Abstract
Human tracheobronchial epithelial cells grown in air-liquid interface culture have emerged as a powerful tool for the study of airway biology. In this study, we have investigated whether this culture system produces “mucus” with a protein composition similar to that of in vivo, induced airway secretions. Previous compositional studies of mucous secretions have greatly underrepresented the contribution of mucins, which are major structural components of normal mucus. To overcome this limitation, we have used a mass spectrometry-based approach centered on prior separation of the mucins from the majority of the other proteins. Using this approach, we have compared the protein composition of apical secretions (AS) from well-differentiated primary human tracheobronchial cells grown at air-liquid interface and human tracheobronchial normal induced sputum (IS). A total of 186 proteins were identified, 134 from AS and 136 from IS; 84 proteins were common to both secretions, with host defense proteins being predominant. The epithelial mucins MUC1, MUC4, and MUC16 and the gel-forming mucins MUC5B and MUC5AC were identified in both secretions. Refractometry showed that the gel-forming mucins were the major contributors by mass to both secretions. When the composition of the IS was corrected for proteins that were most likely derived from saliva, serum, and migratory cells, there was considerable similarity between the two secretions, in particular, in the category of host defense proteins, which includes the mucins. This shows that the primary cell culture system is an important model for study of aspects of innate defense of the upper airways related specifically to mucus consisting solely of airway cell products.
Keywords: mucus, mucin, innate immunity, proteomics, human tracheobronchial epithelial cell culture
normal human tracheobronchial epithelium protects the airways by providing a barrier against injury from external insult. In the large airways, this barrier is augmented by a layer of mucus, which is essential for airway integrity and pulmonary defense. Normal induced sputum (IS), a model of mucus, is composed of 1–3% solids by weight, including ions, and is thus ∼97–99% water. This complex extracellular matrix sequesters particulates, toxins, and pathogens, aiding their removal from the airway by mucociliary transport and/or cough clearance (19). The composition of the mucus in the human respiratory tract is determined mainly by the release of products from goblet and ciliated cells of the surface epithelium and from mucus and serous cells of the submucosal glands (15). However, plasma proteins, inflammatory cell products, and salivary proteins also contribute to expectorated mucus. Respiratory mucus is thus a complex secretion, as highlighted by a recent proteomic analysis that identified 191 different proteins in human IS (25). The biological properties of mucus, therefore, result from the combination of these different components. In normal physiology, mucus provides an effective biological and physical barrier to protect the airway surface. However, in diseases such as cystic fibrosis (CF), chronic obstructive pulmonary disease, and asthma, accumulation of mucus results in compromised host defense and generally correlates with morbidity and mortality (for reviews see Refs. 1 and 30). Whether pathology results simply from accumulation of “normal” mucus or from the production of mucus with abnormal composition and properties is not known. In an effort to address these important issues, we and others are employing primary cell cultures derived from normal and diseased human airways, since there are considerable ethical, technical, and financial difficulties in investigating the biochemistry of mucus from humans or large animal models. The development of heterogeneous air-liquid interface cultures of airway epithelial cells (containing ciliated, secretory, and basal cells) has provided a robust model system (12) to study many aspects of airway epithelial biology and pathogenesis, such as enzymatic regulation (3), mucin characterization (14, 41) and secretion (26), signaling (20), inflammatory response (21), CF pathogenesis (22, 23), gene transfer (27), and viral pathogenesis (38, 46).
Studies of sputum have shown that the gel-forming mucins MUC5B and MUC5AC (18, 34), together with other glycoproteins, proteoglycans, host defense proteins, serum proteins, nucleic acids, glycolipids, ions, and water (6, 9, 10, 13, 17, 45), are principal constituents of expectorated mucus. Although the air-liquid interface cultures secrete a “mucus” that can be transported by the ciliated cells (12, 22), it is not known whether the mucus produced accurately reflects the protective properties of airway mucus produced in vivo and, thus, whether it is a valid model for the study of innate defense of the airways. Therefore, as a first step toward addressing this issue, the aim of this study was to analyze the protein composition of mucus produced by human tracheobronchial cells in air-liquid interface cultures and compare this with normal human IS. Special emphasis was placed on development of an approach that would include the mucins, inasmuch as mucins are the major structural components of the mucus matrix and have been dramatically underrepresented in proteomic analyses of mucous secretions (7, 8, 25, 33, 36, 37, 42–44). The reason for this is clearly methodological and correlates with the physical size and composition of these molecules. To this end, we have devised a mass spectrometry (MS)-based approach that relies on prior separation of mucins (epithelial and gel-forming) and the other nonmucin proteins present in the two secretions.
MATERIALS AND METHODS
Cell culture.
Human tracheobronchial epithelial cells were obtained from airways resected from normal donor tissue from the University of North Carolina (UNC) lung transplant program or from the National Disease Research Interchange under UNC Institutional Review Board-approved protocols. Primary airway epithelial cells from normal donors (3 men and 3 women, 32–44 yr of age) with no history of lung disease were isolated by the UNC CF Center Tissue Culture Core, expanded on plastic to generate passage 1 cells, and plated at a density of 6 × 105 cells per well on permeable 24-mm-diameter supports (T-Col, Transwell) (12). Air-liquid interface culture for 4–6 wk formed well-differentiated, polarized human tracheobronchial epithelial cultures that resemble in vivo pseudostratified mucociliary epithelium (27). The conditions for harvesting the culture secretions are described in some detail elsewhere (14). Briefly, we obtained mucous secretions by incubating 1 ml of PBS on the apical surface of the cultures for 30 min at 37°C, removing the PBS with a large-caliber pipette, and repeating the procedure, thus obtaining 2 ml per wash per culture. Such washings, obtained from cultures from six different donors, were pooled, placed immediately on ice, and subsequently centrifuged at 300 g for 10 min to remove cells. To disperse the sample, we added solid guanidine HCl (GuHCl) to bring the solution to 4 M GuHCl. This procedure was repeated on a number of occasions to obtain ≥40 ml of solution. The sample was subjected to isopycnic centrifugation (see below).
Induction and initial processing of sputum.
