Abstract
Ubiquilins (UBQLNs) are adaptor proteins thought to deliver ubiquitinated substrates to proteasomes. Here, we show a role for UBQLN in autophagy: enforced expression of UBQLN protects cells from starvation-induced death, whereas depletion of UBQLN renders cells more susceptible. The UBQLN protective effect requires the autophagy-related genes ATG5 and ATG7, two essential components of autophagy. The ubiquitin-associated domain of UBQLN mediates both its association with autophagosomes and its protective effect against starvation. Depletion of UBQLN delays the delivery of autophagosomes to lysosomes. This study identifies a new role for UBQLN in regulating the maturation of autophagy, expanding the involvement of ubiquitin-related proteins in this process.
Keywords: ubiquilin, ubiquitin-like, PLIC, autophagy, autophagosomes
Introduction
The family of proteins known as proteins linking IAP (CD47) to cytoskeleton (PLIC), or ubiquilins (UBQLNs), have partial homology to ubiquitin. They have a ubiquitin-like (UBL) domain at the amino terminus, which binds to ubiquitin-interacting motifs (UIMs) expressed by proteasomes (Walters et al, 2002) and endocytic adaptors (Hofmann & Falquet, 2001). A carboxy-terminal ubiquitin-associated (UBA) domain interacts with ubiquitin chains present on proteins destined for proteasomal degradation (Kleijnen et al, 2000; Mah et al, 2000; Funakoshi et al, 2002; Feng et al, 2004). Unlike ubiquitin, UBQLNs do not covalently tag proteins, although they are thought to regulate the proteasomal degradation of ubiquitin conjugates (Kleijnen et al, 2000; Mah et al, 2000). UBQLNs also regulate the function of the thrombospondin receptor CD47 (Wu et al, 1999) and G protein signalling (N'Diaye & Brown, 2003), and might also affect endocytosis through interaction with UIM-containing proteins (Regan-Klapisz et al, 2005; N'Diaye et al, 2008); in addition, UBQLNs might interact with the kinase mammalian target of rapamycin (mTOR; Wu et al, 2002). Although mostly cytosolic, a fraction of UBQLN associates with cell membranes (Wu et al, 1999), although the membrane-bound compartments with which UBQLN associates have not been identified.
Autophagic pathways are alternative mechanisms to proteasomes for the degradation of cytoplasmic contents. Macroautophagy (herein called autophagy) allows cells to digest cytoplasmic constituents, including organelles (Klionsky & Emr, 2000). When autophagy is induced, a double membrane formed in the cytoplasm elongates, surrounds and ultimately seals off a region of the cytoplasm. This compartment, called the autophagosome, fuses with lysosomes, leading to the degradation of its content by lysosomal enzymes (Klionsky & Emr, 2000). In addition to soluble proteins and organelles, autophagy can degrade insoluble inclusions (Williams et al, 2006) and even bacteria (Deretic, 2005). Many biological functions have been linked to autophagy, including nutrient recycling for prolonged survival during starvation (Kuma et al, 2004), cytoprotection through the removal of aggregates in some neurodegenerative diseases (Williams et al, 2006), antimicrobial activity and antigen presentation in immunity (Deretic, 2005; Munz, 2006). This study shows that UBQLNs regulate starvation-induced autophagy at the step of lysosomal degradation, and expands the role of ubiquitin-binding proteins in the regulation of autophagy.
