Abstract
ATP-binding cassette (ABC) transporters make up one of the largest classes of proteins found in nature, and their ability to move a variety of substrates across the membrane using energy from the binding or hydrolysis of ATP is essential to an array of human pathologies and to bacterial viability. MsbA is an essential ABC transporter that specifically transports lipid A across the inner membranes of Gram-negative organisms such as Escherichia coli. The exact mechanisms of function during the binding and hydrolysis of ATP at the molecular level remain unclear. The studies presented and summarized in this work directly address the role and local dynamics of specific residues within the conserved ABC motifs in E. coli MsbA using in vivo growth and biochemical activity assays coupled with site-directed spin labeling electron paramagnetic resonance (EPR) spectroscopy motional and accessibility analysis. This first comprehensive analysis of the specific residues in these motifs within MsbA indicates that closure of the dimer interface does not occur upon ATP binding in this transporter.
ATP-binding cassette (ABC)1 transporters constitute one of the largest and most important families of integral membrane proteins. They specifically contribute to drug resistance by using ATP hydrolysis to export delivered drugs back across the cell membrane, and the function or dysfunction of other known ABC transporters results in serious genetic disorders such as cystic fibrosis (1). ABC transporters are typically comprised of two membrane-spanning domains and two intracellular ATP binding domains [or nucleotide binding domains (NBDs)].
MsbA is an ABC transporter that functions as a homodimer with a molecular mass of 130 kDa and is found in the inner membranes of Gram-negative bacteria such as Escherichia coli, Salmonella typhimurium, Vibrio cholera, and Pseudomonas aeruginosa (2). Its role is to transport the negatively charged lipid A across the hydrophobic bacterial inner membrane. Because lipid A is the major component of the outer leaflet of the outer membrane of Gram-negative bacteria, its synthesis and transport are essential for cell growth. Thus, the functional loss of MsbA from the bacterium results in a toxic accumulation of lipid A within the inner membrane (3), making MsbA the only ABC transporter currently know to be required for bacterial viability (2).
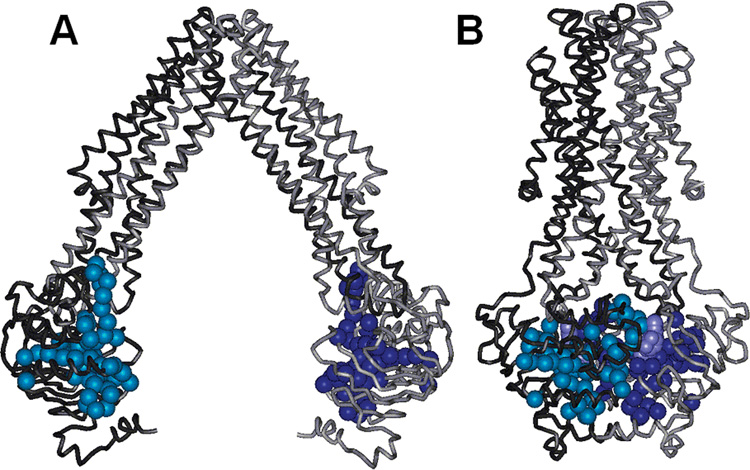
A reanalyzed crystal structure of the E. coli MsbA homodimer is now available at a resolution of 5.3 Å, along with the structures of MsbA from S. typhimurium and V. cholera and homologues such as SAV1866 (4–6). These structures reveal that the MsbA monomer contains a trans-membrane six-helix bundle linked to a soluble nucleotide binding domain (NBD), where ATP is bound and hydrolyzed to transport lipid A across the membrane (see Figure 1). ABC transporters share at least five conserved regions that identify them as ATPases: Walker A and Walker B sequences (7), a C-loop, which is a consensus sequence that contains the ABC transporter signature sequence LSGGQ, and two individual residues termed the H-motif and the Q-loop. Additionally, the conserved sequences known as the P-, D-, and A-loops have been identified as being characteristic of ABC transporter NBDs (8, 9). MsbA contains all of these conserved primary sequences (2), which identify it as an ABC transporter. The Walker A, Walker B, Q-loop, and A-loop sites within one monomer and the C-loop from the opposing monomer are proposed to make contact with ATP on the basis of the crystal structures of ABC transporters (e.g., refs 10–13).
FIGURE 1.
Crystal structures of the MsbA homodimer from (A) E. coli in the open state and (B) S. typhimurium in the ADP/Vi-bound closed state (4). The NBD sites studied here are colored light blue and dark blue on opposite monomers, and the nucleotide is colored purple.
The site-directed spin labeling (SDSL) electron paramagnetic resonance (EPR) spectroscopy technique is able to provide localized information about the environment and location of an individual residue within a protein structure(14–19). This technique allows the direct probing of the local environment, structure, and proximity of individual residues. EPR is not limited by macromolecular size or the optical properties of a sample, making it especially applicable to the investigation of membrane proteins, such as ABC transporters, that are typically difficult to study by other spectroscopic methods yet comprise an important and large class of biologically relevant structures.
In the studies reported here, we have used SDSL EPR analysis coupled with extensive in vivo activity analysis and in vitro kinetic data to systematically analyze the ATP binding and hydrolysis steps for the bacterial ABC transporter MsbA. The studies and conclusions presented directly address the local dynamics and involvement of specific residues within each of the conserved ABC motifs in E. coli MsbA and provide the first extensive collection of functional activity data on the whole array of conserved NBD motif mutants in any ABC transporter.
EXPERIMENTAL PROCEDURES
Site-Directed Mutagenesis and Protein Preparation
Site-directed cysteine mutants were introduced into the C-less (C88S/C315S) MsbA gene in a pET28 (Novagen) plasmid using the QuikChange mutagenesis kit (Stratagene) and verified by sequencing at the Medical College of Wisconsin Protein and Nucleic Acid Facility, as previously described (20). MsbA protein was expressed in E. coli NovaBlue cells (Novagen) and purified by cobalt affinity chromatography using Talon resin (BD Biosciences Clontech) as described previously (20). The locations of each of the sites studied are identified in Figure 1.
