypes suggested that the genomic amplification of B-Myb was not enriched in the basal-like subtype, but instead was enriched in the luminal B subtype (Bergamaschi et al., 2006). Therefore, within luminal tumors, B-Myb amplification and expression is an event that appears to be selected for and portends a poor prognosis (Figures 2B, C). In basal-like tumors the high expression of B-Myb may be due to other regulatory mechanisms, possibly by virtue of their inherently high proliferation rates, by amplification of transcription factor(s) targeting B-Myb, or by selectively enhanced promoter activity. This dual behavior of basal-like tumors, a subtype typical of chemo-responsiveness but with poor patient outcomes, has been described before and termed the “basal-like tumor paradox” (Carey et al., 2006), and the data presented here suggest that B-Myb may be a key regulator of this complex phenotype.
Recently, Schwab et al. 2007, linked the nonsynonymous B-Myb germline variant (S427G, rs2070235) to a decrease in overall cancer risk when combining neuroblastomas, chronic myelogenous leukemia, and colon cancers into a single dataset and comparing this grouping of cases to non-cancer bearing controls. While both studies are technically accurate (i.e. similar allele frequencies in the control populations; CBCS: homozygous major allele 72.6%, heterozygous 24%, homozygous minor allele 3.4%; Schwab et al., 2007: 72.1%, 27.3%, and 0.4%, respectively), the association of B-Myb rs2070235 genotype and basal-like breast cancer susceptibility differs from the previous report and has not been described before. Here, in a population based case-control study, we found that the rs2070235 minor allele was associated with increased risk of basal-like breast cancer but not other subtypes in the CBCS (Table 1). The discrepancy between the Schwab data and what is reported here may be due to differences between breast cancer and the cancers examined in the Schwab paper. Even amongst breast cancers there is significant heterogeneity across the subtypes in terms of the etiologic role of the B-Myb allele, and an analysis of breast cancer without subdivision would have missed the association of rs2070235 with the basal-like breast cancer subtype.
Presently, the function of the S427G variant is unknown. Our in vitro studies in breast epithelial cell lines showed neither a phenotypic difference between the overexpression of B-Myb or its variant on chemosensitivity relative to each other, nor did we observe a difference in their baseline gene expression patterns. It is temping to speculate that rs2070235 has functional consequences and somehow contributes to the etiology of basal-like tumors. The B-Myb variant was found to be nearly ten-fold more frequent in African-Americans (7%) versus non-African-Americans (0.8%) in the CBCS. This is relevant in light of recent data showing that premenopausal African-Americans are approximately twice as likely to develop basal-like tumors compared with premenopausal Caucasians (Carey et al., 2006; Millikan et al., 2008). Of note, Schwab et al., 2007 demonstrated that the B-Myb S427G protein was more stable than the wild-type protein. This increased stability may correspond to elevated B-Myb protein levels, possibly increasing transcriptional activity of G2/M cell cycle genes and leading to higher inherent proliferation rates. This alteration of cell cycle control may contribute to B-Myb’s influence on poor outcome breast cancers. Additional assays are required to more fully investigate the role of rs2070235 and other variants in B-Myb that may also lie in linkage disequilibrium with the G allele. This is an important area of investigation, since it is possible that one or more variants in B-Myb could contribute to the higher frequency of basal-like breast cancer among African-American breast cancer patients and may contribute to the pathophysiology of basal-like breast cancer.
Since chemotherapy is currently the only option for basal-like patients, we explored if increased B-Myb expression had any effect on chemosensitivity in vitro. We observed a statistically significant increase in sensitivity to two TOP2A inhibitors, doxorubicin and etoposide, in the HME-CC and ME16C cells overexpressing B-Myb or its variant, but this phenotype was not observed in the basal-like tumor derived cell lines (Figure 3). Also, there was no significant difference in chemosensitivity to the other tested drugs in any of the cell lines, which included camptothecin, a DNA topoisomerase I (TOP1) inhibitor. TOP2A, a nuclear enzyme that relaxes both negative and positive DNA supercoils by creating double-stranded DNA breaks, is of particular importance for proper DNA duplication during S-phase of the cell cycle (Smith et al., 1994). TOP2A inhibitors cause the enzyme to become trapped on double-strand DNA breakpoints, thereby causing G2 cell cycle checkpoint arrest. Since B-Myb is a G2/M regulating gene, increased B-Myb expression may be promoting the TOP2A-inhibitor treated cells through the G2 checkpoint via induction of downstream B-Myb target genes. By facilitating cells through G2, with less regard for DNA damage, the B-Myb overexpressing cells may attempt to cycle again, eventually leading to the increased sensitivity to TOP2A phenotype observed. TOP2A itself was on the B-Myb induced gene list (Supplementary Table 2) and thus more of the target of doxorubicin and etoposide was present, adding to the observed sensitivity phenotype in B-Myb overexpressing cells.