Results And Discussion
UBQLN regulates starvation-induced cell death
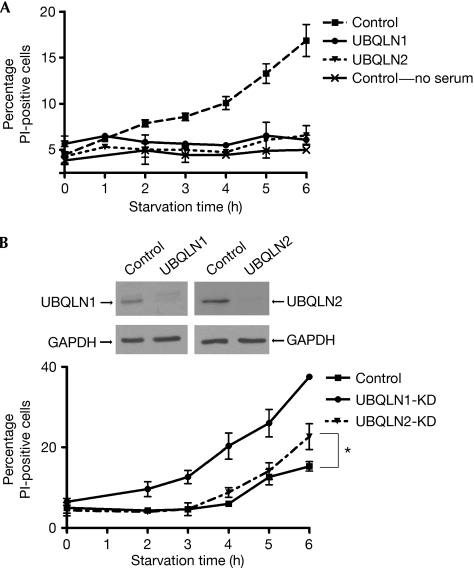
Upon starvation, HeLa cells transfected with green fluorescent protein (GFP; control cells) showed increased propidium iodide (PI) staining (a marker of cell death; Fig 1A). This was not due to the absence of fetal calf serum, as the removal of serum alone did not induce cell death (Fig 1A). In contrast to control cells, UBQLN1- or UBQLN2-transfected cells did not show any increase in PI staining (Fig 1A). However, short interfering RNA (siRNA)-mediated depletion of each protein, which did not significantly affect the other isoform (N'Diaye et al, 2008), rendered cells more susceptible to starvation, as indicated by an increase in cell death compared with control cells (Fig 1B). The effect of UBQLN1 depletion was more pronounced than that of UBQLN2, which might reflect differences in endogenous expression. Nonetheless, both overexpression and depletion experiments indicate that UBQLN regulates cell survival during starvation.
Figure 1.
Ubiquilins protect against starvation-induced cell death. (A) HeLa cells expressing GFP-tagged UBQLN1 or UBQLN2 (or GFP for controls) were transferred to starvation medium (PBS) for the indicated time. Where specified, only serum was removed from the culture medium. Viability of transfected cells was assessed by PI staining and analysed by flow cytometry. The graph represents the mean of three experiments±s.e. (B) HeLa cells were depleted of UBQLN1 or UBQLN2 by siRNA transfection; depletion was verified by Western blot. Cells were starved for increasing periods of time, and their viability was analysed as in (A). The graph represents the mean of three experiments±s.e; *P<0.05 between control and UBQLN2-KD at 6 h (paired Student's t-test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; KD, knockdown; PI, propidium iodide; siRNA, short interfering RNA; UBQLN, ubiquilin.
UBQLN cytoprotective effect requires autophagy
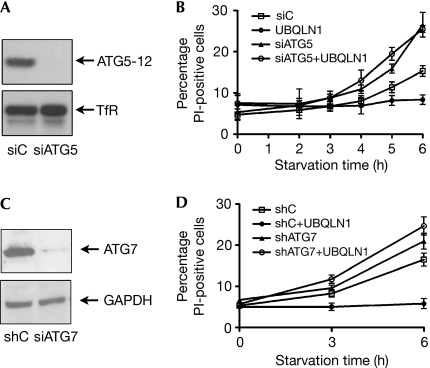
To determine the role of autophagy in the survival of HeLa cells during starvation, we depleted autophagy related gene 5 (ATG5), an essential component of autophagy (Kuma et al, 2004; Fig 2A). Depletion of ATG5 rendered cells more susceptible to starvation (Fig 2B); thus, under these conditions, ATG5 functions as an autophagy-dependent survival factor, rather than as a pro-apoptotic signalling molecule (Yousefi et al, 2006). It is also consistent with the observation that the inhibition of autophagy sensitizes cells to starvation-induced cell death (Boya et al, 2005). Importantly, depletion of ATG5 abolished the UBQLN1-protective effect, as UBQLN1 transfection did not enhance survival in ATG5-knockdown (KD) cells, in contrast to its effect in control cells (Fig 2A,B). Depletion of ATG7 (Fig 2C), another molecule essential for autophagy (Komatsu et al, 2005), also decreased cell resistance to starvation (Fig 2D) and abrogated the ability of UBQLN1 to prolong cell viability (Fig 2D). Thus, UBQLN protection against starvation requires autophagy.
Figure 2.
The ubiquilin-protective effect against starvation requires autophagy. (A) HeLa cells were transfected with siRNAs for autophagy-related gene ATG5 (siATG5) or with a non-silencing siRNA (siC). Depletion of ATG5, which decreased the levels of the ATG5–ATG12 covalent complex, was confirmed by Western blot. Loading control: transferrin receptor (TfR). (B) Control or ATG5-KD cells transfected with UBQLN1 were starved, and the cell viability was assessed as in Fig 1; the graph represents the mean of three experiments±s.e. (C) HeLa cells were stably transfected with control (shC) or ATG7 (shATG7) shRNA; depletion was verified by Western blot (loading control: GAPDH). (D) Control or ATG7-KD cells transfected with UBQLN1 were starved for 3 or 6 h and assessed for their viability. The graph represents the mean of three experiments±s.e. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KD, knockdown; Pl, propidium iodide; shRNA, short hairpin RNA; siRNA, short interfering RNA; UBQLN, ubiquilin.