Purified MsbA cysteine mutants were labeled with a 10-fold molar excess of the sulfhydryl-specific MTSL (2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate spin label) reagent overnight at 4 °C. Unreacted label was removed by extensive dialysis against 50 mM sodium phosphate (pH 7) and 0.01% DM buffer, and protein concentrations were determined by the detergent-compatible BCA protein assay (Pierce) after concentration in Amicon Ultra centrifugal filters. Spin labeled MsbA was reconstituted into inner membrane liposomes [65:25:10 phosphatidylethanolamine (PE):phosphatidylglycerol (PG):cardiolipin (CL) ratio (Avanti Polar Lipids)] at a 250:1 lipid:protein molar ratio, as described previously (21).
In Vivo Growth Assay
Plasmid-containing WT or mutant MsbA was electroporated into electrocompetent WD2 cells [a kind gift from C. Raetz, Duke University (Durham, NC), and W. Doerrler, Louisiana State University (Baton Rouge, LA)] carrying the A270T mutation in the chromosomal copy of MsbA and allowed to incubate at 30 °C before being plated onto tetracycline/kanamycin LB agar plates and incubated overnight at 30 °C. One colony from each plate was grown in a LB/tetracycline/kanamycin mixture at 30 °C until the OD600 reached 0.3–0.5, at which time it was diluted to an OD of 0.03 and added to a 96-well plate; 200 µL of cells was added to each well and grown in triplicate. Plates were monitored for 90 min at 30 °C and then for 10 h at 44 °C in a Thermo Varioskan Flash microplate reader. WT and C-less MsbA, and empty pET28 plasmid, were run as positive and negative controls, respectively. Cells containing plasmid-encoded MsbA with normal function typically grew to an OD600 of greater than ~0.5, while cells containing no MsbA or completely inactive MsbA grew to an OD600 of approximately ~0.3 or less. Each mutant was tested in triplicate per run and also at least twice in separate runs with fresh WD2 cells.
ATPase Assay
MsbA (8.125 µg) in the presence of a 250-fold molar excess of inner membrane lipids containing 1 mol % lipid A (Avanti Polar Lipids) was added to 50 mM HEPES (pH 7), 1 mM dithiothreitol, 7.5% DM, 10 mM MgCl2, and 3 mM ATP buffer containing [γ-32P]ATP (Perkin-Elmer) at 37 °C and the hydrolysis reaction stopped at various time points (typically 0.1–2 min) by addition of perchloric acid and placement of the mixture on ice. Equal volumes of ammonium molybdate and water-saturated isobutanol were added to the reactions, and 200 µL of the 450 µL organic phase was removed for counting. ATP hydrolysis was assessed in triplicate by quantitating the release of γ-32Pi using Cherenkov counting in a Tri-Carb liquid scintillation counter (Perkin-Elmer). Serial dilutions of the [γ-32P]ATP-containing 3 mM stock solution of MgATP were used as standards. Rates of hydrolysis were determined by plotting the nanomoles of Pi released per milligram of protein versus time. As spin labeled controls, V426C and S482C proteins exhibited very similar rates of hydrolysis (within 5%) when compared to unlabeled protein, verifying the spin label does not interfere with ATPase activity at these example sites.
Site-Directed Spin Labeling EPR Spectroscopy
CW EPR spectroscopy was carried out on a Bruker ElexSys 500 X-band spectrometer equipped with a super high Q (SHQ) cavity (Bruker Biospin). Spectra were recorded over a 100 G scan width, with a 100 kHz field modulation of 1 G and under nonsaturating conditions. Samples were typically 10 µL in volume and were contained in a glass capillary. The concentration of the protein reconstituted into liposomes was approximately 200 µM in 20 mM MOPS (pH 7) containing 20 mM ATP, 2 mM EDTA, 20 mM MgCl2, and 2 mM sodium orthovanadate, as appropriate.
RESULTS
In Vivo Activities of Conserved NBD Sites in MsbA
The in vivo growth assay adapted from Doerrler and Raetz (22, 23) for MsbA NBD mutants yields information about whether both the transport and hydrolysis processes are intact and efficient enough to maintain the viability of living cells. This is accomplished by taking advantage of the fact that MsbA is essential to cell survival and that the chromosomal copy of MsbA has been altered so that it is inactivated at high temperatures (>42 °C), allowing the activity of the plasmid-encoded MsbA mutants to be directly assayed. The results of assaying 50 single-cysteine mutants within key regions of the NBD of MsbA are illustrated in Figure 2. Representative growth curves for the Walker B and control proteins are plotted in Figure 3, indicating the range of growth differences over time. Mutants can be readily categorized into those with weak or no activity (OD < 0.3), intermediate activity, and full activity (OD > 0.5).
FIGURE 2.
In vivo growth assay results for 50 single-cysteine mutants within key regions of the NBD of MsbA. WT and C-less MsbA are positive controls, and an empty pET28 plasmid without the MsbA gene serves as a negative control (black bars). Dark gray bars identify 26 sites within known conserved ABC transporter motifs, as identified, and light gray bars correspond to neighboring sites. Error bars represent the standard deviation from the mean.
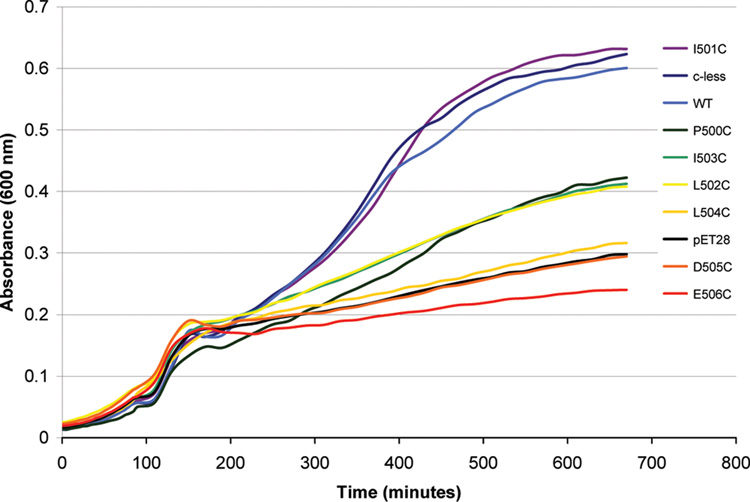
FIGURE 3.
Representative growth curves for the Walker B mutants and control proteins. The graph illustrates the range of growth differences over time as monitored at OD600.
