Abstract
The role of subunit a in promoting proton translocation and rotary motion in the Escherichia coli F1Fo ATP synthase is poorly understood. In the membrane-bound Fo sector of the enzyme, H+ binding and release occur at Asp-61 in the middle of the second transmembrane helix (TMH) of subunit c. Protons are thought to reach Asp-61 at the center of the membrane via aqueous channels formed at least in part by one or more of the five TMHs of subunit a. Aqueous access pathways have previously been mapped to surfaces of aTMH4. Here we have substituted Cys into the second and fifth TMHs of subunit a and carried out chemical modification with Ag+ and N-ethylmaleimide to define the aqueous accessibility of residues along these helices. Access to cAsp-61 at the center of the membrane may be mediated in part by Ag+-sensitive residues 248, 249, 251, and 252 in aTMH5. From the periplasmic surface, aqueous access to cAsp-61 may be mediated by silver-sensitive residues 115, 116, 119, 120, 122, and 126 in aTMH2. The Ag+-sensitive residues in TMH2, -4, and -5 form a continuum extending from the periplasmic to the cytoplasmic side of the membrane. In an arrangement of helices supported by second-site revertant and crosslinking analyses, these residues cluster at the interior of a four-helix bundle formed by TMH2–5. The aqueous access pathways at the interior of subunit a may be gated by a swiveling of helices in this bundle, alternately exposing cytoplasmic and periplasmic half channels to cAsp-61 during the H+ transport cycle.
H+ transporting F1Fo ATP synthases consist of two structurally and functionally distinct sectors termed F1 and Fo. In the intact enzyme, ATP synthesis or hydrolysis takes place in the F1 sector and is coupled to active H+ transport through the Fo sector. Structurally similar F1Fo ATP synthases are present in mitochondria, chloroplasts, and most eubacteria (1). The F1 sector lies at the surface of the membrane and in Escherichia coli consists of five subunits in an α3β3γ1δ1ε1 stoichiometry. The Fo sector spans the membrane and in E. coli consists of three subunits in an a1b2c10 stoichiometry (2). In the complete membranous enzyme, the rotation of subunit γ is proposed to be driven by H+ transport-coupled rotation of a connected ring of c subunits in the Fo sector of the enzyme. The c subunit spans the membrane as a hairpin of two α-helices, and in the case of E. coli, contains the essential Asp-61 residue at the center of the second transmembrane helix (TMH). Asp-61 is thought to undergo protonation and deprotonation as each subunit of the oligomeric ring moves past a stationary subunit a. Subunit a is believed to provide access channels to the proton-binding Asp-61 residue, but the actual proton translocation pathway is only partially defined (3–5).
Subunit a is known to fold with five TMHs (6–8), with aTMH4 packing in parallel to cTMH2, i.e., the helix to which Asp-61 is anchored (9). The interaction of the conserved Arg-210 residue in aTMH4 with cTMH2 is thought to be critical during the deprotonation–protonation cycle of cAsp-61 (3, 4, 10–12). A number of studies support the positioning of subunit a and the two b subunits at the periphery of the c ring (3, 13, 14). Second-site revertant analysis (4, 6, 10, 15, 16) supports an arrangement of helices with aTMH2 and aTMH5 in contact with aTMH4.
The chemical labeling of cysteine side chains introduced by site-directed mutagenesis has been used as a means of mapping aqueous accessible regions on many membrane proteins. Several reagents have been used to modify the genetically introduced Cys to determine accessibility, including N-ethylmaleimide (NEM) (17, 18) and Ag+ (19). Modification of Cys by these reagents is thought to depend on ionization of the Cys sulfhydryl to its thiolate form and thus is expected to occur preferentially in an aqueous environment (17–20). Previously, we used this method to test residues in aTMH4 for aqueous accessibility (5) and identified a silver-sensitive pocket centered around N214C in aTMH4 that extended to the cytoplasmic surface at Ser-206 and to residue H245C in TMH5 at the center of the membrane. In this report, we have focused on TMH2 and aTMH5, using the same methods, and these studies reveal two distinct regions of aqueous accessibility in subunit a. Cys substitutions in residues 115, 116, 119, 120, 122, and 126 of aTMH2 are very sensitive to Ag+ and may probe an aqueous pathway from cAsp-61 to the periplasmic surface of the membrane. In aTMH5, Cys substitutions in residues 248, 249, 251, and 252 of aTMH5 are most sensitive to Ag+ modification and appear to form an aqueous accessible pocket at the center of the membrane. Taken together, these results indicate that subunit a may facilitate aqueous access to cAsp-61 from both sides of the membrane. We propose that the swiveling of helices in subunit a may lead to the alternate exposure of the cytoplasmic and periplasmic half channels to cAsp-61 during the protonation–deprotonation cycle.