The sputum induction procedure is described elsewhere (28). Sputum induction with hypertonic saline has proven to be a safe, valid, and reproducible method to induce secretions from the surfaces of the central airways (2, 29). The use of hypertonic saline has proven more effective than isotonic saline in generating sputum, and hypertonicity does not affect sputum cell composition (4), suggesting that the contents of hypertonic saline-derived IS are probably preexisting and not the result of recruitment to the airways by the hypertonic stimulus. Briefly, three 7-min inhaled doses of 3%, 4%, and 5% hypertonic saline were administered to the subjects via ultrasonic nebulizer (Ultraneb 99, DeVilbiss, Sunrise Medical) after baseline spirometry. The subjects breathed at tidal volume without nasal occlusion. At the end of each 7-min inhalation period, subjects performed a three-step cleansing procedure before a cough attempt: each subject 1) rinsed his/her mouth and gargled with water, 2) cleared the back of his/her throat (but did not cough), and 3) blew his/her nose. Subjects then performed a deep cough, and in a passive fashion (i.e., without clearing the back of the throat) brought the sputum sample up into the throat for expectoration. The sample was expectorated into a sterile specimen cup, capped, and placed on ice for the duration of the procedure. All subjects completed the entire procedure and tolerated it without incident. Recruitment of subjects for sputum induction was done under a UNC Institutional Review Board-approved protocol (no. 98-0799). Informed consent was obtained from all subjects before the IS procedure. Normal subjects (4 men and 6 women, 22–34 yr of age) were healthy, nonsmoking volunteers; they had no history of lung disease and were taking no medications. Sputum samples (the entire expectorant) were pooled, diluted with an equal volume of PBS, placed on a roller shaker for 20 min at 4°C, and then centrifuged at 300 g for 20 min to eliminate cells. Finally, an equal volume of 8 M GuHCl was added to the supernatant.
Sample preparation for MS.
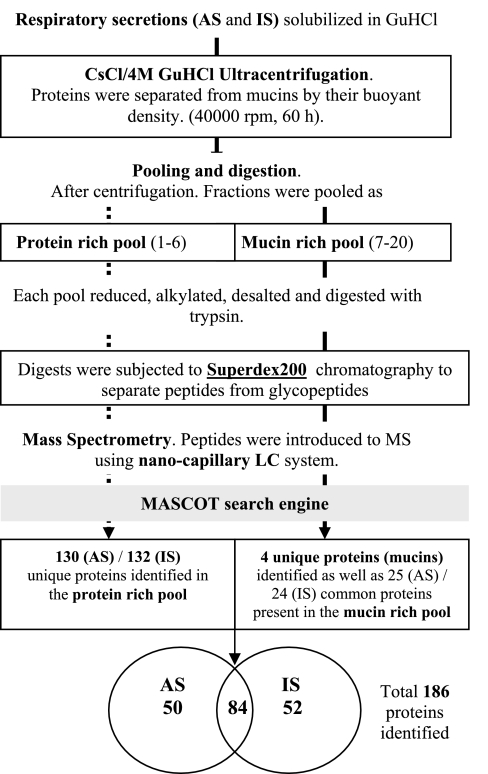
A schematic diagram of the procedure is given in Fig. 1. Collected secretions were brought to 4 M GuHCl by the addition of solid GuHCl, and then CsCl was added to a density of 1.45 g/ml. Isopycnic density-gradient centrifugation was performed for 60 h at 40,000 rpm on a Beckman L8-M ultracentrifuge using a 50.2 TI, 12 × 40 ml rotor. Sample was emptied as 2-ml fractions from the top of the tube, and fractions were then analyzed for mucins (anti-mucin antisera and periodic acid-Schiff), nonmucin proteins (amido black staining), and density. Gradient fractions were pooled to yield protein- and mucin-rich pools, and after reduction of disulfide bonds with 10 mM DTT for 2–3 h at 37°C, free thiols were alkylated with 30 mM iodoacetamide for 1 h in the dark at ambient temperature. Reduced and alkylated samples were subjected to a HiTrap desalting column (Sephadex G-25, Amersham Biosciences) to exchange the buffer to 50 mM NH4HCO3 and then digested with proteomic-grade trypsin (Sigma; 0.5 μg) for 18 h at 37°C. Tryptic digests were chromatographed on a Superdex 200 HR 10/30 column and eluted with 50 mM NH4HCO3 at a flow rate of 0.3 ml/min; the large mucin glycopeptides were separated from the low-molecular-weight peptides by liquid chromatography (LC) using the Ettan LC chromatographic system (Amersham Pharmacia Biotech). The peptide pool was dried down 10 times by volume using a vacuum concentrator (Heto), and then samples were mixed 1:1 with 1% formic acid and subjected to nano-LC-electrospray ionization-tandem MS (MS/MS) analysis.
Fig. 1.
Schematic summary of the approach used in this study. AS, apical secretions; IS, induced sputum; GuHCl, guanidine HCl; MS, mass spectrometry; LC, liquid chromatography.
MS.
Digested samples (2 μl) were introduced into a Waters Q-Tof micro-mass spectrometer via a Waters CapLC system that was configured with a PepMap C18 (LC Packing, 300 μm ID × 5 mm) preconcentration column in series with an Atlantis (Waters) dC18 NanoEase (75 m × 150 mm) nanoscale analytic column. Samples were separated on the column with a gradient of 5% acetonitrile in 0.1% formic acid to 60% acetonitrile in 0.1% formic acid over 45 min. All data were acquired using Masslynx 4.0 software. The MS data-directed analysis acquired MS survey data from mass-to-charge ratio 200–1,500 with the criteria for MS to MS/MS including ion intensity and charge state using a 1-s MS survey scan followed by 1.5-s MS/MS scans, each on three different precursor ions. All analyses were repeated twice for each sample, and peptides identified in the first run were excluded from the second analysis.
Data processing and database searching.