WT and C-less, and empty pET28 plasmid, were run with each set as positive and negative controls, respectively. As seen in Figure 2 and Figure 3, WT and C-less consistently grew to an OD600 of at least 0.5, while pET28 grew to a maximum OD of 0.3. The A-loop mutation identified as a conserved aromatic residue in ABC transporters is at position Y351 in MsbA and after mutation to a cysteine is unable to support growth, consistent with its identified role as an essential aromatic residue.
Cysteine mutations in the highly conserved Walker A region from S378 to T384, which are known to be directly involved in binding the ATP phosphates, were unable to support growth in this assay, suggesting that each site is in fact essential for intact function within the cell, while sites I385–T390 tolerate the cysteine substitutions and are able to support cell growth. It is interesting to note that there appears to be a sharp transition in activity between T384 and I385, which also corresponds fairly well with the disappearance of any ATP-induced changes in the EPR spectra beyond site I385 (discussed below) and very well to the end of the originally defined Walker A region at T384.
Surprisingly, Q-loop mutation Q424C, which is proposed to hydrogen bond to the Mg2+ bound to ATP, is able to support some growth and is not completely inactive in the cell. Surrounding sites S423C and V426C appear to be unaffected by the apparently nonperturbing cysteine substitutions.
The C-loop mutations directly within the LSGGQ signature motif, which is also involved in binding directly to ATP, are well-tolerated from L481 to G483; however, the G484C and Q485C mutations are no longer able to support cell growth. It is interesting that L481C, S482C, and G483C are all able to support growth, though the level of growth of S482C is reduced. Surrounding sites in this key motif are generally accommodating of the cysteine substitutions, although nearby nonconserved mutations E476C and G478C are unable to support normal cell growth.
The P-loop, a single conserved proline at position P500 in MsbA, is located immediately prior to the Walker B motif (residues 501–506). Among these residues, the I501C mutation has no effect on cell growth, while the remaining mutants grow poorly, with decreasing activity toward the C-terminal end of this motif and ending with no growth at all for the E506C substitution. The D-loop (residues 509–512), a somewhat conserved motif with the sequence SALD, follows this region, and growth is affected only by the A510C mutation; the remaining D-loop sites are unaffected by cysteine substitution.
Finally, the sites surrounding the H-motif (H537) are relatively amenable to cysteine substitutions, with the exception of K528C and I542C, the sites farthest from H537 that were studied here. H537C, as expected for a highly conserved histidine important for ATP hydrolysis, was not able to support growth.
In summary, we introduced cysteine mutations within and surrounding each of the currently identified conserved motifs in ABC transporter NBDs. As expected, the level of cell growth was markedly reduced by mutations at many of the sites in these conserved domains, yet the overall in vivo growth assay results show that a surprisingly large number of sites are amenable to substitution with cysteine even when mutations were introduced directly into highly conserved sites. These results suggest that functionality may be due to a particular set of residues being involved in specific interactions with the remaining sites being responsible for global folding or packing interactions within these motifs.
Kinetics of ATPase Activity
To complement the in vivo growth assay data, in vitro kinetics of ATP hydrolysis were obtained for WT, C-less, and 40 single-cysteine mutants within and immediately surrounding the conserved motifs in the MsbA NBD using [γ-32P]MgATP as a substrate. First, WT and C-less (C88S/C315S) proteins were assayed for ATPase activity in the presence of lipid A-containing inner membrane lipids (Table 1). Initial rates of hydrolysis, as measured by the release of Pi over the first 2 min following the addition of 3 mM ATP, are in good agreement at 97 nmol mg−1 min−1 for WT and 102 nmol mg−1 min−1 for C-less protein.
Table 1.
ATPase Kinetic Data for MsbA Mutants
| MsbA | nmol mg−1 min−1a | % WT | |
|---|---|---|---|
| controls | WT | 97 | 100 |
| C-less | 102 | 105 | |
| A-loop | Y351C | 19 | 20 |
| Walker A | S378C | 6 | 7 |
| G379C | 12 | 13 | |
| S380C | 77 | 79 | |
| G381C | 61 | 63 | |
| K382C | 13 | 13 | |
| S383C | 10 | 10 | |
| T384C | 151 | 156 | |
| I385C | 101 | 104 | |
| A386C | 127 | 131 | |
| S387C | 75 | 78 | |
| L388C | 45 | 47 | |
| I389C | 47 | 49 | |
| T390C | 32 | 33 | |
| Q-loop | S423C | 237 | 244 |
| Q424C | 110 | 113 | |
| V426C | 103 | 106 | |
| C-loop | L481C | 91 | 94 |
| S482C | 21 | 22 | |
| G483C | 32 | 33 | |
| G484C | 17 | 18 | |
| Q485C | 75 | 77 | |
| Walker B | P500C | 37 | 38 |
| I501C | 50 | 51 | |
| L502C | 234 | 242 | |
| I503C | 133 | 137 | |
| L504C | 40 | 41 | |
| D505C | 9 | 9 | |
| E506C | 5 | 5 | |
| D-loop | S509C | 131 | 135 |
| A510C | –b | –b | |
| L511C | 4 | 4 | |
| D512C | 10 | 10 | |
| H-loop | V534C | 67 | 69 |
| I535C | 53 | 55 | |
| A536C | 45 | 46 | |
| H537C | 4 | 4 | |
| S540C | 121 | 124 | |
| T541C | 134 | 139 | |
| I542C | 152 | 157 |
Initial rates (vo) at 3 mM ATP.
Not expressed.
The A-loop Y351C mutant is able to hydrolyze ATP at only 20% of the rate of WT, consistent with its inability to sustain cell growth in vivo. Interestingly, the Walker A mutants show a variety of hydrolysis rates, and approximately half of the mutations are able to efficiently hydrolyze ATP (Table 1). Conserved sites S378C, G379C, K382C, and S383C, not surprisingly, exhibit a very limited ability to hydrolyze ATP and release Pi, whereas S380C, G381C, T384C, and I385C mutants are able to support hydrolysis. It is not unexpected that A386C and S387C are fully active because they are technically beyond the conserved Walker A motif; however, sites extended just beyond these residues had unexpectedly diminished ATPase activity with L388C, I389C, and T390C mutations, although their function was sufficient to support cell growth in vivo.