Experimental Procedures
Construction of Cys-Substituted Mutants. The cysteine substitutions generated here were transferred into plasmid pCMA113 (5), which contains a hexahistidine tag on the C terminus of subunit a (6) and genes encoding the entire unc (atp) operon in which all endogenous Cys had been substituted by Ala (21). Most cysteine substitutions were introduced by a two-step PCR method by using a synthetic oligonucleotide, which contained the codon change, and two wild-type primers (22). aTMH5 PCR products were transferred directly into pCMA113 by using unique PflMI (1,136) and BsrGI (1,913) sites (see ref. 23 for nucleotide numbering). aTMH2 products were transferred into pVF196 (6) by using NcoI (1,305) and PstI (1,565) sites and then transferred to pCMA113 with HindIII and BsrGI, as described above. All mutations were confirmed by sequencing the cloned fragment through the ligation sites. All experiments were performed with the mutant whole operon plasmid derivative of pCMA113 in the unc operon deletion host strain JWP292 (2). All plasmid transformant strains were tested for growth on succinate and glucose as described (5).
ATP-Driven Quenching of 9-Amino-6-chloro-2-methoxyacridine (ACMA) Fluorescence. Cells were grown and membranes prepared as described (5). Membranes were suspended in 3.2 ml of HMK buffer [10 mM Hepes·KOH/1 mM Mg(NO3)2/10 mM KNO3, pH 7.5] ACMA was added to a 0.3 μg/ml final concentration, and 30 μl of 0.1 M ATP, pH 7.0, was added to initiate quenching of fluorescence. The reaction was terminated by addition of 8 μl of 288 μM nigericin (0.5 μg/ml final concentration). The level of fluorescence obtained after addition of nigericin was normalized to 100% in calculating the percent quenching due to ATP-driven proton pumping (Table 1). For AgNO3 treatment, 160 μl of membranes at 10 mg/ml was suspended in HMK buffer containing 40 μM AgNO3 and incubated at room temperature for 15 min before carrying out the quenching assay. For NEM treatment, membranes were pretreated at 10 mg/ml in TMG-acetate buffer (50 mM Tris-acetate/5 mM magnesium acetate/10% glycerol, pH 7.5) with 5 mM NEM, and then diluted into HMK-nitrate buffer before carrying out the same quenching assay.
[14C]N-ethylmaleimide Labeling Studies. 14C-NEM (New England Nuclear, 34.2 mCi/mmol) in pentane was added to TMG-acetate buffer and the pentane evaporated by blowing a stream of argon over the solution. The resulting aqueous solution of NEM was then added to 3 mg of membrane vesicles in TMG-acetate buffer such that the final concentration of membranes was 10 mg/ml, and the final concentration of 14C-NEM was 1 mM. The mixture was incubated at room temperature for 1 h, after which the reaction was stopped by addition of 2-mercaptoethanol to 10 mM. SDS was added to 1% and unlabeled NEM to 20 mM. His-tagged subunit a was purified from the SDS-solubilized membranes as described (5). Subunit a purified from 1 mg of membranes was analyzed by SDS/PAGE on a 12% Tris-tricine gel (24), and the resulting gel was stained with Coomassie blue and dried. The intensity of staining of all bands shown falls within the linear range for this dye. The dried gel was exposed to a storage phosphor screen and scanned with a PhosphorImager (Molecular Dynamics) to quantitate radioactivity incorporated into the subunit a band. To determine the amount of protein in each band, the stained dried gel was scanned by using a flatbed scanner linked to a Macintosh computer with TWAIN 5 software (Epson, Long Beach, CA). The scan was quantitated by using the public domain NIH IMAGE program.†
Results
Cys Substitutions in TMH2 and aTMH5 of Subunit a. In this study, Cys substitutions in the second and fifth TMHs of subunit a were transferred into a His-tagged version of subunit a in a plasmid that coded the entire unc (atp) operon wherein all of the endogenous Cys in F1 and Fo had been substituted by Ala (5). These plasmids were transformed into JWP292, a strain with a chromosomal deletion of the entire unc operon. The transformant strains were tested for growth yield on glucose and relative colony size on succinate-containing minimal medium. The Cys substitutions in TMH2 and -5 of subunit a reported here had little effect on growth (see Table 1, which is published as supporting information on the PNAS web site). Additionally, ATP-driven quenching of ACMA fluorescence was performed on inside-out membrane vesicles of the substituted strains to evaluate ATPase-coupled H+ pumping function (Table 1). Of the 45 mutants tested, only D119C in TMH2 and Q252C in TMH5 exhibited substantial reductions in the quenching response (Fig. 1).
