SUMMARY
Large, long-lived species experience more lifetime cell divisions and hence a greater risk of spontaneous tumor formation than smaller, short-lived species. Large, long-lived species are thus expected to evolve more elaborate tumor suppressor systems. In previous work, we showed that telomerase activity coevolves with body mass, but not lifespan, in rodents: telomerase activity is repressed in the somatic tissues of large rodent species but remains active in small ones. Without telomerase activity, the telomeres of replicating cells become progressively shorter until, at some critical length, cells stop dividing. Our findings therefore suggested that repression of telomerase activity mitigates the increased risk of cancer in larger bodied species but not necessarily longer-lived ones. These findings imply that other tumor suppressor mechanisms must mitigate increased cancer risk in long-lived species. Here, we examined the proliferation of fibroblasts from 15 rodent species with diverse body sizes and lifespans. We show that, consistent with repressed telomerase activity, fibroblasts from large rodents undergo replicative senescence accompanied by telomere shortening and overexpression of p16Ink4a and p21Cip1/Waf1 cycline dependent kinase inhibitors. Interestingly, small rodents with different lifespans show a striking difference: cells from small shorter-lived species display continuous rapid proliferation, whereas cells from small long-lived species display continuous slow proliferation. We hypothesize that cells of small long-lived rodents, lacking replicative senescence, have evolved alternative tumor-suppressor mechanisms that prevent inappropriate cell division in vivo and slow cell growth in vitro. Thus, large-bodied species and small but long-lived species have evolved distinct tumor suppressor mechanisms.
Keywords: Lifespan, body mass, senescence, evolution, cancer
INTRODUCTION
Replicative senescence was first described in human cells over 40 years ago (Hayflick & Moorhead, 1961). The major mechanism underlying replicative senescence is progressive telomere shortening, which occurs with every cell division in cells lacking telomerase activity (Harley et al., 1990; Shay & Wright, 2000). Replicative senescence is thought to be an anti-tumor mechanism that limits cell proliferation (Campisi, 2001; Sedivy, 2007) and contributes to long cancer-free human lifespan. Replicative senescence also occurs in the fibroblasts of several nonhuman primates (Gardner et al., 2007; Herbig et al., 2006; Steinert et al., 2002), and large farm animals such as cow (Hornsby et al., 1986), sheep (Davis et al., 2005), and horse (Argyle et al., 2003), reviewed in (Davis & Kipling, 2005; Gorbunova & Seluanov, 2008). But we still know little about senescence, or the lack thereof, in other mammalian species.
Senescence of mouse cells differs from replicative senescence of human cells in many ways (Parrinello et al., 2003; Sedivy, 1998; Wright & Shay, 2000). First, mouse somatic cells express telomerase and have very long telomeres (Prowse & Greider, 1995). Second, when cultured under 20% oxygen mouse fibroblasts slow their growth after approximately 10 population doublings (PDs), but after a few days immortal clones emerge and continue rapid growth (Parrinello et al., 2003). Such spontaneous immortalization does not occur in cultures of human fibroblasts (Wright & Shay, 2000). Third, under physiological oxygen concentration mouse fibroblasts do not display senescent phenotype (Parrinello et al., 2003). Thus, “senescence” of mouse cells appears to be a stress response, which is more akin to stress-induced premature senescence than replicative senescence (Parrinello et al., 2003; Sherr & DePinho, 2000; Wright & Shay, 2000).
The difference in telomere biology between humans and mice is intriguing. We recently undertook a study aiming to understand the evolutionary basis of this difference. To test the hypothesis that somatic repression of telomerase activity might have evolved as a tumor-suppressor adaptation in large or long-lived species we analyzed telomerase activity in fresh tissues of 15 rodent species with diverse lifespans and body masses (Seluanov et al., 2007). Our comparative analysis showed that telomerase activity coevolves with body mass but not lifespan. The largest rodents, capybara and beaver, showed a near complete repression of telomerase activity, as in humans. In contrast, all small rodents showed high levels of telomerase activity in somatic tissues. Surprisingly, telomerase activity was detected in somatic tissues of both short-and long-lived small species, including the longest-lived rodents, the naked mole-rat and the Eastern grey squirrel.
Here we address several questions raised by our previous study. First, does the repression of telomerase activity in the somatic tissues of large rodents mean that their fibroblasts will display replicative senescence? Second, the presence of telomerase activity in somatic tissues of small long-lived rodents is puzzling: these animals remain largely tumor-free during their long lives despite the increased cancer risk of persistent telomerase activity. We therefore asked if fibroblasts from these species display the same rapid proliferation in culture as fibroblasts from short-lived, cancer-prone mice.
To answer these questions we studied replicative lifespan of fibroblasts from 15 rodent species. First, we show that fibroblasts from large rodent species that lack telomerase activity enter typical replicative senescence, as expected. Second, we demonstrate that fibroblasts from small short-lived rodent species that maintain telomerase activity, lack replicative senescence, and display rapid continuous growth. Last, and most surprising, we show that fibroblasts from small long-lived rodents also lack replicative senescence and instead display unusually slow growth. We hypothesize that cells of small long-lived rodents have evolved alternative tumor-suppressor mechanisms — ones distinct from regulation of telomerase activity — that protect them from cancer in vivo and slow their proliferation in vitro.
RESULTS
Rodent species display three distinct patterns of in vitro cell proliferation
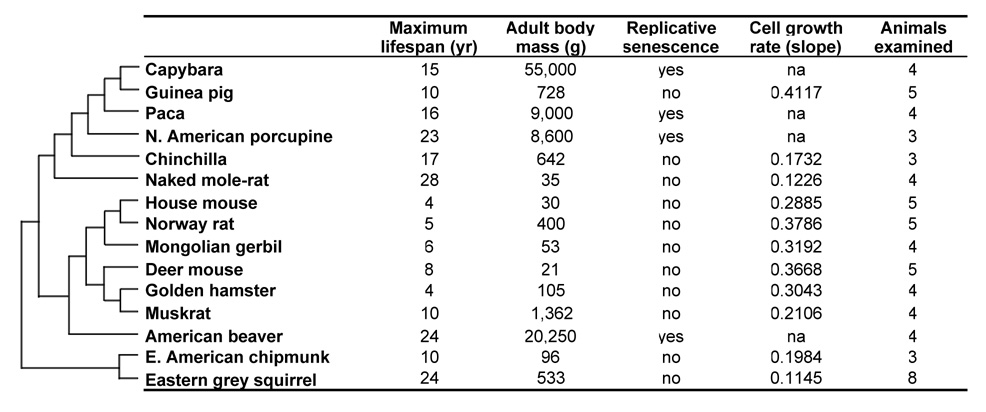
To examine growth characteristics of primary fibroblasts from long- and short-lived rodent species we isolated primary lung and skin fibroblasts from 15 species with diverse lifespans and body masses (Figure 1). Tissues were obtained from at least three young adults of each species, and one lung and one skin fibroblast culture was established from each animal. In total, at least 5 primary fibroblast cultures were analyzed for each species, as some cultures were lost at early stages due to contamination (Figure 1).
Figure 1. Phylogeny and phenotypic data for 15 rodent species.
The tree topology is based on molecular phylogenies inferred from (Adkins et al., 2003; Martin et al., 2000; Michaux et al., 2001; Montgelard et al., 2002; Murphy et al., 2001; Steppan et al., 2004). Body mass and maximum lifespan data is obtained from the following (Buffenstein & Jarvis, 2002; de Magalhaes et al., 2005; Nowak, 1999; Turturro et al., 1999; Weigl, 2005). The data on replicative senescence and growth rates is derived from the growth curves shown in Figure 2. The growth curve for each species represents an average of at least 5 independent cultures.