Strikingly, the ATPase activities of the conserved Q-loop (Q424) and two neighboring sites are unaffected when substituted with cysteine, which indicates that these side chains may not play a direct role in ATP hydrolysis. In addition, two of the sites within the C-loop exhibit hydrolysis rates close to that of WT, while S482C, G483C, and G484C exhibit significantly reduced rates, as expected for such highly conserved amino acids.
Mutations within the Walker B motif showed a significant loss of activity for D505C and E506C. The role of E506 has been well-studied, and its charge swap to E506Q is known to inhibit hydrolysis in other transporters (24, 25). However, L502C and I503C are significantly more active than WT and are clearly not negatively affected by the mutation to cysteine. P500C, I501C, and L504C each show approximately 50% of the ATP hydrolysis activity of WT. While most of the D-loop mutants are inactive with cysteines, the S509C mutant retains ATPase function in vitro, possibly due to the steric similarity of serine and cysteine. Finally, the H-motif H537C mutation is completely inactive; the sites preceding it are somewhat active, and those following the conserved histidine are unaffected or even more efficient than WT at hydrolyzing ATP.
Overall, it is notable that the ability to hydrolyze ATP remains relatively intact for many of the mutants in this set of conserved motifs within MsbA, suggesting that while some side chains play direct and specific roles in the binding, hydrolysis, and release of ADP and Pi, others appear to play indirect or local structural roles in the overall function of this ABC transporter.
EPR Spectroscopy Results
To assess the local changes in the dynamics of the Walker B sites (I501–E506) and the Q-loop (Q424), each single-cysteine mutant was spin labeled and reconstituted into inner membrane lipids, and the spectrum of each was recorded at each stage of ATP binding and hydrolysis. Additionally, two sites surrounding the Q-loop (S423 and V426), along with the H-motif (H537) and a nearby site (V534), were also analyzed by EPR spectroscopy motional analysis. We have previously reported similar EPR analysis of sites in the Walker A motif, C-loop, and just beyond the H-motif (20, 21).
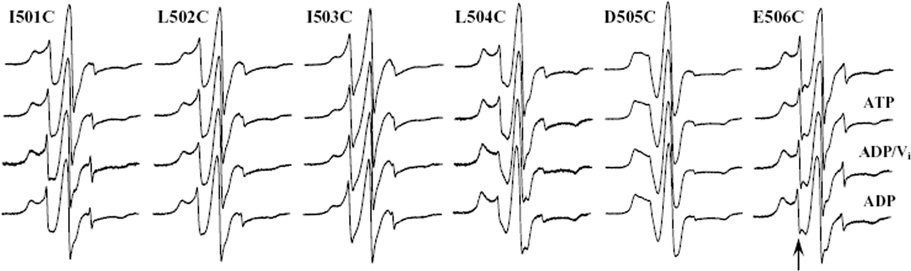
Interestingly, the EPR data for I501C, L502C, and I503C all exhibit somewhat similar spectra containing two motional components, one population of relatively slow motion and the other of relatively fast motion, indicative of either tertiary contacts within the protein fold or two different conformational populations of the protein (Figure 4). These three sites show no change in spectral motion upon addition of ATP or posthydrolysis, intact function in the in vitro assay, and somewhat decreasing activity in the in vivo growth assay. Thus, the absence of changes in the EPR spectra likely reflects their lack of direct involvement with ATP either upon binding or during hydrolysis and is consistent with data from both of the functional assays presented above and with crystallographic data for other NBDs (e.g., refs 26 and 27). The EPR data for L504C and E506C also exhibit two component spectra and show no changes upon addition of ATP, MgATP, or MgATP and vanadate. L504C contains a larger population of immobilized spins than the spin labels at other positions in this motif, and its is the only spectrum to show any changes in its EPR spectrum at any stage; a small change is observed posthydrolysis for this site. L504C has moderate activity in both functional assays, while E506C is completely inactive in both assays, as expected given its established role in the hydrolysis cycle. Finally, D505C does not show any changes in response to substrate binding or hydrolysis, likely due to the lack of function of this mutation either in vivo or in vitro.
FIGURE 4.
CW X-band EPR spectra of spin labeled cysteine MsbA mutants from the Walker B motif. Proteins were reconstituted into lipid membranes and spectra recorded in the absence of substrate (top), in the presence of ATP and EDTA (ATP) prior to hydrolysis, in the presence of MgATP followed by immediate addition of Vi (ADP/Vi) to trap the posthydrolysis state, and in the presence of MgATP (ADP) to allow hydrolysis to proceed. The arrow indicates the fast-motion component observed in many of the spectra shown.
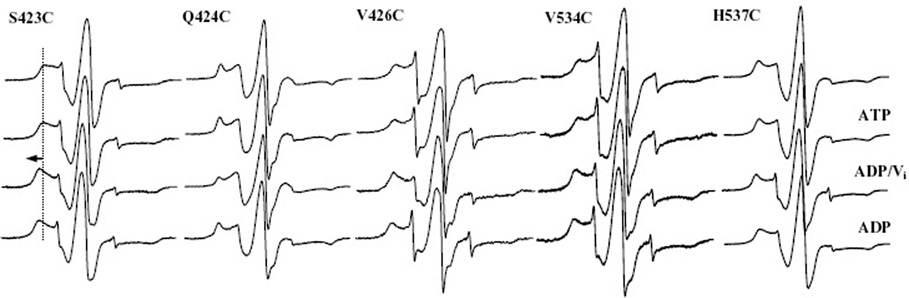
EPR spectra for Q-loop mutant Q424C and two flanking sites also showed no changes upon addition of ATP but did show changes upon addition of vanadate in the posthydrolysis state (Figure 5). Q424C exhibits two motional components in its spectrum, and the populations of these components shift toward the more mobile population at this step; this is the only site studied here that becomes more mobile at the posthydrolysis stage. In contrast, the adjacent site S423C undergoes a significant immobilization posthydrolysis, as evidenced by an increase of 3 G between the outer spectral lines. V426C shows a slight decrease in its more mobile component posthydrolysis but, otherwise, shows little change throughout the catalytic cycle. Activities indicate that Q424C itself is only somewhat active in vivo, yet the observed changes in the EPR spectra are consistent with the functional in vitro activity data; the flanking sites are both fully functional in both assays. The combined results support a model in which this motif undergoes a conformational change upon transition to the posthydrolysis state.