UBQLNs interact with autophagosomes
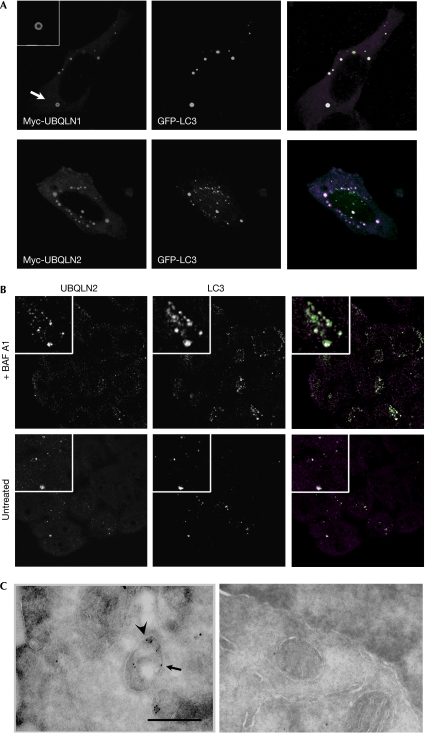
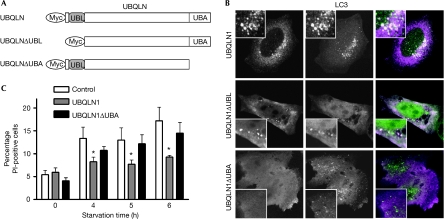
Immunostaining showed that both Myc-tagged UBQLN1 and UBQLN2 had concentrated into ring-like structures (Fig 3A, insets), suggesting that these were vesicles; this was supported further by the occurrence of homotypic fusion events (supplementary Movie S1 online). Staining for endocytic markers was performed, as we and others have shown association of UBQLN with endocytic proteins (Regan-Klapisz et al, 2005; N'Diaye et al, 2008). UBQLN2 vesicles did not localize with the early endosomal marker EEA1 (supplementary Fig S2 online); however, they were often found close to lysosome-associated membrane protein 1 (LAMP1)- or Ras-like protein in rat brain 7 (RAB7)-positive compartments (supplementary Fig S2 online, insets), suggesting their interaction with lysosomes. No colocalization was found with RAB11 or Golgi markers (data not shown); by contrast, extensive colocalization was found with vesicular GFP-tagged microtubule-associated protein 1 light chain 3 (GFP-LC3), a marker for autophagosomes (Fig 3A). UBQLNs and LC3 also colocalized in MCF-10A cells stably expressing GFP-LC3 (Fung et al, 2008; data not shown) and at endogenous levels (Fig 3B), which ruled out an artefact of overexpression. Immunoelectron microscopy also showed double membrane compartments labelled with both LC3 and UBQLN2 (Fig 3C). Deletion of the UBQLN1 UBL domain (Fig 4A) had no effect on its interaction with LC3 (Fig 4B); by contrast, deletion of the UBA domain abolished its association with LC3 (Fig 4B). Similar observations were made with UBQLN2 mutants (supplementary Fig S3 online). The UBA domain, which is highly conserved between UBQLN1 and UBQLN2, interacts with ubiquitinated proteins (Kleijnen et al, 2000; Mah et al, 2000; Funakoshi et al, 2002; Feng et al, 2004) and might also mediate homodimerization (Feng et al, 2004). Therefore, its requirement for UBQLN interaction with autophagosomes suggests that ubiquitinated proteins might be involved and that multimerization might be required. Deletion of the UBA domain also abrogated the UBQLN1-protective effect against starvation (Fig 4C), suggesting that resistance to starvation-induced cell death requires the interaction of UBQLN with autophagosomes.
Figure 3.