In support of this hypothesis, the gene list identified by microarray analysis as being highly expressed in doxorubicin-treated, B-Myb overexpressing cells (Supplementary Table 2) was significantly enriched in genes required for G2/M progression. For example, this list included the newly described B-Myb interacting protein LIN-9, a protein required along with B-Myb for the transcription of G2/M genes (Osterloh et al., 2007). In addition, our B-Myb target gene list contained many previously identified B-Myb targets including CDC2, Cyclin B1, and BIRC5 (Zhu et al., 2004; Osterloh et al., 2007). This gene list represents putative mammary B-Myb target genes and again suggests that B-Myb has its greatest effect on G2/M genes. In agreement with the gene list enrichment results, when B-Myb overexpressing cells were treated with low doses of doxorubicin or etoposide, more cells accumulated in G1 versus controls, suggesting that the control cell line was appropriately inhibiting cell cycle progression at G2, whereas B-Myb overexpressing lines were bypassing this checkpoint (Figure 5).
The results of this study enhance our understanding of the role of B-Myb in breast cancer by identifying new B-Myb target genes, by showing that this gene is highly expressed in basal-like breast cancers, and by showing it is of prognostic value for survival and predictive value for pathological complete response. Also, we have described a significant correlation between a B-Myb variant and an increased risk of basal-like breast cancer. These findings point to B-Myb as a biomarker that is of potential clinical importance for determining disease risk and for guiding treatment. In addition to its role in basal-like cancers, B-Myb may also be of great importance in luminal tumors, a breast cancer subtype with relatively good prognosis, since B-Myb expression was capable of stratifying the poor from the good actors within this group, and it is within this group where B-Myb is occasionally amplified on the DNA level. The link between B-Myb high expression and increased chemotherapy sensitivity in vitro is mirrored by similar findings in vivo where we and others have shown that basal-like tumors are, on average, the most sensitive to multi-agent chemotherapy regimens that contain an anthracycline (Rouzier et al., 2005; Carey et al., 2007). It is unlikely that B-Myb expression alone is responsible for the chemotherapy sensitivity of basal-like tumors, but when coupled with the loss of TP53 function, which is known to occur in basal-like tumors (Sorlie et al., 2001), and the loss of RB function (Derenzini et al., 2008; Herschkowitz et al., 2008), multiple important checkpoints are deficient in basal-like tumors and it is this lack of control that may ultimately prove to be their Achilles’ heel.
Materials and Methods
Cell Lines
hTERT-immortalized, human mammary epithelial cell lines (HME-CC, ME16C), and basal-like breast cancer-derived lines (SUM102, SUM149) were cultured as described (Troester et al., 2004; Hoadley et al., 2007). Full-length, human B-Myb cDNA (GenBank NM_002466) was cloned into the pBabe.puro.GWrfA (Gateway Reading Frame A) vector using Gateway® Cloning Technology (Invitrogen, Carlsbad, CA, USA). Retrovirus was produced in Phoenix 293T cells by transfecting with ten micrograms of vector using Lipofectamine 2000 (Invitrogen), as per manufacturer’s instructions. Media was changed 24 hours post-transfection and supernatants collected 12 hours later. Seventy-five micrograms of polybrene were added to the collected supernatants and applied to the mammary cell lines. Stable populations were selected by culturing in 1 ug/mL puromycin for HME-CC and ME16C, or 0.5 ug/mL puromycin for SUM102 and SUM149.
B-Myb variant (S427G) was created in the pBabe.puro.B-Myb expression vector using QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene, Los Angeles, CA, USA).
Western Blot Analysis
Cells were grown in 10 cm tissue culture-treated dishes until 80 percent confluence, followed by harvest, protein isolation and quantification as previously described (Troester et al., 2004). Membranes were probed for B-Myb (sc-725; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), and β-actin (AC-15; Abcam, Cambridge, MA, USA), followed by anti-rabbit or anti-mouse IgG horseradish peroxidase-linked whole antibody (Amersham Biosciences) and detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, USA).
Cell Cycle Analysis
Cells were treated with a range of doxorubicin (0-70 nM) or etoposide (0-2 nM) doses and DNA content was analyzed using a modified propidium iodide staining assay. Briefly, one million cells were collected by trypsinization, washed in 1x PBS, and fixed in 70% ethanol at 4C overnight. Cells were washed with PBS/0.2% BSA and resuspended in 500 microliters of PBS/0.2% BSA and 100 micrograms RNaseA. Propidium iodide was added to a final volume of 50 micrograms, and cells incubated at 37C for 30 minutes. DNA content analysis was performed using a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). To determine fractions of cells in G1 and G2/M phases, histograms of DNA content were analyzed using Summit v4.3 software (Dako, Carpinteria, CA, USA) gating around 2N and 4N DNA content. Analyses were done on three separate days and mean values were calculated.