FIGURE 5.
CW X-band EPR spectra of spin labeled MsbA cysteine mutants in and around the H-motif (H537) and the Q-loop (Q424). Sample conditions for each series are the same as those described in the legend of Figure 3.
Three sites just past the H-loop (H537), S540C, T541C, and I542C, were previously analyzed by EPR spectroscopy (21), and all exhibited significant changes and immobilization in the vanadate-trapped posthydrolysis state. Here, we analyzed spectra from H537C and V534C, located on the opposite side of the H-loop. The small change observed at H537C posthydrolysis indicates that a very small population of the protein is undergoing structural changes upon MgATP binding within the time frame required for collection of its EPR spectrum or that a conformational change is occurring within the protein that does not result in the release of Pi and transport of lipid A. The small change at V534C provides further evidence of a conformational change in the region of the H-loop following MgATP binding and ATP hydrolysis by MsbA.
The Walker A sites extended from position 378 to 390 were previously analyzed by EPR, with conserved sites 378–385 all showing motional changes upon vanadate trapping (20). Sites 386–390 did not show significant changes at any step. Most strikingly, Walker A sites S378C, S380C, K382C, T384C, and I385C revealed motional changes in the presence of ATP. Additionally, LSGGQ sites 481–485 each showed significant changes in their EPR spectra solely upon vanadate trapping (21) and a small change in S482C upon ATP binding. Neither T384C nor S423C was affected by the addition of MgCl2 or MgCl2 and Vi in the absence of ATP (data not shown), verifying that the changes observed in the presence of MgATP or MgATP/Vi are due to hydrolysis-induced changes in the spectra.
Summary of All Data Collected for Sites in the Conserved NBDs of MsbA
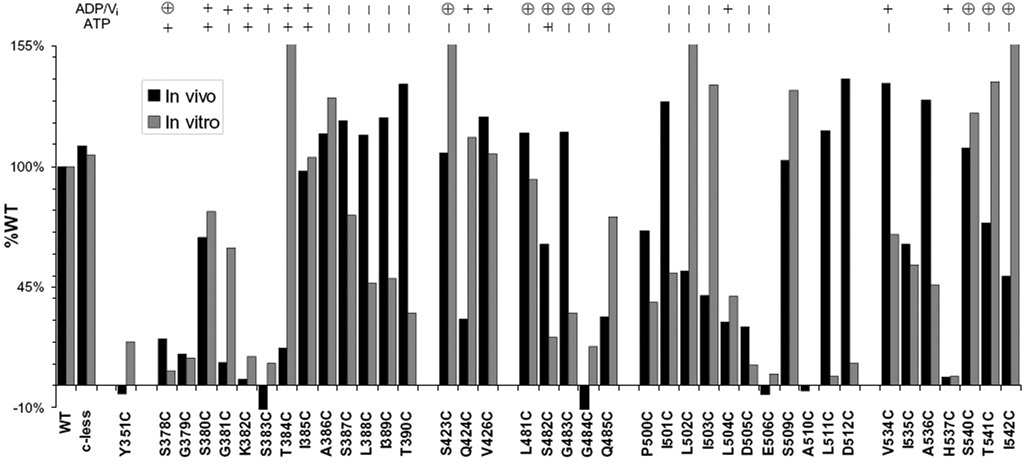
The collective set of data based on in vivo assays representing full functionality of ATPase and transport activity in MsbA mutants, in vitro kinetic data for ATP hydrolysis on each mutant, and in vitro localized motional changes for each site are summarized in Figure 6. This set includes the activity and spectral data presented here, along with summaries of EPR data previously published on sites in the Walker A region, C-loop, and just beyond the H-motif (20, 21). The in vivo and in vitro data as a percent of the WT values are graphed side by side for a direct comparison of the activities of the studied sites within and surrounding key NBD motifs. In addition, the results of each of the experiments are illustrated for each site in Figure 7. For 25 of the 40 sites studied here, the results of the two functional assays correspond very well. However, several sites stand out that show very different results between the two assays; six sites, at least one in each of the motifs except the A-loop, are unable to sustain good growth rates but retain high ATPase rates, and nine sites spread mainly throughout the C- and D-loops were able to sustain growth within the cells but displayed reduced in vitro ATPase activities.
FIGURE 6.
In vitro ATPase activity data and in vivo growth data for 40 single-cysteine mutants within the NBD of MsbA plotted side by side as a percentage of the values obtained for WT MsbA. Sites S423C, L502C, and I542C exhibited values greater than 155% of that of WT (see Table 1), and the A510C protein did not express for purification. EPR motional changes are summarized across the top with symbols for no change (−), small changes (+), and large changes (⊕) upon ATP binding (ATP) and vanadate trapping (ADP/Vi).
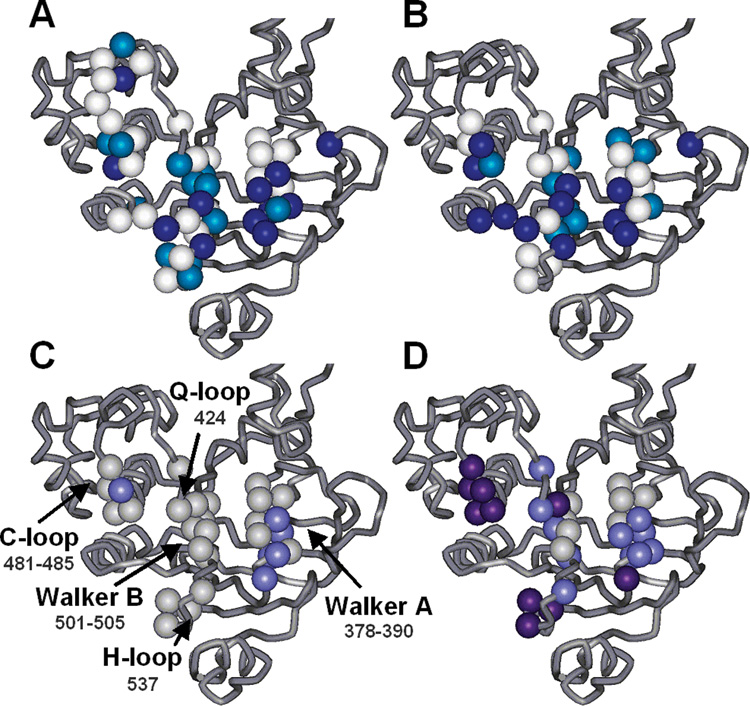
FIGURE 7.