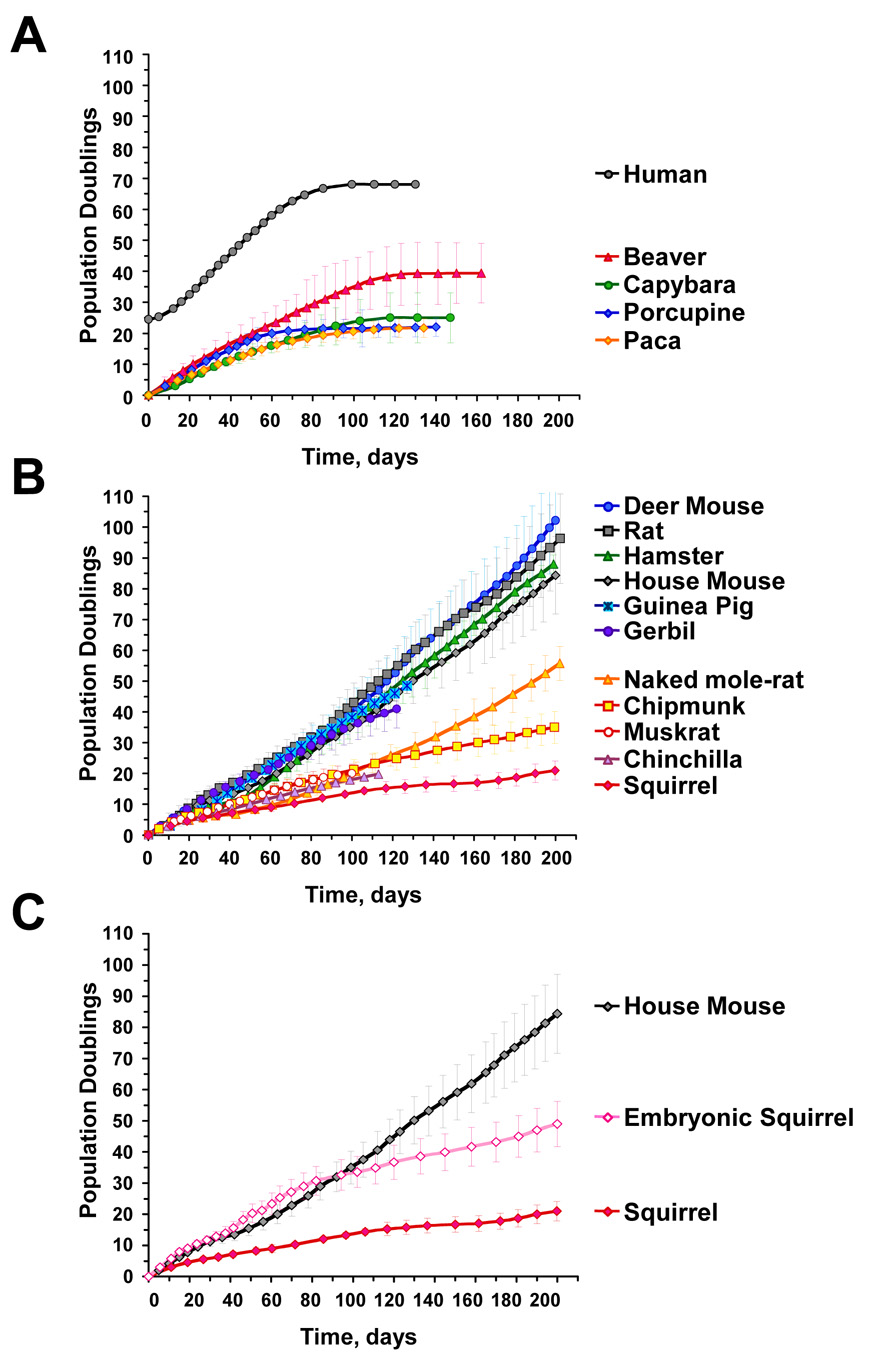
Cells were passaged for 100 to 200 days under physiological oxygen concentration (3%) to avoid inducing premature senescence due to oxidative stress (Parrinello et al., 2003). Cell numbers were counted at every passage, and growth curves were determined for each species (Figure 2). The growth curves of skin and lung fibroblasts were similar; therefore the skin and lung cell data were combined for each species. Qualitatively, the fibroblast growth curves of rodent species fell into three groups: (1) replicative senescence (Figure 2A); (2) continuous fast growth (Figure 2B); and (3) continuous slow growth (Figure 2B).
Figure 2. Growth curves of primary rodent fibroblasts.
Cells were isolated from skin and lung tissue and grown at 3% oxygen. The number of donors for each species is shown in Figure 1. (A) Species that use replicative senescence. Each curve is an average of at least 5 independent cultures, and error bars show s.d. Beaver cultures entered senescence at PD39±10; porcupine and paca cultures entered senescence at PD22±3; and capybara cultures senesced at PD25±8. Human primary fibroblasts culture HCA2 is shown as a reference. (B) Rodents that do not use replicative senescence fall into two groups: fast growing and slow growing. Each curve is an average of at least 5 independent cultures, and error bars show s.d. (C) Growth of grey squirrel embryonic fibroblasts (SEFs). Cells grow rapidly for the first 30 PDs, and then slow down and attain the growth rate of adult cells. SEFs were isolated from 4 embryos derived from 2 pregnant females.
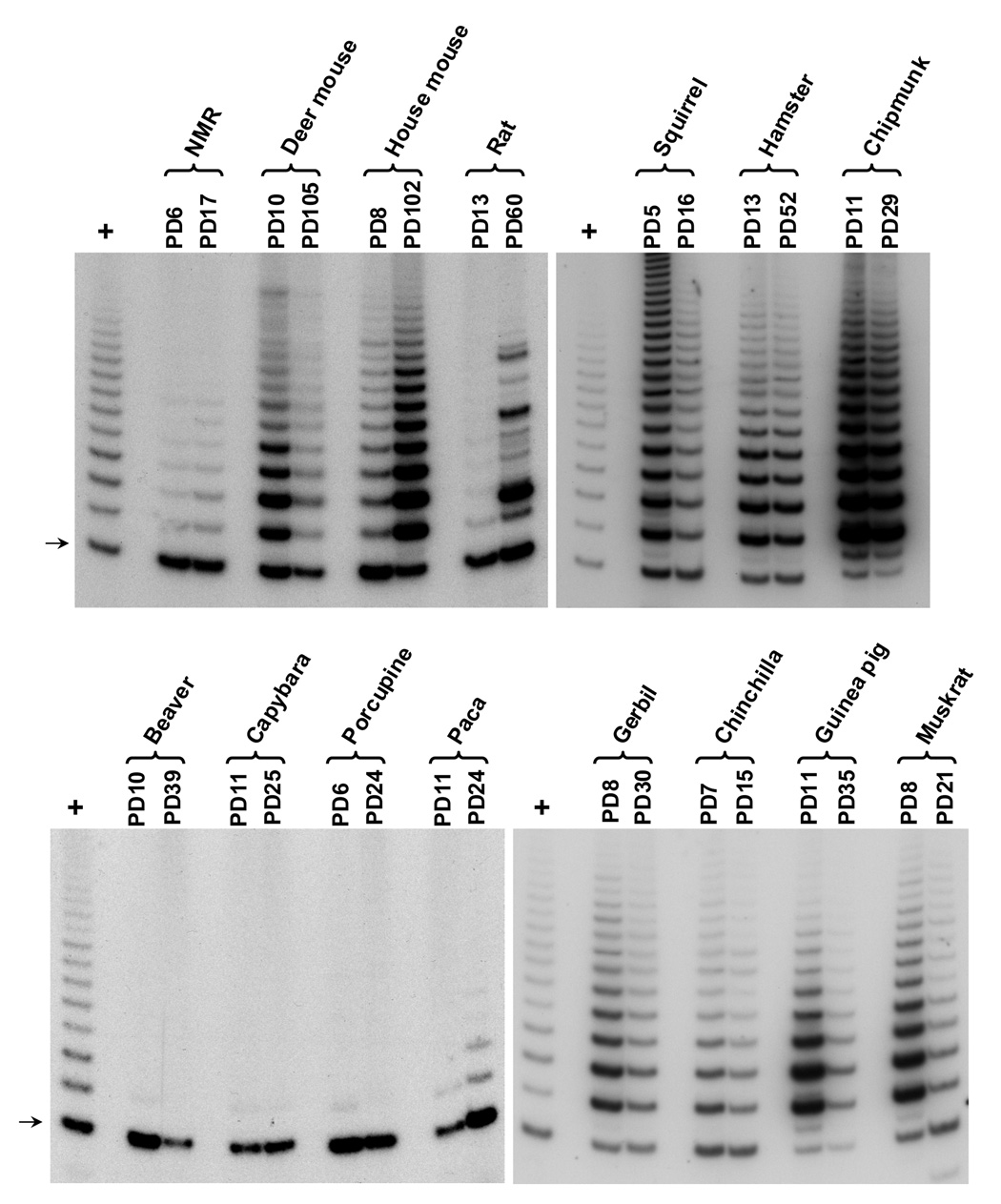
To understand the cell biological basis of these contrasting fibroblast growth curves, we next studied telomere biology in these cells. Telomerase activity and telomere length were examined at early and late cell passages. Telomerase activity was assayed using a modified telomeric repeat amplification protocol (TRAP) (Szatmari & Aradi, 2001) (Figure 3). The TRAP assay measures the extension of the telomeric sequence by telomerase in whole cell extracts. Fibroblasts from beaver, porcupine, and capybara had no detectable telomerase activity. Early passage paca fibroblasts were telomerase negative but started to express weak telomerase activity at late passages. Fibroblasts from all other animals expressed high levels of telomerase activity.