The raw data were processed using the Proteinlynx module of Masslynx 4.0 to produce *.pkl (peaklist) files. The peptide Qa filter was set to 30 to eliminate poor-quality spectra, and the minimum peak width at half-height was set to 4 to eliminate background noise peaks. Smoothing (×2 Savitzky-Golay) and polynomial fitting were performed on all peaks, and the centroid was taken at 80% of the peak height. The processed data were searched against the National Center for Biotechnology Information nonredundant protein database (version 20080123, 5,869,585 sequences) and Swiss-Prot (release 48.7, 190,255 sequences) using the in-house MASCOT (Matrix Science) search engine (version 2.0). Parameters used for the MASCOT search were as follows: taxonomy human, 0.2-Da mass accuracy for parent ions and 0.3-Da accuracy for fragment ions, allowance for one missed cleavage, and use of carbomidomethyl-cysteine and methionine oxidation as fixed and variable modifications, respectively. MASCOT probability-based Mowse individual ion scores >40 were accepted as indicating identity or extensive homology (P < 0.05). MS/MS spectrum scores between 20 and 40 were examined individually with the acceptance criteria as follows: the parent and fragment ion masses were within the calibrated tolerance limits, and the spectrum contained an extended series of consecutive y or b ions. If a peptide was assigned to multiple isoforms or multiple entries of the protein, we specify only the major form of the protein unless a specific peptide points to a region of the protein that exists only in one of the isoforms. Identified proteins were checked for a secretory signal with SignalP (http://www.cbs.dtu.dk/services/SignalP/).
SDS-PAGE analysis.
An aliquot from the low-density protein-rich pool from the AS and IS samples was dialyzed against water and freeze-dried. Samples were resolubilized in 8 M urea reduction buffer [25 mM Tris, 2% SDS (pH 8.0), and 5 mM DTT], boiled for 5 min, and then separated on a precast 4–20% SDS-polyacrylamide gel (Pierce) using an electrophoresis apparatus (Hoefer SE 260, Amersham Biosciences). After electrophoresis, the gel was stained with Coomassie blue stain. Certain bands were excised and digested with trypsin essentially as described previously (35).
Quantitation of protein- and mucin-rich pools by refractive index measurement.
Absolute values of concentration were determined by gel chromatography and differential refractometry (dn/dc): dn/dc of 0.165 ml/g was used for mucins, which we measured previously at 650 nm (23, 39). Briefly, aliquots from the protein- and mucin-rich pools or unfractionated secretions were chromatographed on a Sephadex G25 (2.5 × 2 cm, for total biomolecule measurement) or a Sepharose CL-2B (15 × 2.5 cm, for total mucin measurements) column and eluted with 0.2 M NaCl at a flow rate of 500 μl/min. The column effluent was passed through an in-line enhanced optimal system laser photometer (Dawn, Wyatt Technologies) coupled to a digital signal-processing inferometric refractometer (Wyatt/Optilab) to measure light scattering and sample concentration, respectively. Light scattering and refractive index were measured continuously; light scattering was used only to identify and define the mucin peak. The captured data were integrated and analyzed with the Astra software provided with the Dawn laser photometer. Optimum dilutions were sought by trial and error, and then paired values were obtained to check reproducibility, which was within 5% on average. In general, protein- and mucin-rich pools contain on average ∼650 and 350 μg/ml of total protein (Sephadex G25), respectively, as assessed by refractometry. Typically, unfractionated culture secretions contain on average ∼1,000 μg/ml of total biomolecules (Sephadex G25) and 250–300 μg/ml of total mucin macromolecules (Sepharose CL-2B) as assessed by refractometry.
RESULTS
Other studies on the composition of mucous secretions (induced sputum, saliva, and cervical mucus) have highlighted their complexity. However, the mucins, which are major components of these secretions and key determinants of mucus properties, are not well represented in these analyses (8, 25, 33, 36, 37, 43, 44). Therefore, we sought to gain a more comprehensive analysis of mucin-containing secretions from the apical surface of human airway cells in culture (AS) and sputum induced from the airways of normal individuals (IS). To this end, CsCl/4 M GuHCl density-gradient centrifugation was performed to separate mucins from the other proteins in the AS and IS. A typical density-gradient profile (Fig. 2) shows that the majority of the proteins were present in the low-density fractions (Fig. 2A; assessed by amido black staining) while the gel-forming mucins were present in the higher-density fractions (Fig. 2; assessed by periodic acid-Schiff staining and immunoreactivity with mucin-specific antisera). Fractions were pooled to yield a protein- and a mucin-rich fraction, and the weight of material in each fraction was determined by refractometry after gel filtration chromatography. This analysis showed that, for AS, ∼62% of the total biomolecules are found in the protein-rich fraction and 38% are found in the mucin-rich fraction. Similar values were observed for IS: 65% in the protein-rich fraction and 35% in the mucin-rich fraction. The proteins in the two pools from each sample were digested with trypsin and identified by MS/MS (see materials and methods).
Fig. 2.
Group separation of mucins and proteins by density-gradient centrifugation. After CsCl/4 M GuHCl density-gradient centrifugation (60 h, 40,000 rpm), fractions (2 ml) were emptied from the top and assayed for protein by amido black (○) and carbohydrate by periodic acid-Schiff (PAS, ▴) stain (A) and for the mucins MUC5B (□) and MUC5AC (•) by immunodetection with mucin-specific antisera (B). Dashed lines, density gradient. Fractions were pooled to yield a protein-rich (fractions 1–6) and a mucin-rich (fractions 7–20) pool. Typical relative mass recoveries in 2 pools of the gradient are shown (measured by refractometry).
Analysis of the protein-rich fraction.
SDS-PAGE analysis of IS and AS protein-rich fractions revealed that the major bands in each sample were not the same (Fig. 3). These major bands (Fig. 3A) were excised from the gel, digested with trypsin, and identified by MS/MS (Fig. 3B). This analysis revealed that polymeric IgG receptor, serum albumin, LPLUNC1, actin, and cytokeratin were major proteins common to both pools and consistent with high peptide coverage (Fig. 3B). In contrast, other major proteins, for example, circulating immunoglobulins and leukocyte elastase, were identified only in the IS sample; however, these two proteins are most likely derived from cell types (B cells and neutrophils) that are not represented in the cell cultures. MS/MS of the tryptic peptides derived from the AS and IS protein-rich fractions identified a total of 130 proteins in AS and 132 in IS (Table 1); 80 proteins are common to AS and IS. Table 1 omits 14 known components of saliva (42, 44) and 24 potential inflammatory cell and serum-derived proteins (according to the Swiss-Prot protein database annotation) found in IS (see the list of all the proteins found in the supplemental Table 1 in the online version of this article).
Fig. 3.