Illustration of results for each site studied overlaid onto the open face of the E. coli MsbA NBD. (A) In vivo growth assay results. Mutants with little or no growth (<25% of the WT rate) are colored dark blue, those with intermediate growth light blue, and those with full growth (>75% of the WT rate) white. (B) In vitro ATPase activity results using the same color scheme as in panel A but corresponding to hydrolysis rates as a percentage of WT rather than growth rates. (C and D) Changes in spin label mobility as observed by EPR (C) upon addition of ATP/EDTA and (D) upon addition of MgATP/Vi. Cases in which there are no changes in the mobility of the spin label side chain compared to the resting state are colored gray, cases in which there are small changes light purple, and cases in which there are large changes dark purple. The sites that do not show changes upon vanadate trapping include L505C and E506C, which are considered inactive in each of the functional assays.
The full activities of S423C and V426C, flanking the Q-loop at Q424, and the six sites studied flanking the H-motif at H537 strongly indicate that the glutamine and histidine side chains are single sites that act alone in their role within MsbA function and that the surrounding sites play roles in local structure or folding interactions. It is striking to note that Q424 appears to be important for global conformational changes within MsbA or lipid A flipping rather than ATP hydrolysis since it retains ATPase activity but supports only limited growth in the in vivo assay. Y351 is clearly a vital single aromatic residue within MsbA, as characterized for P-glycoprotein (9, 28).
One could argue that the introduction of a cysteine disrupts interaction of the C-loop with ATP; however, the activities are all at least somewhat intact with the exception of that of G484C. We have previously shown that motional changes for each of these sites are observed for the posthydrolysis state, which is strong evidence that ATP is still able to bind to MsbA containing cysteine mutations within this motif. Activities of mutants in the Walker A motif are varied but overall indicate the importance of this motif to ATP binding and hydrolysis. Similarly, the combined data for the Walker B motif confirm that D505 and E506 are key sites within MsbA and indicate that the preceding hydrophobic residues are less important, with the exception of L504, which is also worth noting as an important player in hydrolysis. Finally, the D-loop data clearly indicate that the S509C mutation has no effect while the A510C is critically important. Although L511 and D512 are known to be dysfunctional as a proline and a glycine in vivo (29), respectively, the cysteine substitutions exhibit full functionality in vivo. These results are overall consistent with the previous suggestions of sites and motifs important for MsbA function but highlight the fact that some identified sites are not specifically required for function while others outside of the defined regions of importance are in fact critical for function in this transporter.
EPR Spectroscopy Data Indicate ATP Binding Does Not Close the MsbA NBDs
Sites 534 and 540–542 surrounding the H-motif at H537 show no motional changes as studied by EPR upon addition of ATP but significant changes upon vanadate trapping, indicating that this motif is involved in a significant rearrangement posthydrolysis, but not upon ATP binding. The C-loop, or the LSGGQ signature motif, sites exhibit characteristics similar to those of the H-motif, where no significant changes are observed in motion for these critical residues upon addition of ATP, whereas significant immobilization occurs upon vanadate trapping, suggesting that this motif picks up significant tertiary contacts posthydrolysis, but not upon ATP binding (21). And, again, the Q-loop sites also show no changes upon addition of ATP and significant changes upon vanadate trapping, also indicating that structural changes in this motif are not triggered until hydrolysis occurs.
The spin labels at the Walker B sites show no motional changes at any stage of hydrolysis with the exception of a small change that can be observed for L504C immediately posthydrolysis. For those hydrophobic sites (501–503) supporting the critical D505 and E506 sites, no changes are observed, and this likely indicates that this motif does not undergo a large conformational rearrangement as a whole.
The Walker A motif is the only region examined so far showing notable changes in its EPR spectra upon addition of ATP (20). Specifically, sites 378, 380, 382, 384, and 385 show ATP-induced changes in their motion, suggesting that these sites are directly affected by the binding of ATP or undergo a conformational change upon substrate binding. Given the lack of changes in the motion of the other motifs upon ATP binding, the direct changes observed in the spectra in this motif, and the crystallographic evidence from ATP-bound transporters (e.g., refs 26 and 30) and AMP-PNP-and ADP/Vi-bound S. typhimurium MsbA (4), we conclude that these sites do in fact directly interact with ATP in the liquid-phase environment. This is the first spectroscopic verification of the sites directly involved in ATP interaction. Once vanadate trapping has occurred, each of the sites between positions 378 and 385 undergoes further structural changes, indicating a conformational rearrangement of this motif. The additional changes seen in the Walker A motif and the fact that significant changes are seen in the other motifs only upon vanadate trapping suggest that the NBDs do not dimerize solely upon ATP binding in the absence of Mg2+. In fact, the EPR motion data indicate the motifs do not come into contact or undergo significant conformational rearrangement until hydrolysis takes place, or Mg2+ is present.
On the basis of our results presented here, we propose that in a liquid-phase membrane environment, MsbA sites 378–385 (Walker A) directly bind ATP on each monomer without affecting the other motifs within the NBD and that the remaining motifs, including the C-loop from the opposing monomer, come together as a closed NBD dimer only upon addition of Mg2+ or upon ATP hydrolysis.
DISCUSSION
This is the first comprehensive study of the effects of mutagenesis on the function of all of the conserved NBD sites within E. coli MsbA and the first extensive set of either in vivo or in vitro functional assessments of this bacterial ABC transporter. Overall, specific conserved, and several nonconserved, sites within the MsbA NBD are confirmed to be directly essential to protein function, while others are shown to play a more supporting or structural role. All of the mutants studied, with the exception of A510C, have been successfully purified from the membrane fraction, thus showing a sufficient level of expression, proper folding, and insertion into the inner membrane.