Ubiquilins colocalize with autophagosomes. (A) Immunolocalization of transfected Myc-UBQLN (1 or 2) and GFP-tagged microtubule-associated protein 1 light chain 3 (GFP-LC3) was analysed by microscopy. Inset: magnification of UBQLN1 vesicular staining (arrow), which, after acquisition of Z-stacks, was found in all UBQLN structures analysed. (B) Cells were starved for 2 h, with (top row) or without BAF A1 (bottom row). Cells were stained for endogenous UBQLN2 (magenta) and LC3 (green). (C) Control cells (untransfected, right panel) or cells overexpressing Myc-UBQLN2 and GFP-LC3 (left panel) were transferred into starvation medium for 2 h. Localization of Myc-UBQLN2 and GFP-LC3 was analysed by immunoelectron microscopy using Myc (arrow) and GFP (arrowhead) antibodies coupled to 10 and 15 nm gold particles, respectively. Double-membrane vacuoles (autophagosomes) in transfected cells (left panel) show increased staining by both antibodies compared with autophagosomes in untransfected cells (right panel). Scale bar, 0.5 μm. BAF A, bafilomycin A1; GFP, green fluorescent protein; UBQLN, ubiquilin.
Figure 4.
The UBA domain of ubiquilin is required for interaction with autophagosomes and a protective effect against starvation. (A) Generation of Myc-tagged UBQLN1 deletion mutants. (B) Each mutant was co-transfected with GFP-tagged microtubule-associated protein 1 light chain 3 (GFP-LC3) and the localization of each protein was analysed by microscopy. (C) Starvation-induced cell death was assessed in cells expressing UBQLN1 or UBQLN1ΔUBA. The graph represents the mean of three experiments±s.e., with *P⩽0.05 (P=0.0017 by one-way ANOVA test, comparing control, UBQLN1 and UBQLN1ΔUBA). No difference was found between control and UBQLN1ΔUBA at any time point. ANOVA, analysis of variance; GFP, green fluorescent protein; Pl, propidium iodide; UBA, ubiquitin-associated; UBQLN, ubiquilin.
The association of UBQLN with autophagy was substantiated further by its interaction with protein aggregates, which are known substrates of autophagy. Aggregated huntingtin (Arrasate et al, 2004) and GFP-250 (an aggregate-prone chimaeric protein formed by the fusion of GFP to a mutant of the membrane transporter p115; Garcia-Mata et al, 1999) both recruited UBQLN (supplementary Fig S4 online). Elegant studies in animal models have shown a protective effect for UBQLN1 against the cytotoxicity of huntingtin (HTT) aggregates (Wang et al, 2006); however, the mechanism underlying the effect of UBQLN was not explained. This study identifies UBQLN as a link between autophagy and its substrates; furthermore, GFP-250 does not get ubiquitinated (Garcia-Mata et al, 1999), whereas HTT does (Jana et al, 2005). This suggests that although ubiquitination might mediate the association of UBQLN with autophagosomes, either directly or through a protein localized on autophagosomes, ubiquitination of the substrate is not required in all cases.
Depletion of UBQLN inhibits autophagosome degradation
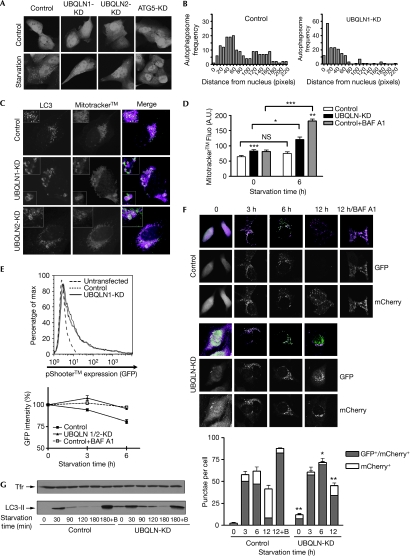
Next, we studied the role of UBQLN in the formation and transport of autophagosomes during starvation. Depletion of each UBQLN did not prevent autophagosome formation, which was in contrast to the depletion of ATG5 used as a control (Fig 5A). However, depletion of UBQLN redistributed LC3 vesicles to the perinuclear region (Fig 5A,B) similar to the effect of bafilomycin A1 (BAF A), which inhibits autophagosome–lysosome fusion (Yamamoto et al, 1998). This was not due to a smaller size of UBQLN-KD cells (supplementary Fig S5 online). Interestingly, BAF A also triggers apoptosis in nutrient-deprived cells (Boya et al, 2005). To test this hypothesis, we monitored the degradation