Identification of proteins in protein-rich pool by SDS-PAGE in combination with tandem MS (MS/MS). A: aliquots from AS and IS protein-rich pools after density-gradient centrifugation were dialyzed against water, freeze-dried, and solubilized in 8 M urea. Proteins were separated by SDS-PAGE on a 4–20% gradient gel under reducing conditions and visualized by Coomassie blue staining. Positions of molecular mass markers (kDa) are indicated at left. Major bands (1–7 for AS and 1–6 for IS) were digested in-gel with trypsin, and proteins were identified by MS/MS (B). First number in parentheses is number of peptides identified by gel digestion, and second number represents number of peptides identified for the same protein by shotgun analysis. Albumin detected in AS is from a bovine source and is derived from culture medium.
Table 1.
Proteins identified in the protein-rich pool from AS and IS
| Protein Name | SwissProt Accession No. | Protein Name | SwissProt Accession No. | |||
|---|---|---|---|---|---|---|
| Common proteins in protein-rich pool | ||||||
| 14-3-3 protein-ξ/δ | P29312 | Guanine nucleotide-binding protein β-subunit | P04901 | |||
| Actin, cytoplasmic 1 | P02570 | Heat shock 70-kDa protein 1 | P08107 | |||
| Aldehyde dehydrogenase | P43353 | Heat shock protein 90α | P07900 | |||
| α-Enolase, lung specific | Q05524 | Histone H2B.s | P57053 | |||
| α1-Antichymotrypsin | P01011 | Histone H4 | P02304 | |||
| α-Actinin 1 | P12814 | Insulin-like growth factor-binding protein 7 | Q16270 | |||
| Annexin A1 | P04083 | Keratin type I, cytoskeletal 1 | P04264 | |||
| Annexin A2 | P07355 | Keratin type II, cytoskeletal 8 | P05787 | |||
| Annexin A5 | P08758 | Keratin type I, cytoskeletal 19 | P08727 | |||
| Antileukoproteinase 1 | P03973 | Keratin type II, cytoskeletal 5 | P13647 | |||
| β2-Microglobulin | P01884 | Keratin type II, cytoskeletal 6A | P02538 | |||
| β-Microseminoprotein | P08118 | Lactotransferrin | P02788 | |||
| Brain acid-soluble protein 1 | P80723 | LPLUNC1 | Q8TDL5 | |||
| Calcium and integrin-binding protein | Q99828 | Lysozyme | P00695 | |||
| Calcyclin | P06703 | Macrophage migration inhibitory factor | P14174 | |||
| Calcyphosine | Q13938 | Mucin 1 | P15941 | |||
| Calgizzarin | P31949 | Neutrophil defensin 1 | P59665 | |||
| Calgranulin A | P05109 | Neutrophil gelatinase-associated lipocalin | P80188 | |||
| Calgranulin B | P06702 | PLUNC | Q9NP55 | |||
| Calmodulin | P02593 | Peptidyl-prolyl cis-trans isomerase A | P05092 | |||
| Cathepsin D | P07339 | Peroxiredoxin 1 | Q06830 | |||
| CD59 glycoprotein | P13987 | Peroxiredoxin 5 | P30044 | |||
| Ceruloplasmin | P00450 | Phosphoglycerate kinase 1 | P00558 | |||
| Chloride intracellular channel protein 1 | O00299 | Pigment epithelium-derived factor | P36955 | |||
| Clara cell phospholipid-binding protein | P11684 | Polymeric Ig receptor | P01833 | |||
| Clusterin | P10909 | Proactivator polypeptide | P07602 | |||
| Cofilin nonmuscle isoform | P23528 | Prolactin-inducible protein | P12273 | |||
| Complement C3 | P01024 | Prominin 1 | O43490 | |||
| Complement C4 | P01028 | Pyruvate kinase | P14618 | |||
| Complement factor H | P08603 | Rab GDP dissociation inhibitor-β | P50395 | |||
| Cystatin C | P01034 | Serum albumin | P02768 | |||
| Deleted in malignant brain tumors 1 | Q96DU4 | SOD (Cu-Zn) | P00441 | |||
| Dermcidin | P81605 | Tetraspanin 1 | O60635 | |||
| Dipeptidyl-peptidase I | P53634 | Transketolase | P29401 | |||
| Ezrin | P15311 | Tubulin α-chain | Q9BQE3 | |||
| Ezrin-radixin-moesin-binding phosphoprotein 50 | O14745 | Tubulin β-chain | P05217 | |||
| Galectin-3-binding protein | Q08380 | Uteroglobin-related protein 2 | Q96QR1 | |||
| Gelsolin | P06396 | Vimentin | P08670 | |||
| Glutathione S-transferase | P09211 | WAP 4-disulfide core domain protein 2 | Q14508 | |||
| GAPDH | P04406 | Zinc-α2-glycoprotein | P25311 | |||
| Proteins found only in AS | ||||||
| 10-kDa heat shock protein 10790 | P61604 | Guanine nucleotide-binding protein Gyα | P29992 | |||
| Adenylate kinase isozyme 1 | P00568 | Insulin-like growth factor-binding protein 2 | P18065 | |||
| Agrin | O00468 | Insulin-like growth factor-binding protein 3 | P17936 | |||
| Aldose reductase | P15121 | Kallikrein 11 | Q9UBX7 | |||
| Annexin A11 | P50995 | Keratin type I, cytoskeletal 9 | P35527 | |||
| Arylsulfatase A | P15289 | Leucine-rich α2-glycoprotein | P02750 | |||
| β-1,4-Galactosyltransferase 1 | P15291 | Leukocyte elastase inhibitor | P30740 | |||
| β-Hexosaminidase α-chain | P06865 | l-Lactate dehydrogenase A chain | P00338 | |||
| Bile salt-activated lipase | P19835 | Metalloproteinase inhibitor 1 | P01033 | |||
| Carcinoembryonic antigen-related cell adhesion molecule 8 | P31997 | Myosin light chain alkali | P24572 | |||
| Cathepsin B | P07858 | Nucleobindin 2 | P80303 | |||
| Cathepsin H | P09668 | Potassium-transporting ATPase α-chain | P54707 | |||
| Cathepsin S | P25774 | Prostate stem cell antigen | O43653 | |||
| Complement component C6 | P13671 | Protein UNQ6350/PRO21055 | Q7Z5L0 | |||
| Complement factor B | P00751 | Retinoic acid receptor responder protein 1 | P49788 | |||
| Complement factor D | P00746 | S-100P protein | P25815 | |||
| Connective tissue growth factor | P29279 | Selenium-binding protein 1 | Q13228 | |||
| Dipeptidyl-peptidase II | Q9UHL4 | Sodium-dependent phosphate transporter | Q9P0V7 | |||
| Dystroglycan | Q14118 | Squamous cell carcinoma antigen 1 | P29508 | |||
| Ephrin-A1 | P20827 | Syntenin-1 | O00560 | |||
| Epididymal secretory protein E1 | P61916 | Tenascin | P24821 | |||
| Erythrocyte band 7 integral membrane protein | P27105 | Thioredoxin | P10599 | |||
| Fructose bisphosphate aldolase A | P04075 | Transcobalamin I | P20061 | |||
| Galectin-3 | P17931 | Transgelin 2 | P37802 | |||
| Growth/differentiation factor 15 | Q99988 | Triosephosphate isomerase | P00938 | |||
| Proteins found only in IS* | ||||||
| α1-Acid glycoprotein 1 | P02763 | Profilin-1 | P07737 | |||
| Azurocidin | P20160 | Protein FAM3D | Q96BQ1 | |||
| Cornifin A | P35321 | Pulmonary surfactant-associated protein A1 | Q8IWL2 | |||
| Heat shock protein β1 | P04792 | Pulmonary surfactant-associated protein B | P07988 | |||
| Histone H3.1 | P68431 | S100 calcium-binding protein A4 | P26447 | |||
| Keratin type I, cytoskeletal 16 | P08779 | Trefoil factor 3 | Q07654 | |||
AS, human tracheobronchial cell culture apical secretion; IS, induced sputum.