It is remarkable that a number of residues within conserved motifs of this protein can be mutated to cysteine and still retain activity. For example, S380C of the Walker A motif is 68–79% active in both assays and exhibits EPR changes, which makes this mutation stand out among an entire motif that otherwise does not tolerate substitution with cysteine. Similarly, L481C in the highly conserved C-loop and S509C in the D-loop are both essentially unaffected by cysteine substitution. This suggests that L481 is not essential as a leucine even though it is highly conserved across many ABC transporter NBDs and supports the fact that S509 is only somewhat conserved across transporters.
On the other hand, L504 in the Walker B motif appears to play a considerable role in MsbA function, as does its D505 neighbor, and sites 388–390, which are outside of the traditional Walker A motif, displayed reduced ATPase activities, indicating that amino acid side chains beyond the well-conserved key regions can also be important in binding to ATP, stabilizing ATP hydrolysis, or indirectly supporting key structural rearrangements within the transporter.
In this study, we have also identified several sites showing high ATPase rates and low growth rates, indicating an inability to bind or transport lipid A. It is unlikely that sites within the NBDs could have a direct effect on the inability of lipid A to bind or be flipped, suggesting that although these mutants readily hydrolyze ATP, they are unable to induce the global protein change necessary for lipid A binding and transport by the membrane-spanning domains in vivo. Another possibility is that the detergent environment in the ATPase assay allows the protein to compensate structurally for the mutation, whereas the forces imposed on the protein within its natural membrane do not.
In the opposite scenario, where the proteins are more active in vivo than in vitro, there is the possibility that the cells compensate for poorly functioning proteins by overexpressing them and yielding normal growth results or that certain mutations cause the protein to function poorly after going through the purification process but are able to remain intact within the cell. As an example of these possibilities, the activity differences observed for the Q-loop (Q424C) in MsbA, where this mutation had no effect on the ATPase activity in vitro but was able to support only limited growth, align well with the proposal for HlyB that the Q-loop is one of the conserved sites responsible for communication between the two NBDs or between the NBDs and the transmembrane domains and is not directly involved in ATP binding or hydrolysis (30).
Many of the spectra exhibit small populations of spins undergoing very fast rotational motion, which is likely due to a small fraction of unfolded protein. These populations vary by protein preparation and tend to be only a few percent of the total protein. It is worth noting that these denatured protein peaks disappear somewhat upon vanadate trapping, which is possibly due to this fraction of the protein being only partially unfolded and still affected by changes in the protein structure induced by hydrolysis and vanadate trapping.
It is possible that the introduction of sulfhydryl groups into a symmetrical homodimer could generate disulfide bonds in the protein that may affect the function of the protein. In vivo, the cytoplasm is a reducing environment, so we do not expect potential disulfide bonds between the MsbA monomers to have affected the in vivo growth data presented for any of the mutations. In agreement with this belief, of those mutants demonstrating weak growth in the in vivo assay, only S378C and G379C exhibited significant disulfide-bonded dimers after purification (data not shown). Both of these mutations also exhibit significantly reduced activity in the ATPase activity assay, which includes a large excess of the reducing agent, DTT, suggesting that the amino acid change itself is responsible for each dysfunction and not the formation of disulfide-bonded dimers in vivo.
The proposal that ATP binding alone does not trigger the MsbA NBDs to come into contact with each other as a dimer is not in line with most data presented for other transporters. However, the majority of the evidence for dimer closure is based on crystal structures containing unnatural ATP analogues or dysfunctional mutations. Although there is no crystal structure of MsbA with ATP bound, the structure of MsbA from S. typhimurium containing the unnatural analogue AMP-PNP is in the closed state (4). Also, SAV1866, which is highly homologous to MsbA especially in the NBD regions, crystallized in a closed conformation with ADP bound (5). Interestingly, MsbA from V. cholera without substrate was also crystallized in a closed state (4), further illustrating the possibility that the crystallization conditions may favor a closed structure that is not present under more physiologically relevant conditions. In the cell, ATP binds to the ABC transporters in the presence of Mg2+, but it is difficult to study the direct effect of Mg2+ in the presence of ATP without introducing an unnatural nonhydrolyzable analogue or a dysfunctional mutation. Our previous studies on the Walker A region of MsbA showed that MgAMP-PNP did not induce the same changes in the EPR spectra as ATP and thus did not serve as a functionally relevant substitute (20), a conclusion also reached for other ABC transporters (11, 25, 31). Other closed crystal structures with ATP bound are the result of the use of dysfunctional mutations such as E506Q and H537A (MsbA notation), which allows MgATP to be trapped within the protein prior to hydrolysis (26, 30, 32). The addition of MgCl2 in the absence of ATP does not alter the EPR spectra.
There is evidence that substrate may need to be present to close the NBD dimer interface. In the maltose transporter, it was recently discovered that maltose binding protein (MBP), which delivers the transported maltose ligand to the periplasmic side of the MalFGK2 complex, is required for the MalK monomers (NBDs) to dimerize in the presence of ATP alone (33). In the absence of MBP, ATP binding alone does not bring the two MalK proteins within the proximity of each other, as we see for MsbA in the absence of substrate. Thus, it is possible that MsbA also requires its lipid A substrate to be present for its NBDs to dimerize. However, the ability of MsbA to hydrolyze ATP in the absence of lipid A contradicts this idea, unless dimerization is not necessary for hydrolysis and only for lipid A transport. Simulation studies of BtuCD indicate that one of the NBDs closes in the presence of ATP but only when its periplasmic binding protein BtuF is bound to the complex (34). Our data do not preclude the potential for such a semiopen state for the MsbA NBDs in the presence of ATP.
The significant tertiary contacts made between the two monomers in the closed dimer shown in the ADP/Vi-bound S. typhimurium crystal structure indicate that if the structure is valid in solution, we should observe significant changes in the motion of those side chains physically located at the dimer interface (see Figure 1). In fact, we observe significant changes only in the EPR spectra of the Walker A mutants upon addition of ATP and not in any of the mutants on the interface. As two examples, sites S423 and L481 are functional in both assays and located on the dimer interface in the closed crystal structure but do not show any motional changes by EPR until hydrolysis is allowed to occur. If the MsbA NBDs approach each other sufficiently closely in the presence of ATP alone to form the dimer found in the crystal structure, then these specific example sites would be expected to exhibit significant changes in motion due to their direct locations on the dimer interface. These types of changes are not observed in our studies using ATP but are observed once hydrolysis has occurred. In addition, S540 is the only site that becomes more mobile upon vanadate trapping, and this change does not occur in the presence of ATP alone. Other work on a single site within the MsbA NBD using the SDSL technique shows that the NBDs come much closer together when moving from the open resting state to the posthydrolysis state (by addition of MgATP/Vi), though the study does not address the ATP-bound state (35).