Figure 3. Telomerase activity in early and late passage rodent fibroblasts.
Telomerase activity was examined using modified TRAP assay. Lanes labeled with “+” contains a sample of telomerase positive human cancer cell line. Arrow indicates a primer dimer, which is not dependent on telomerase activity. NMR, naked mole-rat. The mouse sample shown is from a of 129/Sv×BL6 F1 hybrid. The rat sample shown is from a Fischer F344 rat.
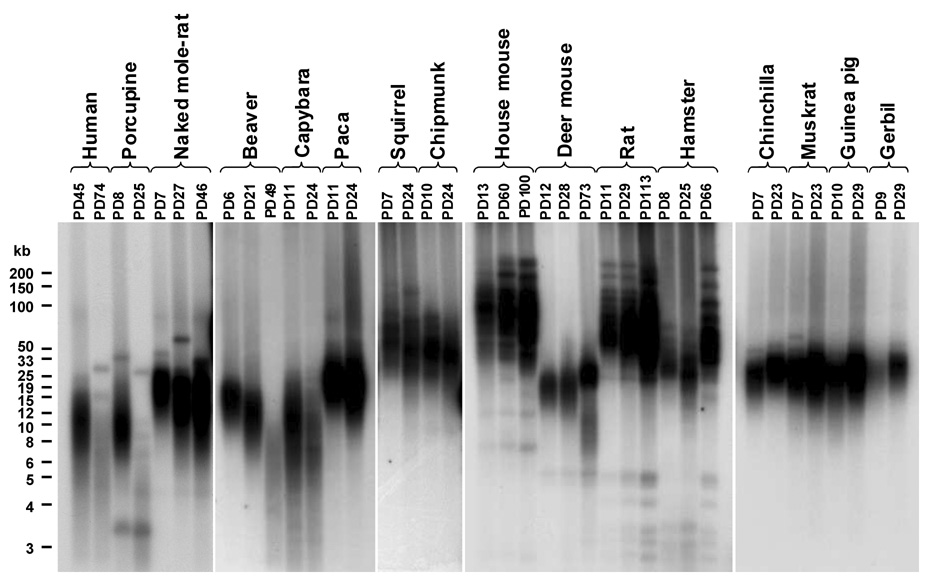
Telomere length was examined using the terminal restriction fragment (TRF) method (Figure 4). Beaver, porcupine, and capybara displayed telomere shortening with increasing passages, which is consistent with the absence of telomerase activity in these cells. Paca telomeres did not shorten. Cells from the species that express telomerase activity, did not show appreciable telomere shortening during in vitro culture.
Figure 4. Telomere length in early and late passage rodent cultures.
Telomere length was analyzed using a terminal restriction fragment assay as described in Experimental procedures. Five µg of genomic DNA is loaded in each lane. The mouse sample shown is from a 129/Sv×BL6 F1 hybrid. The rat sample shown is from a BN rat.
Inbred mouse strains have been reported to have longer telomeres than outbred strains (Bickle et al., 1998; Hemann & Greider, 2000; Manning et al., 2002). Since our collection included inbred as well as wild-caught house mice and Norway rats we compared telomere biology in these animals (Supplementary Figure 1). Fibroblasts of both inbred and wild-caught animals expressed telomerase activity, but the activity was somewhat higher in wild-caught animals. In agreement with the previous reports telomeres of the wild-caught mouse were shorter than telomeres of inbred mouse. However, telomeres of the wild-caught rat were longer than the telomeres of inbred rat. Importantly, cell growth rates did not differ between inbred and wild-caught animals indicating that using inbred mice and rats does not confound the analysis of cell growth characteristics.
Fibroblasts from large species undergo replicative senescence
Fibroblasts from beaver, porcupine, capybara, and paca entered replicative senescence (Figure 2A). Beaver cells senesced at PD39±10 (population doublings mean±range), and porcupine and paca cells senesced at PD22±3, and capybara cells senesced at PD25±8. Senescent rodent fibroblasts attained flat and enlarged morphology typical of senescent human cells, and showed positive SA-β-galactosidase staining (data not shown). As discussed above, beaver, porcupine, and capybara fibroblasts expressed no detectable telomerase activity (Figure 3), and senescence in these cells was accompanied by telomere shortening (Figure 4), strongly suggesting that the observed replicative senescence is mediated by telomere shortening. Telomere length at the time of senescence was not uniform across the species. None of the beaver, porcupine, or capybara cultures spontaneously immortalized. Paca fibroblasts did not express telomerase activity at early passages, but later became telomerase positive. Interestingly, 2 out of 5 paca cultures formed immortal clones after being kept in senescence for 3 weeks. Thus, replicative senescence in paca is less stringent than in beaver, porcupine, and capybara.