Potential salivary- and plasma-sourced proteins were excluded (see supplemental Table 2).
Analysis of the mucin-rich fraction.
MS/MS analysis of tryptic peptides derived from the mucin-rich fraction identified a total of 29 proteins in AS and 28 in IS (Table 2); as expected, this fraction was dominated by mucins. The same five mucins, namely, MUC1, MUC4, MUC16, MUC5B, and MUC5AC, were identified in AS and IS (Fig. 4A), together with another glycoprotein, DMBT1/gp340, which has been shown to copurify with mucins under these conditions (40) and is also considered to be a mucin (24). Four of these mucins (MUC4, MUC5AC, MUC5B, and MUC16) were unique in this pool; MUC1 and DMBT1 were also present in the protein-rich pool. The nonmucin proteins identified in this fraction were also present in the lower-density, protein-rich pool. To compare the mucin peptide coverage, we have only considered the nonglycosylated regions of the protein. In the AS sample, the gel-forming mucin MUC5B was, from this perspective, the dominant mucin (61 peptides, 45% coverage); lesser amounts of MUC5AC (32 peptides, 21% coverage) and the epithelial mucins MUC1 (5 peptides, 19% coverage), MUC4 (8 peptides, 10% coverage), and MUC16 (21 peptides 11% coverage) were found. In the IS sample, MUC5B was again the major mucin detected (51 peptides, 38% coverage), but MUC5AC appeared to be present at a comparable level (47 peptides, 30% coverage). The epithelial mucins MUC1 (2 peptides, 7% coverage), MUC4 (1 peptide, 1% coverage), and MUC16 (6 peptides, 4% coverage), although identified in IS, contributed fewer peptides and less coverage (of the nonglycosylated regions) than in the AS sample, suggesting that they were present at a lower level in IS. A measure of the enrichment of the mucins against all other proteins may be taken as the ratio of total protein peptide to mucin peptide in the two pools: 72 in pool 1 and 0.8 in pool 2 (AS).
Table 2.
Proteins identified in the mucin-rich pool from AS and IS
| Protein Name | SwissProt Accession No. | |
|---|---|---|
| Common proteins in mucin-rich pool | ||
| Mucin 1 | P15941 | |
| Mucin 4* | Q99102 | |
| Mucin 5AC* | P98088 | |
| Mucin 5B* | Q9HC84 | |
| Mucin 16* | Q96RK2 | |
| Actin, cytoplasmic 1 | P02570 | |
| Antileukoproteinase 1 | P03973 | |
| β2-Microglobulin | P01884 | |
| β-Microseminoprotein | P08118 | |
| CD59 glycoprotein | P13987 | |
| Clara cell phospholipid-binding protein | P11684 | |
| Clusterin | P10909 | |
| Deleted in malignant brain tumors 1 | Q96DU4 | |
| Dermcidin | P81605 | |
| Glutathione S-transferase | P09211 | |
| Heat shock protein 90α | P07900 | |
| Keratin type I, cytoskeletal 1 | P04264 | |
| Keratin type II, cytoskeletal 6A | P02538 | |
| Lactotransferrin | P02788 | |
| LPLUNC1 | Q8TDL5 | |
| Neutrophil defensin 1 | P59665 | |
| PLUNC | Q9NP55 | |
| Polymeric Ig receptor | P01833 | |
| Serum albumin | P02768 | |
| Tetraspanin 1 | O60635 | |
| WAP 4-disulfide core domain protein 2 | Q14508 | |
| Proteins found only in AS | ||
| Complement C3 | P01024 | |
| Insulin-like growth factor-binding protein 2 | P18065 | |
| Leucine-rich α2-glycoprotein | P02750 | |
| Proteins found only in IS** | ||
| Lysozyme | P00695 | |
| Azurocidin | P20160 | |
Unique proteins (mucins) identified in this pool.
Potential salivary- and plasma-sourced proteins were excluded (see supplemental Table 2).
Fig. 4.
Mucins (A) and protein groups (B) in AS and IS. Values in A are averages over 4 independent experiments; error bars represent SD. All peptides from proteins from extraneous sources, e.g., saliva, serum, and lung, were excluded from B (see supplemental Table 2 for excluded proteins).