of mitochondria (mitophagy) on starvation (Komatsu et al, 2005), a process that has been shown to protect cells by degrading damaged organelles (Lemasters, 2005), and an indicator of autophagic activity (Kawai et al, 2006; Rodriguez-Enriquez et al, 2006). A substantial fraction of UBQLN-KD cells showed increased entrapment of mitochondria into autophagosomes compared with control cells, as more mitochondria dye (Mitotracker™) localized in the lumen of enlarged LC3-positive vacuoles (Fig 5C). Flow cytometry analysis confirmed Mitotracker accumulation in UBQLN-KD cells (Fig 5D). A decrease in Mitotracker fluorescence intensity on starvation could not be shown, possibly due to the stability of the fluorophore. Hence, a mitochondrial GFP cDNA (supplementary Fig S6 online) was expressed in control and UBQLN-KD cells (Fig 5E) and GFP intensity was measured by flow cytometry (Fig 5E, graph). Starvation induced a decrease in GFP: around 20% of initial fluorescence was lost after 6 h (Fig 5E) and around 80% was lost after 24 h (data not shown). Degradation of GFP was inhibited by BAF A (Fig 5E), confirming that mitophagy occurred through lysosomal activity. By contrast, no significant reduction in GFP was observed in UBQLN-KD cells, indicating that depletion of UBQLN inhibits lysosomal degradation of mitochondria during starvation.
Figure 5.
Depletion of ubiquilin inhibits the degradation of autophagosomes by lysosomes. (A) Autophagosome subcellular localization during starvation was analysed by microscopy in UBQLN- or autophagy-related gene ATG5-KD cells expressing GFP-tagged microtubule associated protein 1 light chain 3 (GFP-LC3). (B) Autophagosome spatial distribution was quantified by measuring the distance of each LC3 vesicle from the nucleus (in pixels) and by analysing the frequency distribution. (C) Control or UBQLN-KD cells expressing GFP-LC3 were starved and stained with the mitochondria dye Mitotracker™. Dual staining of LC3 (green) with Mitotracker (magenta) was analysed by microscopy. (D) Control and UBQLN-KD cells stained with Mitotracker were starved for 6 h; BAF A was added where indicated. Mitotracker fluorescence was measured by flow cytometry. Unless specified otherwise, values for UBQLN-KD and control+BAF A were compared with that of controls at the same time points, with ***P<0.001, **P<0.01 and *P<0.05 (paired Student's t-test). (E) Cells were transfected with pShooter™ together with control or UBQLN (1+2) siRNA. Expression of pShooter was comparable in control and UBQLN-KD cells (top). Cells were starved for 3 or 6 h, and the intensity of GFP was quantified by flow cytometry (bottom). The fluorescence value obtained with controls (no starvation) was considered as 100%, and the value obtained for the 3- and 6-h time points was normalized to controls. The graph represents the mean±s.e. of four assessments. (F) Cells expressing a chimaeric LC3 fused to both GFP and mCherry were transfected with control or UBQLN siRNA, and starvation was induced for the indicated times. Puncta from 25 cells were scored and analysed by microscopy for their fluorescence colour. The graph represents the mean of three experiments±s.e. The number of mCherry single-positive puncta in UBQLN-KD cells was compared with that of controls for each time point; **P<0.01 and *P<0.05 (paired Student's t-test). (G) Control or UBQLN-KD cells were starved for increasing periods. Where indicated, BAF A was added to the starvation medium (+B). Cell lysates were analysed by Western blot for their content in LC3-II. Loading control: transferrin receptor (TfR). BAF A, bafilomycin A1; GFP, green fluorescent protein; KD, knockdown; siRNA, short interfering RNA; UBQLN, ubiquilin.