Clearly, the role of lipid A and Mg2+ in the dimerization of the NBDs in MsbA remains an important unanswered question and is the focus of our current investigations. Overall, there is overwhelming evidence observed in all motifs studied that a structural change occurs in the MsbA NBDs upon transition to the posthydrolysis state that is important for initiating lipid A transport and/or Pi release.
ACKNOWLEDGMENT
We thank John Paul Savaryn for assistance on this project and Dr. Jimmy Feix for critical reading of the manuscript.
Footnotes
This work was supported by the National Institutes of Health (GM070642).
Abbreviations: ABC, ATP-binding cassette; EPR, electron paramagnetic resonance; SDSL, site-directed spin labeling; CW, continuous wave; NBD, nucleotide binding domain; MTSL, 2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate spin label; WT, wild-type; DM, dodecyl maltopyranoside; PE, phosphatidylethanolamine; PG, phosphotidylglycerol; CL, cardiolipin; DTT, dithiothreitol.
REFERENCES
- 1.Cotten JF, Welsh MJ. Covalent Modification of the Regulatory Domain Irreversibly Stimulates Cystic Fibrosis Transmembrane Conductance Regulator. J. Biol. Chem. 1997;272:25617–25622. doi: 10.1074/jbc.272.41.25617. [DOI] [PubMed] [Google Scholar]
- 2.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 4.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 6.Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 7.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt L, Benabdelhak H, Blight MA, Holland IB, Stubbs MT. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: Identification of a variable region within ABC helical domains. J. Mol. Biol. 2003;330:333–342. doi: 10.1016/s0022-2836(03)00592-8. [DOI] [PubMed] [Google Scholar]
- 9.Ambudkar SV, Kim IW, Xia D, Sauna ZE. The A-loop, a novel conserved aromatic acid subdomain upstream of the Walker A motif in ABC transporters, is critical for ATP binding. FEBS Lett. 2006;580:1049–1055. doi: 10.1016/j.febslet.2005.12.051. [DOI] [PubMed] [Google Scholar]
- 10.Zaitseva J, Oswald C, Jumpertz T, Jenewein S, Wiedenmann A, Holland IB, Schmitt L. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 2006;25:3432–3443. doi: 10.1038/sj.emboj.7601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oswald C, Holland IB, Schmitt L. The motor domains of ABC-transporters. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2006;372:385–399. doi: 10.1007/s00210-005-0031-4. [DOI] [PubMed] [Google Scholar]
- 12.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 13.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klug CS, Feix JB. Methods and Applications of Site-Directed Spin Labeling EPR Spectroscopy. In: Correia JJ, Detrich HW, editors. Methods in Cell Biology, Biophysical Tools for Biologists, Volume One: In Vitro Techniques. New York: Academic Press; 2008. pp. 617–658. [DOI] [PubMed] [Google Scholar]
- 15.Klug CS, Feix JB. SDSL: A Survey of Biological Applications. In: Berliner LJ, Eaton SS, Eaton GR, editors. Biological Magnetic Resonance Volume 24. Hingham, MA: Kluwer Academic/Plenum Publishers; 2004. pp. 269–308. [Google Scholar]
- 16.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 17.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 18.Isas JM, Langen R, Haigler HT, Hubbell WL. Structure and dynamics of a helical hairpin and loop region in annexin 12: A site-directed spin labeling study. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 19.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Buchaklian AH, Klug CS. Characterization of the Walker A motif of MsbA using site-directed spin labeling electron paramagnetic resonance spectroscopy. Biochemistry. 2005;44:5503–5509. doi: 10.1021/bi047568v. [DOI] [PubMed] [Google Scholar]
- 21.Buchaklian AH, Klug CS. Characterization of the LSGGQ and H motifs from the Escherichia coli lipid A transporter MsbA. Biochemistry. 2006;45:12539–12546. doi: 10.1021/bi060830a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerrler WT, Reedy MC, Raetz CR. An Escherichia coli Mutant Defective in Lipid Export. J. Biol. Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 23.Doerrler WT, Gibbons HS, Raetz CR. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 24.Orelle C, Dalmas O, Gros P, Di Pietro A, Jault JM. The Conserved Glutamate Residue Adjacent to the Walker-B Motif Is the Catalytic Base for ATP Hydrolysis in the ATP-binding Cassette Transporter BmrA. J. Biol. Chem. 2003;278:47002–47008. doi: 10.1074/jbc.M308268200. [DOI] [PubMed] [Google Scholar]
- 25.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oldham ML, Khare FA, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 27.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 28.Kim IW, Peng XH, Sauna ZE, FitzGerald PC, Xia D, Muller M, Nandigama K, Ambudkar SV. The Conserved Tyrosine Residues 401 and 1044 in ATP Sites of Human P-Glycoprotein Are Critical for ATP Binding and Hydrolysis: Evidence for a Conserved Subdomain, the A-Loop in the ATP-Binding Cassette. Biochemistry. 2006;45:7605–7616. doi: 10.1021/bi060308o. [DOI] [PubMed] [Google Scholar]
- 29.Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 1996;20:1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 30.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdon G, Albers SV, Dijkstra BW, Driessen AJM, Thunnissen AM. Crystal Structures of the ATPase Subunit of the Glucose ABC Transporter from Sulfolobus solfataricus: Nucleotide-free and Nucleotide-bound Conformations. J. Mol. Biol. 2003;330:343–358. doi: 10.1016/s0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12837–12842. doi: 10.1073/pnas.0803799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivetac A, Campbell JD, Sansom MS. Dynamics and function in a bacterial ABC transporter: Simulation studies of BtuCDF system and its components. Biochemistry. 2007;46:2767–2778. doi: 10.1021/bi0622571. [DOI] [PubMed] [Google Scholar]
- 35.Borbat PP, Surendhran K, Bortolus M, Zou P, Freed JH, Mchaourab HS. Conformational Motion of the ABC Transporter MsbA Induced by ATP Hydrolysis. PLoS Biol. 2007;5:2211–2219. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]