DISCUSSION
We have compared the protein composition of pooled AS from air-liquid interface cultures of normal human bronchial epithelial cells with IS from normal healthy donors. Such primary cell cultures are commonly employed as a model system for a host of biological studies, from CF pathogenesis to viral infection (3, 14, 20–23, 26, 27, 38, 46), and our particular focus is the innate protective properties of mucus, specifically, the contribution of mucins to the structure of this complex environment. As a consequence, our approach was developed to take into account these major host defense components, which we show (by refractometry) contribute 20–30% of the mass of the total biomolecules in IS and AS from cell culture. This is an important finding, since proteomic analyses of human IS (25), as well as other mucous secretions such as cervicovaginal (33, 36), nasal (8, 37), saliva (42, 44), and airway surface liquid (7), have either not identified mucins or have identified few peptides. For instance, a proteomics study by Candiano et al. (7) on the airway surface liquid of the bronchial epithelial cell culture reports no mucins. The study by Nicholas et al. (25) on sputum reports the presence of MUC5B (13 peptides), MUC5AC (6 peptides), and MUC1 (1 peptide). In contrast, we have identified 98 peptides from MUC5AC and MUC5B from IS (see supplemental Fig. 1 for comparison); 5 of these peptides may have arisen from either MUC5B or MUC5AC, since these mucins share some identical sequence. The reason for the discrepancy is likely to be in the approaches taken to separate the components in the mucous secretions before MS-based identification. Previous studies have typically employed one- and two-dimensional PAGE methods, in which large glycoproteins, such as mucins, would be largely excluded and, therefore, not represented in subsequent MS analyses. However, the approach taken here separates the molecules in solution on the basis of their buoyant density; therefore, they are not excluded from the MS analysis. Thus future proteomic studies on mucus, from whatever tissue, should ensure that the methodology does not exclude these important glycoproteins.
Mucins represent a large proportion of the mass of biomolecules in the cell culture and in vivo secretions, but using the refractometry-based analysis alone, we cannot discriminate between the gel-forming mucins and the large epithelial mucins MUC1, MUC4, and MUC16. However, the MS data of the mucin-rich pool show, on the basis of the number of peptides identified from each mucin, that the gel-forming mucins MUC5AC and MUC5B are more abundant than the epithelial mucins. Furthermore, the cell culture secretion contains more MUC5B than MUC5AC, whereas IS contains similar levels of these two mucins. It is noteworthy that MUC2, which has been shown to be present in trace amounts in sputum (18), was not identified in AS or IS, indicating that this mucin is not a major component of either secretion.
No specific functional association has been identified for MUC5AC and MUC5B in airway mucus, but the MUC5B-rich mucus produced in culture is at least able to be transported by cilia (22, 23). Although no distinct function has been attributed to each mucin type, they are produced in distinct locations within the normal airways: MUC5B is mainly a product of the submucosal glands, and MUC5AC is produced by the surface epithelium (15). Thus our data suggest that the culture may actually have aspects of a ductal/glandular phenotype, rather than simply representing the epithelial surface, where the normal goblet cell mucin is shown to be MUC5AC. In support of this notion, other proteins such as DMBT1 (40) and a number of other protective antibacterial proteins, such as lysozyme (11) and PLUNC and LPLUNC1 (5), found in this study are also products of glandular cells. The presence of known serous cell-derived proteins, such as lysozyme, α1-antichymotrypsin, neutrophil gelatinase-associated lipocalin, and heat shock cognate 71 (16), suggests that the cultures possess a mixed mucous and a serous secretory phenotype.
The contribution of epithelial mucins (MUC1, MUC4, and MUC16) to both secretions is minor, but, interestingly, their contribution (based on the number of peptides identified and coverage of the nonglycosylated regions) to the cell culture secretions is greater, as confirmed by Western blotting after agarose electrophoresis (unpublished data). Why this should be the case is not clear; it might result from the different methods used to collect the two samples. The cell culture sample was collected from the apical surface of the cells after 24–48 h, whereas in vivo mucus elicited by the hypertonic saline treatment was collected within minutes of stimulation. Thus the in vivo secretions may contain a higher level of freshly secreted material; furthermore, the mucus is not confined to a culture dish but can be moved from the local area by mucociliary clearance. Thus the culture sample may contain a higher level of cell surface molecules as a result of epithelial cell turnover or increased proteolysis-induced shedding as a consequence of its inability to be moved from the local area.
Overall, the composition of the in vivo and in vitro secretions is similar. However, there are many differences in the constituent proteins, likely because of the diversity of cell types contributing to the in vivo mucus. Whereas the cell culture mucus is dominated by molecules released from ciliated epithelial cells and mucin-secreting cells (13), IS contains secretions from these cells and extra contributions from cells in the submucosal glands and peripheral lung (e.g., surfactant-related proteins derived from alveolar type II cells). Furthermore, it will also contain proteins from plasma exudate and saliva (see supplemental Table 2). Once the contribution of salivary, immune cell, and blood-related proteins has been taken into account, the similarity of the protein composition of the two secretions becomes increasingly evident. Given that the culture secretions have been shown to form flowing mucus (22), we infer that the cohort of proteins identified is sufficient to make a gel with properties that enable transport by cilia. However, we do not know if it provides an effective biological barrier and how, or even if, these properties differ from those of mucus produced in vivo. One might speculate that the “extra” components in the sputum confer different biological properties to mucus; for example, the role of plasma-derived proteins as complementary factors to epithelium-derived proteins in the physiology of normal mucus is likely to be important. Furthermore, the extra components are likely to make a greater contribution to the secretion in disease, e.g., albumin and fibrinogen in asthmatic sputum (31). Thus, although we acknowledge some limitations of this in vitro model system, the secretions from these cultures, with their absence of proteins from plasma and migratory cells, provide a good starting model to address questions relating to the innate defense of the airway epithelium. In particular, the cultures will provide a unique insight into the role of airway cell-derived proteins, many of which have no proposed function within the context of mucus in airway defense.
Removing the mucins as a separate category, we divided the remainder of the proteins into nine categories according to their proposed general functions by the UniProtKB/Swiss-Prot database (Fig. 4B). The proportion assigned to each category is based on the total number of peptides of all the proteins identified and contributing to that category; although this method is not completely rigorous, it does permit comparison of the two samples. Clearly, proteins in the innate immune category form the major group. In support of this notion, transcript profiling data of air-liquid interface cultures of CF and non-CF airway epithelial cells show that transcripts for host defense proteins, some of which are major proteins identified in our analysis (polymeric immunoglobulin receptor, SPLUNC, and LPLUNC1) are highly abundant (32). From a comparison of all proteins considered to be important for innate immunity, we found that 33 such proteins were common to IS and AS, 41 are present in AS, and 39 are present in IS (putative serum and saliva-derived host defense proteins are not included in these numbers). The preponderance of such proteins in the cell culture secretion suggests that it will be a useful tool for study of innate immunity.