Cytotoxicity Assay
Cell line sensitivities to drugs were assessed using a modified mitochondrial dye conversion assay (Cell-Titer 96, Promega #G4100, Madison, WI, USA) as described (Troester et al., 2004; Hoadley et al., 2007). Chemotherapeutics (carboplatin, doxorubicin, 5-fluorouracil, paclitaxel, etoposide, camptothecin) were purchased from Sigma (St. Louis, MO, USA). The 72-hour inhibitory concentration that caused a 50% reduction in MTT dye conversion (IC50) was determined using nonlinear regression (SAS Statistical Software, Cary, NC, USA) (Vanewijk & Hoekstra, 1993). Differences in the IC50 estimates were tested by a traditional ANOVA test of nested models was performed using the R system for statistical computing (R Development Core Team, 2006 http://www.R-project.org).
Microarray Analysis
Five replicates each of HME-CC B-Myb-overexpressing and vector control cell lines were treated with the 72-hour IC50 dose of doxorubicin. Poly-A(+) RNA was collected (Micro-FastTrack2.0 mRNA Isolation Kit, Invitrogen) from treated control cells and B-Myb overexpressing lines, reverse transcribed and labeled using the Agilent Low RNA Input Linear Amplification Kit (Agilent Technologies, Santa Clara, CA, USA), and hybridized to Agilent Human 44K Custom Oligo microarrays as described (Hu et al., 2005). An untreated HME-CC cell line reference was co-hybridized to all arrays (e.g. untreated HME-CC vs. doxorubicin-treated empty vector HME-CC). Microarrays were scanned on an Axon Genepix 4000B microarray scanner and analyzed using GenePix Pro 5.1 software (Molecular Devices, Sunnyvale, CA, USA). Data was normalized using Lowess normalization on the Cy3 and Cy5 channels. Microarray data is available at the UNC Microarray Database [http://genome.unc.edu] and at the Gene Expression Omnibus (GSE11429).
Microarray Statistical Analyses
Supervised microarray analysis was performed by selecting genes with an absolute signal intensity of at least 30 units in both dye channels and data present in at least 70% of experimental samples. A two-class, unpaired Significance Analysis of Microarrays (SAM) was performed to identify significant genes associated with B-Myb expression with a false discovery rate (FDR) of less than 3% (Tusher et al., 2001).
The Netherlands Cancer Institute breast cancer dataset (NKI-295, n=295) was used for analysis of B-Myb expression across breast cancer subtypes (van de Vijver et al., 2002); however, only locally-treated tumors (no chemotherapy) were used in survival analyses (n=165). Association of B-myb expression states (rank ordered and split in halves: low/high) relative to survival was tested using the Cox-Mantel log-rank test and results visualized using Kaplan-Meier survival plots (WinSTAT v.2007.1). Testing the association of B-myb expression versus subtypes was performed using ANOVA. Three other published datasets were analyzed, as above, for survival (Miller et al., 2005; Wang et al., 2005) or pathological response (Hess et al., 2006) by chi-square using the R system for statistical computing.
B-Myb Genotyping (CBCS)
The Carolina Breast Cancer Study (CBCS) is a population-based, case-control study of breast cancer conducted in 24 counties of central and eastern North Carolina between 1993 and 2001 (Newman et al., 1995; Millikan et al., 2003). B-Myb genotyping (rs2070235) was conducted using DNA extracted from peripheral blood lymphocytes for 1256 cases with subtype information (500 African-American, 756 Caucasian) and 1814 controls (679 African-American, 1135 Caucasian). For complete details see Supplementary Materials.
Supplementary Material
Acknowledgements
We thank Drs. Melissa A. Troester and William K. Kaufmann for comments and discussion. This work was supported by funds from the NCI Breast SPORE program (P50-CA58223-09A1), by RO1-CA-101227-01, by the Breast Cancer Research Foundation and the V Foundation for Cancer Research.