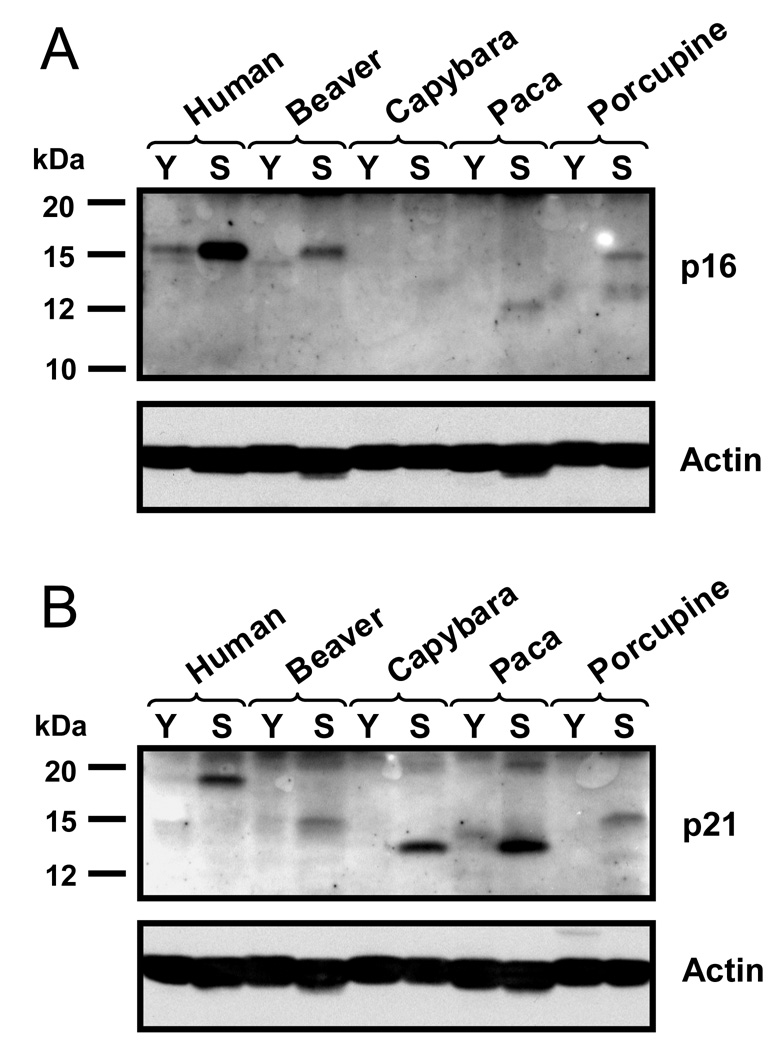
We next tested whether replicative senescence in beaver, porcupine, capybara, and paca cells is accompanied by up-regulation of p16Ink4a (p16) and p21Cip1/Waf1 (p21) cyclin dependent kinase inhibitors (CDKIs), as in human cells. Upregulation of p16 is believed to signal cell stress, while upregulation of p21 is more specific to telomere-mediated senescence (Herbig et al., 2004). Young and senescent beaver, capybara, paca, and porcupine fibroblasts were subjected to Western blot analysis with antibodies to conserved regions of p16 and p21 (Figure 5). Each protein was tested with two different antibodies, to confirm the protein identity. p16 and p21 proteins are not highly conserved, and the sizes varied among different species. p16 was strongly up-regulated in senescent beaver, paca, and porcupine fibroblasts. A very faint band was visible in senescent capybara cells, which could result from either a low amount of p16 or from poor antibody affinity. p21 was up-regulated in senescent cells of all four species. These results further confirm that large rodents use telomere-mediated replicative senescence.
Figure 5. Activation of p16 (A) and p21 (B) in senescent rodent fibroblasts.
Western blot was performed using antibodies to conserved regions of the proteins. p16 and p21 are poorly conserved and molecular weights differ between species. Y, young cells; S, senescent cells.
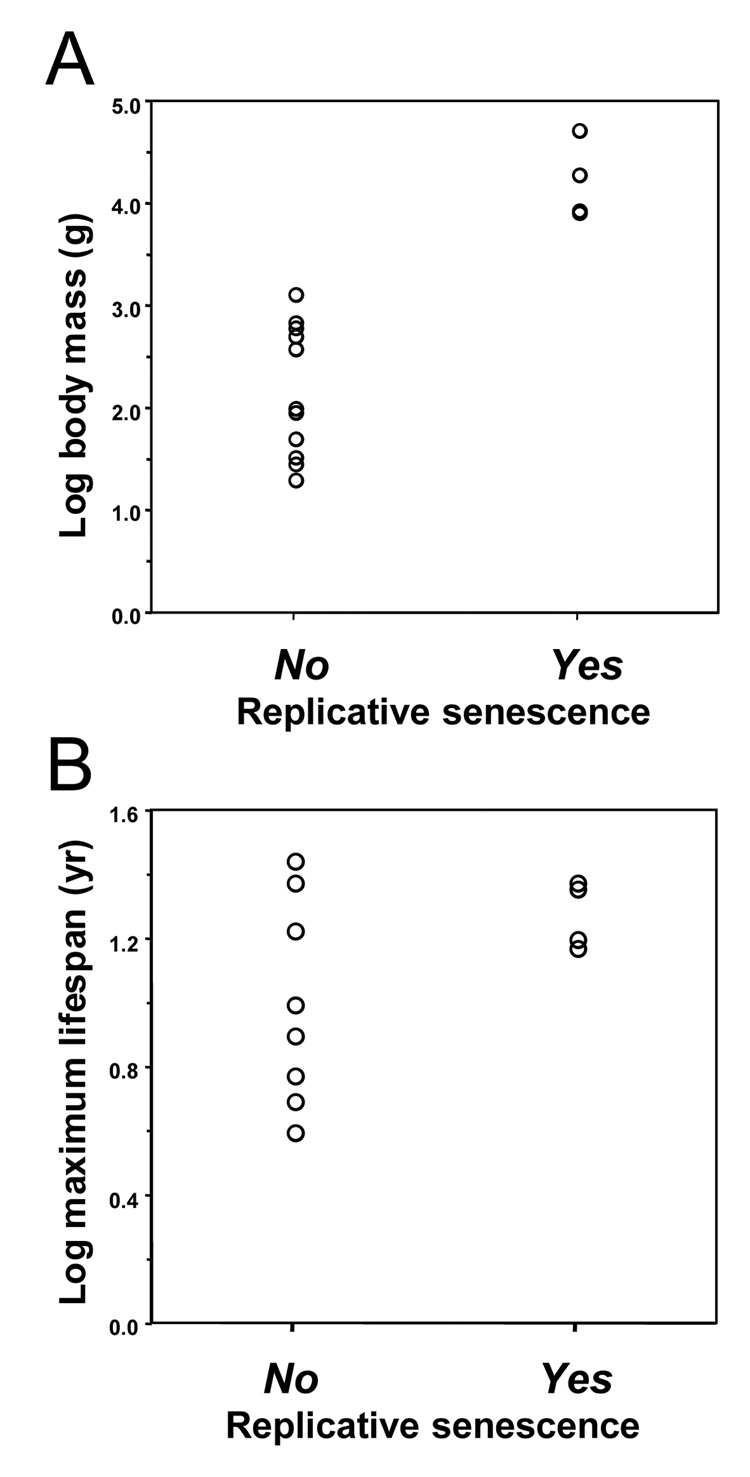
The common feature of the four rodent species, beaver, porcupine, capybara, and paca, that use replicative senescence is their large body mass (Figure 1). We therefore statistically evaluated whether the presence of replicative senescence correlates with body mass or lifespan across all 15 rodent species. These analyses reveal that replicative senescence is correlated with body mass (t=−5.87, df=13, P<0.0001) but not with maximum lifespan (t=−2.06, df=13, P=0.0606; Figure 6). To correct these analyses for phylogenetic non-independence, we analyzed phylogenetically independent contrasts (Felsenstein, 1985; Harvey & Pagel, 1991; Purvis et al., 1994). These analyses confirm that the evolutionary change in body mass is correlated with the evolution of replicative senescence (t=4.86, df=2, P=0.0398) while change in the lifespan is not (t=1.93, df=2, P=0.1939).
Figure 6. Species that display replicative senescence have larger body mass.
Relationship of replicative senescence with body mass (A), and maximum lifespan (B) was analyzed. Replicative senescence correlates with body mass (t=−5.87, df=13, P<0.0001) but not with maximum lifespan (t=−2.06, df=13, P=0.06). “No” and “Yes” indicate presence or absence of replicative senescence in the species. Plots use species data uncorrected for phylogeny.