Proteins identified in the culture secretion and the IS were further analyzed to determine whether they were secretory proteins; for this analysis, proteins were checked for signal peptide cleavage sites with the SignalP 3.0 server (Fig. 5). In AS secretion 73 (54%) proteins were found to have signal peptide cleavage sites compared with 70 (52%) in IS. The remainder of the proteins not considered secreted proteins by this analysis comprise cytoplasmic, membrane, lysosomal, Golgi, and cytoskeletal-related components according to the UniProtKB/Swiss-Prot database annotations. A substantial proportion of the cell-derived proteins in the nonsecreted category are cytoskeletal in origin. Of these, proteins such as actin and ezrin are mainly associated with the glycocalyx, especially around the microvilli on ciliated cells. This raises the following questions: 1) How do these proteins arise in the secretion? 2) What is the impact of these proteins on the protective properties of the secretion? Since no major cellular organelle proteins, such as cytochrome c from mitochondria, glucose-6-phosphatase from endoplasmic reticulum, or ribosomal proteins, were detected, it seems possible that these proteins, of epithelial origin, may arise as part of the secretion process from the cells, rather than a generalized disruption process. Whether these proteins are a further part of the protective function of mucus is not clear but warrants further investigation.
Fig. 5.
Analysis and comparison of cellular origin of proteins in AS and IS. Presence of secretory signals was checked by SignalP 3.0 and SecretomeP 2.0 server. Nonsecreted proteins were classified according to UniProtKB/Swiss-Prot database annotations. Potential salivary- and blood-sourced proteins were excluded.
Summary and conclusions.
We have described a compositional comparison of proteins from normal primary human respiratory cell culture washings and IS. These data suggest that the culture is a valid model for study of innate immune protection functions of mucins and mucus (derived wholly from airway cells) on a ciliated epithelium. Except for the mucins, the sputum compositional data presented here are in good agreement with data from other compositional studies (23) and provide a set of reference data for comparison of secretory proteins from pathological states of the lung, such as chronic obstructive pulmonary disease, asthma, CF, and lung cancer, in culture and sputum. This study was motivated by two questions: 1) Is the protein and mucin composition of the cell culture secretion similar to that of in vivo mucus? If we exclude the proteins that could not originate in the airway epithelia, there is a high level of identity (90%) of proteins; thus we take the answer to be yes. 2) Is it a valid model to investigate aspects of innate defense of the airways? The percentage of shared proteins associated with innate immunity (87%) also indicates a positive answer.
GRANTS
This work was partially supported by a gift from an anonymous donor for research targeted to proteomics of CF lung disease (J. K. Sheehan), National Heart, Lung, and Blood Institute Grants HL-080098, HL-066973, and HL-084934-02, and a grant from the Wellcome Trust (D. J. Thornton).
Supplementary Material
Acknowledgments
The authors thank the directors and teams of the UNC CF Center Tissue Culture Core and the Morphology and Morphometry Core for supplying reagents and technical expertise. The authors also thank Drs. Richard Boucher, William Davis, and Scott Randell for useful discussion.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Adler KB, Li Y. Airway epithelium and mucus: intracellular signaling pathways for gene expression and secretion. Am J Respir Cell Mol Biol 25: 397–400, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Alexis NE, Soukup J, Nierkens S, Becker S. Association between airway hyperreactivity and bronchial macrophage dysfunction in individuals with mild asthma. Am J Physiol Lung Cell Mol Physiol 280: L369–L375, 2001. [DOI] [PubMed] [Google Scholar]
- 3.Atherton HC, Jones G, Danahay H. IL-13-induced changes in the goblet cell density of human bronchial epithelial cell cultures: MAP kinase and phosphatidylinositol 3-kinase regulation. Am J Physiol Lung Cell Mol Physiol 285: L730–L739, 2003. [DOI] [PubMed] [Google Scholar]
- 4.Bacci E, Cianchetti S, Paggiaro PL, Carnevali S, Bancalari L, Dente FL, Di Franco A, Giannini D, Vagaggini B, Giuntini C. Comparison between hypertonic and isotonic saline-induced sputum in the evaluation of airway inflammation in subjects with moderate asthma. Clin Exp Allergy 26: 1395–1400, 1996. [PubMed] [Google Scholar]
- 5.Barnes FA, Bingle L, Bingle CD. Pulmonary genomics, proteomics, PLUNCs. Am J Respir Cell Mol Biol 38: 377–379, 2008. [DOI] [PubMed] [Google Scholar]
- 6.Boat TF, Cheng PW. Biochemistry of airway mucus secretions. Fed Proc 39: 3067–3074, 1980. [PubMed] [Google Scholar]
- 7.Candiano G, Bruschi M, Pedemonte N, Musante L, Ravazzolo R, Liberatori S, Bini L, Galietta LJ, Zegarra-Moran O. Proteomic analysis of the airway surface liquid: modulation by proinflammatory cytokines. Am J Physiol Lung Cell Mol Physiol 292: L185–L198, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Casado B, Pannell LK, Iadarola P, Baraniuk JN. Identification of human nasal mucous proteins using proteomics. Proteomics 5: 2949–2959, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Cavaliere F, Masieri S, Vagnoni S, Proietti R, Magalini SI. Airway secretion electrolytes: reflection of water and salt states of the body. Crit Care Med 17: 891–894, 1989. [PubMed] [Google Scholar]
- 10.Coles SJ, Bhaskar KR, O'Sullivan DD, Neill KH, Reid LM. Airway mucus: composition and regulation of its secretion by neuropeptides in vitro. Ciba Found Symp 109: 40–60, 1984. [DOI] [PubMed] [Google Scholar]
- 11.Dajani R, Zhang Y, Taft PJ, Travis SM, Starner TD, Olsen A, Zabner J, Welsh MJ, Engelhardt JF. Lysozyme secretion by submucosal glands protects the airway from bacterial infection. Am J Respir Cell Mol Biol 32: 548–552, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107: 183–206, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Gerson C, Sabater J, Scuri M, Torbati A, Coffey R, Abraham JW, Lauredo I, Forteza R, Wanner A, Salathe M, Abraham WM, Conner GE. The lactoperoxidase system functions in bacterial clearance of airways. Am J Respir Cell Mol Biol 22: 665–671, 2000. [DOI] [PubMed] [Google Scholar]