REFERENCES
- Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes and Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The Triple Negative Paradox : Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Davidson CJ, Tirouvanziam R, Herzenberg LA, Lipsick JS. Functional Evolution of the Vertebrate Myb Gene Family: B-Myb, but Neither A-Myb nor c-Myb, Complements Drosophila Myb in Hemocytes. Genetics. 2005;169:215–229. doi: 10.1534/genetics.104.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Donati G, Mazzini G, Montanaro L, Vici M, Ceccarelli C, et al. Loss of Retinoblastoma Tumor Suppressor Protein Makes Human Breast Cancer Cells More Sensitive to Antimetabolite Exposure. Clin Cancer Res. 2008;14:2199–2209. doi: 10.1158/1078-0432.CCR-07-2065. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumor suppressor is a common event in Basal-like and Luminal B breast carcinomas. Breast Cancer Reserach. 2008. [DOI] [PMC free article] [PubMed]
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic Predictor of Sensitivity to Preoperative Chemotherapy With Paclitaxel and Fluorouracil, Doxorubicin, and Cyclophosphamide in Breast Cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- Hoadley K, Weigman V, Fan C, Sawyer L, He X, Troester M, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack GDDA, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4 doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh D, Marron J, He X, Qaqish B, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Troester MA, Perou CM. High reproducibility using sodium hydroxide-stripped long oligonucleotide DNA microarrays. Biotechniques. 2005;38:121–4. doi: 10.2144/05381MT02. [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, et al. G2/M-Phase-Specific Transcription during the Plant Cell Cycle Is Mediated by c-Myb-Like Transcription Factors. Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer K-H, Gonda TJ, Bishop MJ. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell. 1982;31:453–463. doi: 10.1016/0092-8674(82)90138-6. [DOI] [PubMed] [Google Scholar]
- Leprince D, Gegonne A, Coll J, de Taisne C, Schneeberger A, Lagrou C, et al. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306:395. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- Malaterre J, Carpinelli M, Ernst M, Alexander W, Cooke M, Sutton S, et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. PNAS. 2007;104:3829–3834. doi: 10.1073/pnas.0610055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proceedings of the National Academy of Sciences. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan R, Eaton A, Worley K, Biscocho L, Hodgson E, Huang W-Y, et al. HER2 Codon 655 Polymorphism and Risk of Breast Cancer in African Americans and Whites. Breast Cancer Research and Treatment. 2003;79:355. doi: 10.1023/a:1024068525763. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Newman B, Tse C-K, Moorman Patricia G., Conway K, Smith LV, et al. Epidemiology of basal-like breast cancer. Breast Cancer Research and Treatment. 2008;109:123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Ness SA. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells, Molecules, and Diseases. 2003;31:192. doi: 10.1016/s1079-9796(03)00151-7. [DOI] [PubMed] [Google Scholar]
- Newman B, Moorman P, Millikan R, Qaqish B, Geradts J, Aldrich T, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Research and Treatment. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, et al. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucl. Acids Res. 1988;16:11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh L, vonEyss B, Schmit F, Rein L, Hübner D, Samans B, et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. The EMBO Journal. 2007;26:144–157. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular Evolution of the Myb Family of Transcription Factors: Evidence for Polyphyletic Origin. Journal of Molecular Evolution. 1998;46 doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast Cancer Molecular Subtypes Respond Differently to Preoperative Chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. European Journal of Cancer. 2005;41:2479–2484. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Schwab R, Bussolari R, Corvetta D, Chayka O, Santilli G, Kwok JMM, et al. Isolation and functional assessment of common, polymorphic variants of the B-Myb proto-oncogene associated with a reduced cancer risk. Oncogene. 2007. [DOI] [PubMed]
- Smith P, Soues S, Gottlieb T, Falk S, Watson J, Osborne R, et al. Etoposide-induced cell cycle delay and arrest-dependent modulation of DNA toposisomerase II in small-cell lung cancer cells. British Journal of Cancer. 1994;70:914–921. doi: 10.1038/bjc.1994.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is Required for Inner Cell Mass Formation at an Early Stage of Development. J. Biol. Chem. 1999;274:28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, et al. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997;386:713. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- Trauth K, Mutschler B, Jenkins N, Gilbert D, Copeland N, Klempnauer K. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. The EMBO Journal. 1994;13:5994–6005. doi: 10.1002/j.1460-2075.1994.tb06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troester MA, Hoadley KA, Sorlie T, Herbert B-S, Borresen-Dale A-L, Lonning PE, et al. Cell-Type-Specific Responses to Chemotherapeutics in Breast Cancer. Cancer Res. 2004;64:4218–4226. doi: 10.1158/0008-5472.CAN-04-0107. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van ’t Veer LJ, Dai H, Hart AAM, Voskuil DW, et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Vanewijk PH, Hoekstra JA. Calculation of the EC50 and Its Confidence Interval When Subtoxic Stimulus Is Present. Ecotoxicology and Environmental Safety. 1993;25:25. doi: 10.1006/eesa.1993.1003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JGM, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. The Lancet. 2005;365:671. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of Genes Periodically Expressed in the Human Cell Cycle and Their Expression in Tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. The EMBO Journal. 2004;23:4615–26. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.