In mammals, body mass correlates positively with longevity (Austad, 2005; Austad & Fischer, 1991; Harvey et al., 1989). It would therefore be interesting to test whether evolutionary change in body mass correlates with replicative senescence independently of lifespan (Speakman, 2005). However, since in our data set there are only three phylogenetically independent transitions from absence of replicative senescence to the presence of senescence, we do not have the statistical power to perform the appropriate multivariate analyses.
Fibroblasts from small long-lived species have a slow proliferation rate in culture
Fibroblasts from house mouse, deer mouse, Norway rat, hamster, guinea pig, gerbil, naked mole-rat, muskrat, chipmunk, chinchilla, and Eastern grey squirrel grew continuously in culture and did not enter replicative senescence (Figure 2B). Consistent with the absence of replicative senescence in these species, their fibroblasts express telomerase activity (Figure 3) and do not display appreciable telomere shortening with increasing passages (Figure 4).
These small-bodied species appear to fall into two groups according to cell proliferation rate (Figure 2B). The first group includes species with rapid in vitro cell proliferation rate: house mouse, deer mouse, Norway rat, hamster, guinea pig, and gerbil. The second group includes species with slow in vitro cell proliferation rates: naked mole-rat, muskrat, chipmunk, chinchilla, and Eastern grey squirrel. The two groups differ statistically in lifespan (Mann-Whitney test, PMWU=0.0097), with the fast group containing shorter-lived species (mean lifespan ± s.d. = 6.2 ± 2.4 years ) and the slow group containing longer-lived species (17.8 ± 8.1 years).
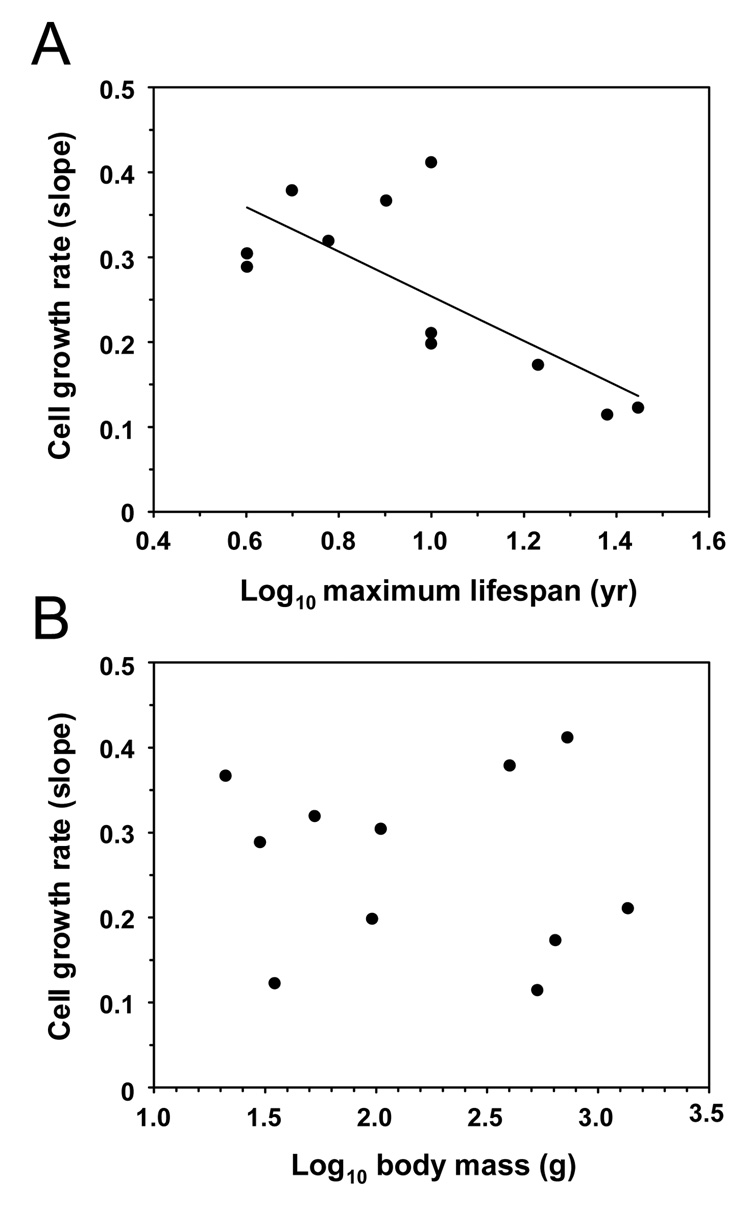
We next statistically evaluated whether fibroblast proliferation rate correlates with lifespan or body mass. Cell proliferation rates for each species were estimated from the slopes of lines fitted to the data from the first 100 days of cell culture in Figure 2B. The slopes were derived from early passages because rodent cells may undergo aneuploidization and clonal selection upon extended passaging. Fibroblast proliferation rate showed a significant negative correlation with maximum lifespan (F1,9=10.68, r2=0.54, P=0.0097) but not with body mass (F1,9=0.11, r2=0.012, P=0.746) (Figure 7). Phylogenetically independent contrasts also showed a significant negative correlation between fibroblast growth rate and maximum lifespan (F1,9=6.7, r2=0.428, P=0.029) but not body mass (F1,9=0.31, r2=0.033, P=0.593). To account for the potential contribution of the correlation between body mass and lifespan we analyzed the residuals of phylogenetically independent contrasts. Evolutionary change in lifespan controlled for body mass was significantly related to growth rate (F1,9=5.898, r2=0.396, P=0.038). (The P-value from the latter analysis should, however, be treated with caution because the heterogeneity of variances assumption was violated and could not be remedied by transformation of the data.) Thus, all three analyses of uncorrected data, of phylogenetically independent contrasts, and of residuals of phylogenetically independent contrasts confirm a significant negative correlation between in vitro fibroblast growth rate and lifespan.
Figure 7. Relationships of cell growth rate with maximum lifespan (A) and body mass (B).
Cell growth rate correlates with maximum lifespan (F1,9=10.68, r2=0.54, P=0.0097) but not body mass (F1,9=0.11, r2=0.012, P=0.746). The analysis is done for the small-bodied species only, and the species having replicative senescence are not included. Plots use species data uncorrected for phylogeny.
During in vitro culture, cells are forced into continuous cell division under non-physiological conditions. Normal cells resist this continuous growth by employing various anti-tumor mechanisms, such as replicative senescence. We propose that long-lived rodents, which do not use replicative senescence, have evolved alternative mechanisms to restrict cell proliferation that make these cells acutely sensitive to their environment, and slow their in vitro growth.
Squirrel embryonic fibroblasts have fast proliferation rate at early passages, but slow down later
To test whether cells from the third group of species are capable of rapid growth under our standard culture conditions we examined the growth of embryonic fibroblasts. We chose Eastern grey squirrel because cells from this species had the slowest growth rate, and we were able to obtain two pregnant females. Squirrel embryonic fibroblasts grew rapidly, similar to mouse fibroblasts up to PD30, after which the proliferation rate slowed to a rate similar to that of an adult squirrel culture (Figure 2C). We hypothesize that the growth control mechanisms, which restrict proliferation of squirrel cells are activated late during squirrel development. This result strongly suggests that the in vitro proliferation rates of adult cell cultures reflect physiological properties of these cells.