- 14.Holmen JM, Karlsson NG, Abdullah LH, Randell SH, Sheehan JK, Hansson GC, Davis CW. Mucins and their O-glycans from human bronchial epithelial cell cultures. Am J Physiol Lung Cell Mol Physiol 287: L824–L834, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Hovenberg HW, Davies JR, Carlstedt I. Different mucins are produced by the surface epithelium and the submucosa in human trachea: identification of MUC5AC as a major mucin from the goblet cells. Biochem J 318: 319–324, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joo NS, Lee DJ, Winges KM, Rustagi A, Wine JJ. Regulation of antiprotease and antimicrobial protein secretion by airway submucosal gland serous cells. J Biol Chem 279: 38854–38860, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Kaliner M, Shelhamer JH, Borson B, Nadel J, Patow C, Marom Z. Human respiratory mucus. Am Rev Respir Dis 134: 612–621, 1986. [DOI] [PubMed] [Google Scholar]
- 18.Kirkham S, Sheehan JK, Knight D, Richardson PS, Thornton DJ. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J 361: 537–546, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest 109: 571–577, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krunkosky TM, Fischer BM, Martin LD, Jones N, Akley NJ, Adler KB. Effects of TNF-α on expression of ICAM-1 in human airway epithelial cells in vitro. Signaling pathways controlling surface and gene expression. Am J Respir Cell Mol Biol 22: 685–692, 2000. [DOI] [PubMed] [Google Scholar]
- 21.Marano F, Boland S, Bonvallot V, Baulig A, Baeza-Squiban A. Human airway epithelial cells in culture for studying the molecular mechanisms of the inflammatory response triggered by diesel exhaust particles. Cell Biol Toxicol 18: 315–320, 2002. [DOI] [PubMed] [Google Scholar]
- 22.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, Boucher RC. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell 95: 1005–1015, 1998. [DOI] [PubMed] [Google Scholar]
- 23.Matsui H, Verghese MW, Kesimer M, Schwab UE, Randell SH, Sheehan JK, Grubb BR, Boucher RC. Reduced three-dimensional motility in dehydrated airway mucus prevents neutrophil capture and killing bacteria on airway epithelial surfaces. J Immunol 175: 1090–1099, 2005. [DOI] [PubMed] [Google Scholar]
- 24.Mollenhauer J, Herbertz S, Helmke B, Kollender G, Krebs I, Madsen J, Holmskov U, Sorger K, Schmitt L, Wiemann S, Otto HF, Grone HJ, Poustka A. Deleted in malignant brain tumors 1 is a versatile mucin-like molecule likely to play a differential role in digestive tract cancer. Cancer Res 61: 8880–8886, 2001. [PubMed] [Google Scholar]
- 25.Nicholas B, Skipp P, Mould R, Rennard S, Davies DE, O'Connor CD, Djukanovic R. Shotgun proteomic analysis of human induced sputum. Proteomics 6: 4390–4401, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Park J, Fang S, Crews AL, Lin KW, Adler KB. MARCKS regulation of mucin secretion by airway epithelium in vitro: interaction with chaperones. Am J Respir Cell Mol Biol 39: 68–76, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickles RJ, McCarty D, Matsui H, Hart PJ, Randell SH, Boucher RC. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol 72: 6014–6023, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pin I, Gibson PG, Kolendowicz R, Girgis-Gabardo A, Denburg JA, Hargreave FE, Dolovich J. Use of induced sputum cell counts to investigate airway inflammation in asthma. Thorax 47: 25–29, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popov TA, Pizzichini MM, Pizzichini E, Kolendowicz R, Punthakee Z, Dolovich J, Hargreave FE. Some technical factors influencing the induction of sputum for cell analysis. Eur Respir J 8: 559–565, 1995. [PubMed] [Google Scholar]
- 30.Randell SH, Boucher RC. Effective mucus clearance is essential for respiratory health. Am J Respir Cell Mol Biol 35: 20–28, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers DF Airway mucus hypersecretion in asthma: an undervalued pathology? Curr Opin Pharmacol 4: 241–250, 2004. [DOI] [PubMed] [Google Scholar]
- 32.Scheetz TE, Zabner J, Welsh MJ, Coco J, Eyestone M, Bonaldo M, Kucaba T, Casavant TL, Soares MB, McCray PB Jr. Large-scale gene discovery in human airway epithelia reveals novel transcripts. Physiol Genomics 17: 69–77, 2004. [DOI] [PubMed] [Google Scholar]
- 33.Shaw JL, Smith CR, Diamandis EP. Proteomic analysis of human cervico-vaginal fluid. J Proteome Res 6: 2859–2865, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Sheehan JK, Thornton DJ. Heterogeneity and size distribution of gel-forming mucins. Methods Mol Biol 125: 87–96, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858, 1996. [DOI] [PubMed] [Google Scholar]
- 36.Tang LJ, De Seta F, Odreman F, Venge P, Piva C, Guaschino S, Garcia RC. Proteomic analysis of human cervical-vaginal fluids. J Proteome Res 6: 2874–2883, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Tewfik MA, Latterich M, DiFalco MR, Samaha M. Proteomics of nasal mucus in chronic rhinosinusitis. Am J Rhinol 21: 680–685, 2007. [DOI] [PubMed] [Google Scholar]
- 38.Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza A virus. J Virol 80: 8060–8068, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornton DJ, Carlstedt I, Howard M, Devine PL, Price MR, Sheehan JK. Respiratory mucins: identification of core proteins and glycoforms. Biochem J 316: 967–975, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornton DJ, Davies JR, Kirkham S, Gautrey A, Khan N, Richardson PS, Sheehan JK. Identification of a nonmucin glycoprotein (gp-340) from a purified respiratory mucin preparation: evidence for an association involving the MUC5B mucin. Glycobiology 11: 969–977, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Thornton DJ, Gray T, Nettesheim P, Howard M, Koo JS, Sheehan JK. Characterization of mucins from cultured normal human tracheobronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 278: L1118–L1128, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Identification of human whole saliva protein components using proteomics. Proteomics 4: 1109–1115, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Wattiez R, Falmagne P. Proteomics of bronchoalveolar lavage fluid. J Chromatogr B Analyt Technol Biomed Life Sci 815: 169–178, 2005. [DOI] [PubMed] [Google Scholar]
- 44.Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol Cell Proteomics 4: 1826–1830, 2005. [DOI] [PubMed] [Google Scholar]
- 45.Yeager H Tracheobronchial secretions. Am J Med 50: 493–509, 1971. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 76: 5654–5666, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.