Next, we studied the effect of UBQLN depletion on the stability of LC3. We expressed a chimaeric LC3 fused to both GFP (acid-sensitive) and mCherry (acid stable). This approach has been used to monitor autophagosome transport (Kimura et al, 2007; Pankiv et al, 2007). In control cells, and on starvation, LC3 initially formed puncta that were positive for both fluorescent markers (Fig 5F, 3 h), indicating that most autophagosomes had not yet fused with lysosomes. With increasing periods of starvation, the number of mCherry single-positive LC3 puncta gradually increased, whereas the number of GFP-mCherry double-positive puncta decreased (Fig 5F, 6 and 12 h). This maturation was blocked by BAF A (Fig 5F). On knockdown of UBQLN, a small but significant accumulation of both GFP and mCherry puncta was observed (Fig 5F). During starvation, the total number of punctate LC3 was comparable in control and UBQLN-KD cells; however, the latter showed significantly decreased numbers of mCherry single-positive puncta at all time points (Fig 5F). To strengthen these results, we monitored the cellular levels of LC3-II upon starvation (Fig 5G). Depletion of UBQLN increased baseline levels of LC3-II, consistent with the increased number of LC3 puncta (Fig 5F). On starvation, LC3-II, which initially increased in both control and UBQLN-KD cells, gradually decreased as autophagosomes fused with lysosomes. This maturation was blocked by the addition of BAF A, which stabilized LC3-II (Fig 5G). UBQLN-KD cells, however, showed a slower decrease of LC3-II compared with control cells (Fig 5G), indicating that UBQLN depletion inhibits autophagosome lysosomal degradation. Previous studies have identified a role for p62, another UBA-containing protein, in regulating autophagy (Bjorkoy et al, 2005; Komatsu et al, 2007). Hence, this suggests a broad involvement of ubiquitin-interacting proteins in regulating autophagy.
Methods
Cell culture. HeLa cells were maintained in Dulbecco's modified Eagle medium +10% fetal calf serum. When stably transfected with control or ATG7 short hairpin RNA (Fung et al, 2008), cells were maintained under 2 μg/ml of puromycin selection.
Transient protein depletion. Depletion of UBQLN by siRNA was described previously (N'Diaye et al, 2008). To deplete ATG5, a pool of four siRNAs was transfected at 100 nM. Protein depletion was assessed by Western blot after 72 h.
Immunostaining and confocal microscopy. The following antibody dilutions were used: Myc (5 μg/ml), LAMP1a (1:10), EEA1 (5 μg/ml), UBQLN1 (5 μg/ml), LC3 (1:1,000) and Alexa Fluor secondary antibodies (2 μg/ml). Cells were mounted in Prolong medium (Invitrogen, Carlsbad, CA, USA) and observed under a confocal microscope (LSM 510; Zeiss, Thornwood, NY, USA).
Starvation-induced cell death. Cells were transferred from complete medium to PBS for the indicated times, detached with trypsin and stained with 2 μg/ml PI. Intensity of PI on GFP-positive cells (UBQLN transfection) or on the whole population (siRNA transfection) was quantified by flow cytometry (FACS Calibur; BD Biosciences, San Jose, CA, USA).
Mitochondria staining. Control or UBQLN-KD cells expressing GFP-LC3 were stained with 500 nM Mitotracker for 30 min at 37°C, starved for 6 h, fixed and analysed by microscopy. For quantitative analysis of mitophagy, cells co-transfected with pShooter™ and UBQLN (or control) siRNAs were starved for the indicated times. The GFP fluorescence intensity of each condition was analysed by flow cytometry.
Quantification of microscopy. To quantify the differences in spatial distribution of puncta (Fig 5A), the distance between each LC3 vesicle and the nucleus was determined in pixels using the ImageJ software, and the frequency distribution was determined using Graph prism (bining=10 pixels).
To quantify the maturation of phagosomes (Fig 5F), the number of single-positive (mCherry) and double-positive (GFP/mCherry) puncta from 25 cells was scored using the ImageJ software.