DISCUSSION
Our study shows that fibroblast growth characteristics have coevolved with species body mass and lifespan. Previously we showed that repression of telomerase activity in rodent tissues coevolves with large body mass in rodents (Seluanov et al., 2007). Here we extend these observations to include a correlation between larger body mass and the presence of replicative senescence: the largest rodents — beaver, capybara, porcupine, and paca — have evolved replicative senescence (Figure 8). The sizes of these species suggest that, to the level of resolution of our study, body mass greater than 8,000 g favors the evolution of replicative senescence (Figure 1). Evolutionary increases in body mass increase cancer risk, as larger animals contain more cells in their body, and any cell may potentially turn cancerous. Increased mortality rate due to cancer then drives the adaptive evolution of repression of telomerase activity in species with large body mass. It has been predicted that larger animals should have multiple, redundant tumor-suppressor systems so that their cells require more steps for tumor formation (Graham, 1983; Leroi et al., 2003; Nunney, 1999). Our data on rodents provide experimental support for this theory.
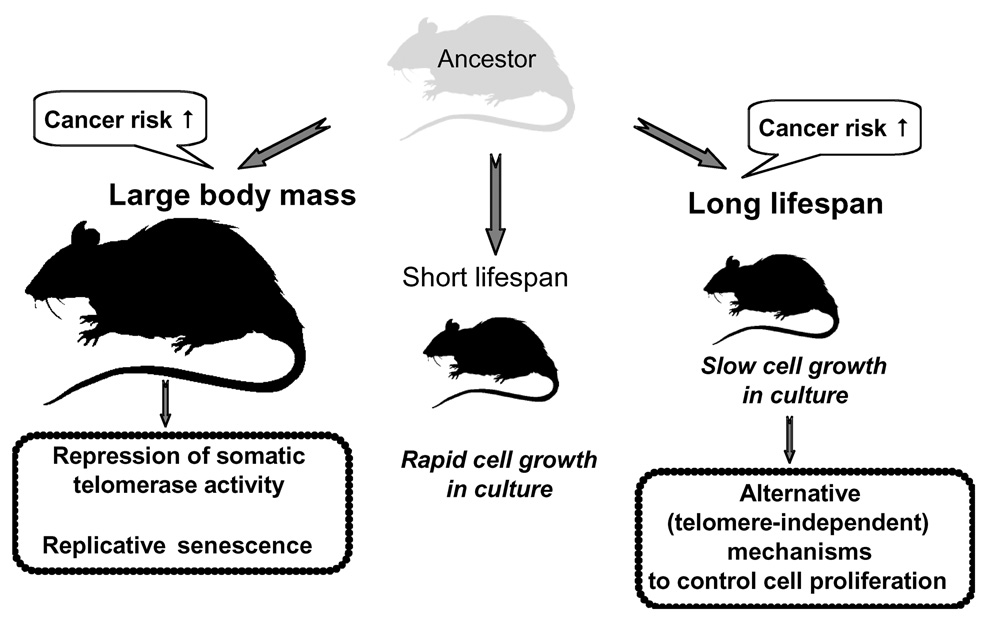
Figure 8. Coevolution of body mass, lifespan and tumor-suppressor mechanisms.
Increase in body mass leads to increased cancer risk and evolution of replicative senescence. Small, short-lived rodent species have lower cancer risk and fewer tumor-suppressors. Fibroblasts from these species grow rapidly in culture. An increase in lifespan in small rodents also increases cancer risk and drives evolution of alternative tumor-suppressor mechanisms, which restrict cell growth in vitro.
Our study is focused exclusively on rodent species and it remains unclear if the same rules apply to other groups of mammals. Telomere biology in mammals studied so far supports the model of coevolution of replicative senescence and body mass (reviewed in (Gorbunova & Seluanov, 2008)). For instance, large mammals such as cow, sheep, and horse do not express telomerase in somatic tissues, and their fibroblasts have finite lifespan in culture (Argyle et al., 2003; Davis et al., 2005; Hornsby et al., 1986). Replicative senescence has also been documented in large primates such as chimpanzee, orangutan, gorilla, baboon, and several macaque species (Gardner et al., 2007; Herbig et al., 2006; Steinert et al., 2002). An intermediate situation where fibroblasts do not express telomerase activity but their cultures fail to undergo growth arrest has been detected in smaller species, such as rabbits and the ring-tailed lemur (Forsyth et al., 2005; Steinert et al., 2002). A general trend seems to be that mammals with a body mass greater than 8,000 g evolve stringent replicative senescence, mammals smaller than 2,000 g do not use replicative senescence, and species with body mass between 2,000 and 8,000 g display a spectrum of intermediate phenotypes.
Even less is known about telomere biology in birds. Long-lived species, such as the storm petrel and common tern, express telomerase throughout their lives (Haussmann et al., 2007). Both of these species are small with an adult body mass below 200 g. Telomerase activity has also been detected in the somatic tissues of a larger bird, the domestic chicken (Venkatesan & Price, 1998). However, chicken fibroblasts do not express telomerase and undergo a clear-cut replicative senescence (Dinowitz, 1977; Ponten, 1970). Little is known about telomere biology in the largest bird species such as ostrich or emperor penguin. The number of population doublings before senescence has been correlated to maximum lifespan (Rohme, 1981), or when phylogenetic correction was applied, to species body mass (Lorenzini et al., 2005). Our data set does not allow for such analysis as only four species undergo replicative senescence with three of them showing similar replicative lifespans.
Long lifespan, like body mass, is expected to increase cancer risk. It may therefore seem puzzling that the long-lived rodents in our study have not evolved replicative senescence. While being a potent tumor suppressor, replicative senescence has many tradeoffs, such as slower wound healing and less robust immune response. Furthermore, replicative senescence is only one of many possible tumor-suppressor mechanisms, and it is plausible that these species rely on other mechanisms to mitigate the cancer risk conferred by their long lifespan. Indeed we found that fibroblasts of small, long-lived species such as grey squirrel, naked mole-rat, chinchilla, musk-rat, and chipmunk exhibit a novel in vitro phenotype: their cells do not enter replicative senescence but instead proliferate slowly in culture (Figure 8). We show that, for small rodent species that have not evolved replicative senescence, in vitro fibroblast proliferation rate negatively correlates with longevity. We hypothesize that the slow in vitro growth rate is a manifestation of those alternative tumor suppressor mechanisms that evolve in small, long-lived species lacking replicative senescence (Figure 8).
Interestingly, embryonic squirrel fibroblasts, proliferated rapidly up to PD30, after which the culture slowed down and attained the adult growth phenotype. Thus, the growth control mechanisms that restrict proliferation of squirrel cells are characteristic of an adult but not embryonic cells. This scenario is reminiscent of the repression of hTERT expression during human embryonic development. Telomerase is expressed in early embryogenesis, but its expression is progressively shut off in later development (Bekaert et al., 2004). Thus, the mechanisms that restrict proliferation of somatic cells are inactive during early development when they would otherwise interfere with rapid cell division.
In vitro culture forces cells to proliferate under non-physiological conditions. In vivo the majority of cells in adult tissues are non-dividing. When placed in culture, cells are stimulated to divide by mitogens provided by fetal serum. The ability of cells to proliferate in culture dishes can be considered a measure of their tumorigenic potential. We hypothesize that long-lived rodents evolve mechanisms that make their cells acutely sensitive to any environmental or physiological imbalances, and arrest cell proliferation in inappropriate conditions. The same mechanisms will prevent inappropriate cell division in vivo, protecting the organism from tumor growth and metastasis.
What are the potential cues that slow proliferation of adult fibroblasts of small long-lived rodents? Perhaps, it could be unrestrained mitogenic stimulation, disrupted cell-cell contacts, sensitivity to DNA damage, or some other as yet undetermined cue. It seems likely that these proliferation control mechanisms differ among species because, as can be inferred from rodent phylogeny (Figure 1), they have evolved with slow aging independently at least three times. In-depth studies of the individual small long-lived species are required to understand the molecular mechanisms responsible for the different anti-cancer adaptations that have evolved among them.