Immunoelectron microscopy. Immunoelectron microscopy was performed on cells untransfected or transfected with Myc-UBQLN2 and GFP-LC3 as described previously (McCaffery & Farquhar, 1995). Thin frozen sections treated with 0.2% glycine, and blocked with 2% fish gelatin, 2% BSA in PBS, pH 7.4, were incubated with 50 μg/ml anti-Myc and 20 μg/ml anti-GFP at 4°C overnight. Controls for nonspecific staining were performed with non-immune serum. Sections were then incubated with 10 nm gold-coupled goat anti-mouse (for Myc) and 15 nm gold-coupled goat anti-rabbit (for GFP) at 1:50 dilution for 60 min. The sections were stained with oxalate uranyl acetate and embedded in 1.5% methyl cellulose (Sigma, St Louis, MO, USA), 0.3% aqueous uranyl acetate (Ted Pella, Redding, CA, USA) and examined under a Philips Tecnal 10 Electron Microscope.
Western blot analysis of LC3. Control or UBQLN-KD cells were starved for increasing periods of time. Cell lysates were analysed on SDS–polyacrylamide gel electrophoresis gel using an LC3 antibody (1:1,000; Axxora, San Diego, CA, USA), which, under these conditions, seems to preferentially bind to the modified form of LC3 (LC3-II).
Statistical analysis. Experiments were performed at least three times, with each condition run in duplicate. Statistical significance was determined by paired Student's t-test and/or with analysis of variance (ANOVA) test. Differences were considered significant with a P-value <0.05.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Fig S1
Supplementary Information
Acknowledgments
We thank Terge Johanssen (University of Tromsø, Norway) for the GFP/mCherry/LC3 cDNA, Nan Chen (University of California, San Francisco; UCSF) for the pBabe-LC3 vector and the ATG7-KD HeLa cells, Phil Stahl (Washington University in St Louis) for the GFP-RAB7 cDNA, Steven Finkbeiner (Gladstone Institute of Neurological Disease) for the GFP-HTT72 cDNA and Elizabeth Sztul (University of Alabama at Birmingham) for the GFP-250 cDNA. This study was supported by the National Institute of Health (NIH) grants to E.J.B. J.D. is supported by an NIH KO8 Award, a Culpeper Scholar Award (Partnership for Cures), and financial support from the UCSF Sandler Program in Basic Sciences.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P et al. (2005) Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V (2005) Autophagy in innate and adaptive immunity. Trends Immunol 26: 523–528 [DOI] [PubMed] [Google Scholar]
- Feng P, Scott CW, Cho NH, Nakamura H, Chung YH, Monteiro MJ, Jung JU (2004) Kaposi's sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol Cell Biol 24: 3938–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H (2002) Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA 99: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J (2008) Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 19: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol 146: 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26: 347–350 [DOI] [PubMed] [Google Scholar]
- Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N (2005) Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem 280: 11635–11640 [DOI] [PubMed] [Google Scholar]
- Kawai A, Takano S, Nakamura N, Ohkuma S (2006) Quantitative monitoring of autophagic degradation. Biochem Biophys Res Commun 351: 71–77 [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell 6: 409–419 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M et al. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Lemasters JJ (2005) Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5 [DOI] [PubMed] [Google Scholar]
- Mah AL, Perry G, Smith MA, Monteiro MJ (2000) Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol 151: 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery JM, Farquhar MG (1995) Localization of GTPases by indirect immunofluorescence and immunoelectron microscopy. Methods Enzymol 257: 259–279 [DOI] [PubMed] [Google Scholar]
- Munz C (2006) Autophagy and antigen presentation. Cell Microbiol 8: 891–898 [DOI] [PubMed] [Google Scholar]
- N'Diaye EN, Brown EJ (2003) The ubiquitin-related protein PLIC-1 regulates heterotrimeric G protein function through association with Gβγ. J Cell Biol 163: 1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- N'Diaye EN, Hanyaloglu AC, Kajihara KK, Puthenveedu MA, Wu P, von Zastrow M, Brown EJ (2008) The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol Biol Cell 19: 1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Regan-Klapisz E et al. (2005) Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM–UBL interaction. J Cell Sci 118: 4437–4450 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ (2006) Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ (2006) Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet 15: 1025–1041 [DOI] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC (2006) Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol 76: 89–101 [DOI] [PubMed] [Google Scholar]
- Wu AL, Wang J, Zheleznyak A, Brown EJ (1999) Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol Cell 4: 619–625 [DOI] [PubMed] [Google Scholar]
- Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, Avruch J (2002) Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta 1542: 41–56 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23: 33–42 [DOI] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8: 1124–1132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig S1
Supplementary Information