In summary, our analysis of fifteen rodent species has uncovered an intricate picture of how increased cancer risk conferred by large body mass or long lifespan drives evolution of tumor suppressor mechanisms (Figure 8). Body mass has previously been linked to the evolution of several characteristics such as repression of telomerase activity (Seluanov et al., 2007), more efficient DNA repair (Promislow, 1994), and the number of population doublings before senescence (Lorenzini et al., 2005). Here we show that both body mass and lifespan contribute to the evolution of tumor suppressor mechanisms, but in two different ways. Large body mass coevolved with replicative senescence, while long lifespan is associated with evolution of alternative mechanisms that increase the sensitivity of the cells to growth conditions and slow cell proliferation in culture.
EXPERIMENTAL PROCEDURES
Animal samples
Capybaras were obtained from Bio Fau Assesoria e Comercio (São Paulo, Brazil). Pacas were from the animal facility at São Paulo State University. Outbred multicolored guinea pigs were purchased from Elmhill Labs. Chinchillas were purchased from Molton Chinchilla Ranch. Naked mole-rats were from the colony of K.C.C. at Vanderbilt University. Two mice were 129/Sv×BL6 F1 hybrids, two mice were BL6, and one mouse was wild-caught in New York State. Two rats were BN, two rats were F344, and one rat was wild caught in New York State. Outbred Mongolian gerbils Crl:Mon(Tum) and outbred hamsters Crl:LVG(Syr) were purchased from Charles River Laboratories. Beavers, deer mice, muskrats, chipmunks, and squirrels were wild-trapped in New York State. All animals used in this study were young adults. Exact age was known for laboratory animals and was estimated for wild-caught animals from body measurements and colour. Live animals were euthanised according to the University of Rochester Animal Care and Use Committee guidelines. Care was taken to minimize pain and discomfort to the animals.
Cell isolation and culture
Primary rodent cells were isolated from lung and under arm skin. We used cell isolation protocol adapted from S. Austad (University of Texas Health Sciences Center, San Antonio). Skin was shaved and cleaned with 70% ethanol. Tissue was minced and incubated in DMEM F-12 media (Gibco) with collagenase (Liberase Blendzyme 3, Roche) at 37°C on a stirrer for 30–90 min. Dissociated cells were then washed, and placed in tissue culture dishes with DMEM F-12 media (Gibco) and antibiotics/antimycotic (Gibco). All cells except naked mole-rat were cultured at 37°C, 5% CO2, 3% O2; naked mole-rat cells were cultured at 32°C, 5% CO2, 3% O2.
After the plates became confluent, cells were replated, and all subsequent culture was performed in EMEM media (ATCC) supplemented with 15% fetal bovine serum (Gibco), nonessential amino acids, 100 units/ml penicillin, and 100 µg/ml streptomycin (Gibco). All the experiments were performed using the same batch of fetal bovine serum. To obtain cell growth curves cells were replated regularly at 70–80% confluence and the numbers of cells plated and harvested were recorded and used to calculate the PDs.
Telomeric Repeat Amplification Protocol
TRAP assay was performed according to a modified protocol described in (Szatmari & Aradi, 2001). Briefly, this protocol uses modified oligonucleotide sequences to ensure that the number of telomeric repeats present in the original telomerase products does not change during PCR amplification. In the first step of the TRAP assay, substrate oligonucleotide is incubated with 0.5 µg of protein extract. If telomerase is present and active, telomeric repeats (GGTTAG) are added to the 3’ end of the oligonucleotide. In the second step, extended products are amplified by PCR in the presence of hot dCTP. A human cancer cell line over-expressing telomerase was used as a reference in each assay.
Telomere lengths
Telomere length was analyzed by Southern blot using the Terminal Restriction Fragment (TRF) method. Genomic DNA was extracted from fibroblasts, 5 µg of DNA was digested overnight with a mixture of AluI, HaeIII, RsaI, and HinfI (restriction enzymes that do not cut within telomeric repeat sequence), and separated using pulse field gel electrophoresis. Dried gels were hybridized with radiolabeled oligonucleotide containing telomeric sequence (TTAGGG)3. Pulse field gels were 1% agarose, and were run in 0.5X TBE using a CHEF-DR II apparatus (Bio-Rad) for 22 h at constant 150V, using ramped pulse times from 1 to 10 s, with a ratio 1.0, at 8°C.
Western Blot
Cells were harvested, washed in PBS, and boiled in 1x Laemmli sample buffer with protease inhibitors (Compleat Mini, Roche) for 10 min with vortexing every 3 min. Extracts were incubated at room temperature for 2 min, and centrifuged at 10,000 g for 10 min. Protein concentrations in the extracts were measured according to modified Lowry procedure (RC DC Protein Assay, Bio-Rad) using bovine serum albumin as a standard. An aliquot (50 µg of proteins) from each sample was then separated on 12% to SDS/PAGE. After electroblotting, the nitrocellulose membrane was incubated with anti-p16 or anti-p21 serum as indicated, washed twice, and incubated with goat anti-rabbit antibodies conjugated to horseradish peroxidase. Identity of each protein was confirmed with two different antibodies: p16, ab14244 and ab17517; p21, ab14061 and ab7960 (Abcam). Equal loading of samples was verified by hybridizing the same membranes with pan-Actin Ab-5 antibodies (Lab Vision).
Statistical analysis
The phylogenetic relationships among the 15 species studied here were inferred from information from six studies (Adkins et al., 2003; Martin et al., 2000; Michaux et al., 2001; Montgelard et al., 2002; Murphy et al., 2001; Steppan et al., 2004), which were based on a sequence analysis of 22 genes. We used Felsenstein’s method (Felsenstein, 1985) of analyzing phylogenetically independent contrasts to test for correlated evolution between replicative senescence, fibroblast proliferation rate, maximum lifespan, and body mass. We generated standardized independent contrasts for phenotypic values using the CRUNCH algorithm of the Comparative Analysis by Independent Contrasts (CAIC) program (Purvis & Rambaut, 1995). We were unable to infer branch lengths for the tree because no common molecular phylogeny exists of all 15 species. We therefore assumed equal branch lengths, consistent with a punctuational model of evolution (Purvis et al., 1994). To test for correlated evolution, linear and multiple regression models forced through the origin were fit to independent contrasts (Harvey & Pagel, 1991). For all analyses, the statistical and evolutionary assumptions of the independent contrasts methods were met (Purvis & Rambaut, 1995). Normality was tested using the Shapiro-Wilk test implemented in R. In analyses of the uncorrected data, body mass was log-transformed to meet the assumption of normality. All statistical tests are two-tailed.
Supplementary Material
Comparison of telomere biology and cell growth kinetics in wild-caught and laboratory mice and rats. (A) Telomerase activity examined by TRAP assay in early and late passage fibroblasts. (B) Telomere length analyzed by terminal restriction fragment assay. (C) Growth curves of primary fibroblasts. Mouse and rat averages (Figure 2B) are compared to the individual growth curves of the wild-caught mouse and rat. LF and SF correspond to lung and skin fibroblasts respectively.
Acknowledgements
We are grateful to Doug Higgins, Hiram Lyon, Linn Sajdak, Lidza Kalifa, Ingrid Sarelius, Coeli Lopes, George Brady, and John Ryan for providing animal tissue and carcasses. This work was supported by grants from US National Institutes of Health, Ellison Medical Foundation, American Federation for Aging Research, and Komen Breast Cancer Foundation to V. G., and Ellison Medical Foundation grant to A.S.
Contributor Information
Andrei Seluanov, Department of Biology, University of Rochester, Rochester, New York 14627, USA.
Christopher Hine, Department of Biology, University of Rochester, Rochester, New York 14627, USA.
Michael Bozzella, Department of Biology, University of Rochester, Rochester, New York 14627, USA.
Amelia Hall, Department of Biology, University of Rochester, Rochester, New York 14627, USA.
Tais H. C. Sasahara, Department of Surgery, LSSCA, College of Veterinary Medicine, University of São Paulo, São Paulo, Brazil.
Antonio A. C. M. Ribeiro, Department of Surgery, LSSCA, College of Veterinary Medicine, University of São Paulo, São Paulo, Brazil.
Kenneth C. Catania, Department of Biological Sciences, Vanderbilt University, Nashville, TN 37232, U.S.A.
Daven C. Presgraves, Department of Biology, University of Rochester, Rochester, New York 14627, USA
Vera Gorbunova, Department of Biology, University of Rochester, Rochester, New York 14627, USA.
REFERENCES
- Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 2003;26:409–420. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- Argyle D, Ellsmore V, Gault EA, Munro AF, Nasir L. Equine telomeres and telomerase in cellular immortalisation and ageing. Mech Ageing Dev. 2003;124:759–764. doi: 10.1016/s0047-6374(03)00104-0. [DOI] [PubMed] [Google Scholar]
- Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Bekaert S, Derradji H, Baatout S. Telomere biology in mammalian germ cells and during development. Dev Biol. 2004;274:15–30. doi: 10.1016/j.ydbio.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bickle C, Cantrell M, Austad S, Wichman H. Effects of domestication on telomere length in Mus musculus and Peromyscus maniculatus. FASEB J. 1998;12:A317. [Google Scholar]
- Buffenstein R, Jarvis JU. The naked mole rat--a new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002;2002:pe7. doi: 10.1126/sageke.2002.21.pe7. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- Davis T, Kipling D. Telomeres and telomerase biology in vertebrates: progress towards a non-human model for replicative senescence and ageing. Biogerontology. 2005;6:371–385. doi: 10.1007/s10522-005-4901-4. [DOI] [PubMed] [Google Scholar]
- Davis T, Skinner JW, Faragher RG, Jones CJ, Kipling D. Replicative senescence in sheep fibroblasts is a p53 dependent process. Exp Gerontol. 2005;40:17–26. doi: 10.1016/j.exger.2004.09.004. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinowitz M. A continuous line of Rous sarcoma virus-transformed chick embryo cells. J Natl Cancer Inst. 1977;58:307–312. doi: 10.1093/jnci/58.2.307. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Forsyth NR, Elder FF, Shay JW, Wright WE. Lagomorphs (rabbits, pikas and hares) do not use telomere-directed replicative aging in vitro. Mech Ageing Dev. 2005;126:685–691. doi: 10.1016/j.mad.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Gardner JP, Kimura M, Chai W, Durrani JF, Tchakmakjian L, Cao X, Lu X, Li G, Peppas AP, Skurnick J, Wright WE, Shay JW, Aviv A. Telomere dynamics in macaques and humans. J Gerontol A Biol Sci Med Sci. 2007;62:367–374. doi: 10.1093/gerona/62.4.367. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A. Coevolution of telomerase activity and body mass in mammals:from mice to beavers. Mech Ageing Dev. 2008 doi: 10.1016/j.mad.2008.02.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. Cancer and evolution: synthesis. J Theor Biol. 1983;101:657–659. doi: 10.1016/0022-5193(83)90021-8. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. Oxford: Oxford University Press; 1991. [Google Scholar]
- Harvey PH, Read AF, Promislow DEL. Life history variation in placental mammals: unifying the data with theory. Oxf. Surv. Evol. Biol. 1989;6:13–31. [Google Scholar]
- Haussmann MF, Winkler DW, Huntington CE, Nisbet IC, Vleck CM. Telomerase activity is maintained throughout the lifespan of long-lived birds. Exp Gerontol. 2007;42:610–618. doi: 10.1016/j.exger.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Greider CW. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- Hornsby PJ, Aldern KA, Harris SE. Clonal variation in response to adrenocorticotropin in cultured bovine adrenocortical cells: relationship to senescence. J Cell Physiol. 1986;129:395–402. doi: 10.1002/jcp.1041290319. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Koufopanou V, Burt A. Cancer selection. Nat Rev Cancer. 2003;3:226–231. doi: 10.1038/nrc1016. [DOI] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 2005;126:1130–1133. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Manning EL, Crossland J, Dewey MJ, Van Zant G. Influences of inbreeding and genetics on telomere length in mice. Mamm Genome. 2002;13:234–238. doi: 10.1007/s003350020027. [DOI] [PubMed] [Google Scholar]
- Martin Y, Gerlach G, Schlotterer C, Meyer A. Molecular phylogeny of European muroid rodents based on complete cytochrome b sequences. Mol Phylogenet Evol. 2000;16:37–47. doi: 10.1006/mpev.1999.0760. [DOI] [PubMed] [Google Scholar]
- Michaux J, Reyes A, Catzeflis F. Evolutionary history of the most speciose mammals: molecular phylogeny of muroid rodents. Mol Biol Evol. 2001;18:2017–2031. doi: 10.1093/oxfordjournals.molbev.a003743. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Bentz S, Tirard C, Verneau O, Catzeflis FM. Molecular systematics of sciurognathi (rodentia): the mitochondrial cytochrome b and 12S rRNA genes support the Anomaluroidea (Pedetidae and Anomaluridae) Mol Phylogenet Evol. 2002;22:220–233. doi: 10.1006/mpev.2001.1056. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Nowak R. Walker's mammals of the world. Baltimore: John Hopkins University Press; 1999. [Google Scholar]
- Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc Biol Sci. 1999;266:493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten J. The growth capacity of normal and Rous-virus-transformed chicken fibroblasts in vitro. Int J Cancer. 1970;6:323–332. doi: 10.1002/ijc.2910060302. [DOI] [PubMed] [Google Scholar]
- Promislow DE. DNA repair and the evolution of longevity: a critical analysis. J Theor Biol. 1994;170:291–300. doi: 10.1006/jtbi.1994.1190. [DOI] [PubMed] [Google Scholar]
- Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci U S A. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Gittleman JL, Luh H-K. Truth or consequences: effects of phylogenetic accuracy on two comparative methods. J. Theor. Biol. 1994;167:293–300. [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrast (CAIC): an Apple Macintosh application for analysing comparative data. Comp. Appl. Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Rohme D. Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc. Natl. Acad. Sci. USA. 1981;78:5009–5013. doi: 10.1073/pnas.78.8.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM. Can ends justify the means? telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc Natl Acad Sci U S A. 1998;95:9078–9081. doi: 10.1073/pnas.95.16.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM. Telomeres limit cancer growth by inducing senescence: long-sought in vivo evidence obtained. Cancer Cell. 2007;11:389–391. doi: 10.1016/j.ccr.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan--two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp Cell Res. 2002;272:146–152. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Szatmari I, Aradi J. Telomeric repeat amplification, without shortening or lengthening of the telomerase products: a method to analyze the processivity of telomerase enzyme. Nucleic Acids Res. 2001;29:E3. doi: 10.1093/nar/29.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–B501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Venkatesan RN, Price C. Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc Natl Acad Sci U S A. 1998;95:14763–14768. doi: 10.1073/pnas.95.25.14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigl R. Longevity of mammals in captivity; from the living collections of the world. Stuttgart: Schweizerbart; 2005. [Google Scholar]
- Wright W, Shay J. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nature Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of telomere biology and cell growth kinetics in wild-caught and laboratory mice and rats. (A) Telomerase activity examined by TRAP assay in early and late passage fibroblasts. (B) Telomere length analyzed by terminal restriction fragment assay. (C) Growth curves of primary fibroblasts. Mouse and rat averages (Figure 2B) are compared to the individual growth curves of the wild-caught mouse and rat. LF and SF correspond to lung and skin fibroblasts respectively.