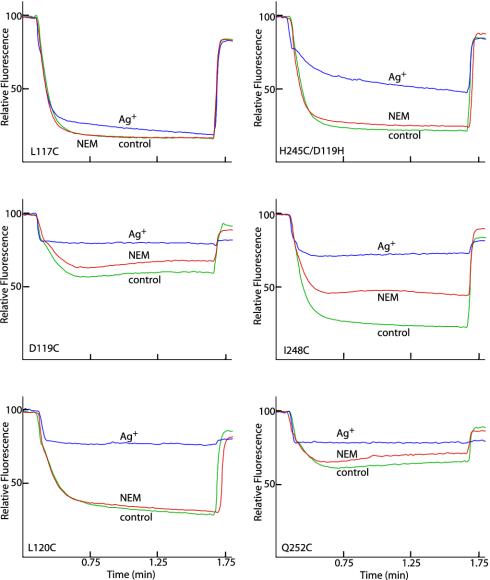
Fig. 1.
Differing sensitivity of Cys substitutions in aTMH2 and aTMH4 to Ag+ and NEM. A 160-μl aliquot of membranes at 10 mg/ml in TMG-acetate was treated with 5 mM NEM for 15 min at room temperature and then diluted into 3.2 ml of HMK buffer containing 0.3 μg/ml ACMA. Alternatively, membranes were diluted into HMK buffer and AgNO3 added to 40 μM for 15 min at room temperature before addition of ACMA. ATP was added to 0.94 mM at 20 s, and the uncoupler nigericin was added to 0.5 μg/ml at 100 s. The traces indicate no treatment (green), NEM treatment (red), or Ag+ treatment (blue). The substitution tested is indicated.
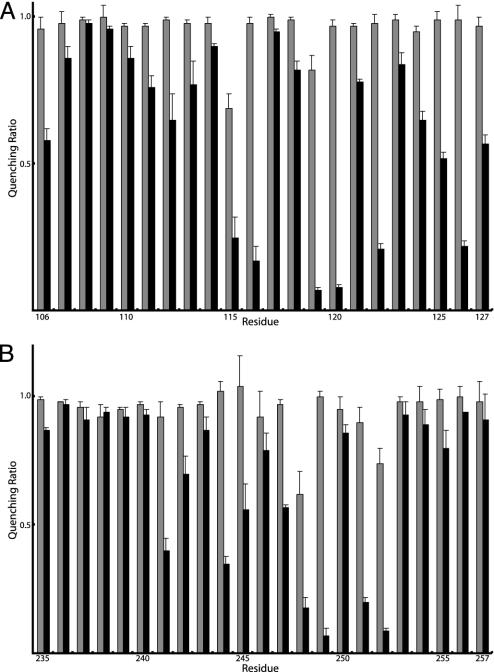
Inhibition of ATPase-Coupled H+ Transport by NEM or Ag+ in aTMH2. Inside-out membrane vesicles with Cys substitutions in residues 106–127 were treated with 5 mM NEM or 40 μM AgNO3 in chloride-free assay buffer, and ATPase-coupled H+ transport was measured by the quenching of ACMA fluorescence. Representative quenching traces for several mutants are presented in Fig. 1. NEM had negligible inhibitory effects on all Cys substitutions in aTMH2, as summarized in Fig. 2A. In contrast, Ag+ inhibited quenching markedly in several of the mutants tested, including Cys substitutions at residues 115, 116, 119, 120, 122, and 126 (Fig. 2A).
Fig. 2.
NEM and Ag+ sensitivity of residues in aTMH2 and aTMH5. Membranes were treated as described in the Fig. 1 legend. The results are presented as the ratios of quenching in the presence of Ag+ or NEM to the quenching in the absence of either reagent. The gray bars represent the quenching ratio ± NEM treatment, and the black bars represent the quenching ratio ± Ag+ treatment. Each bar represents the average ratio from n ≥ 2 determinations ± SD. In nine trials of control membranes lacking Cys, quenching ratios of 0.96 ± 0.06 for NEM and 0.90 ± 0.04 for Ag+ were observed relative to the untreated control.
Inhibition of ATPase-Coupled H+ Transport by NEM or Ag+ in aTMH5. Inside-out membrane vesicles with Cys substitutions in residues 235–257 of the putative aTMH5 and residues 225–234 of the likely periplasmic loop between aTMH4 and aTMH5 were tested for NEM and Ag+ inhibition of ATP-driven ACMA quenching. NEM had little inhibitory effect on any of the 33 residues tested, showing very modest inhibition with only residues 248 and 252 (Figs. 1 and 2B). However, as with aTMH2, a number of mutants were inhibited by Ag+ treatment, the most sensitive being 248, 249, 251, and 252, with other mutants including 241, 244, 245, and 247 showing lesser sensitivity (Fig. 2B).‡
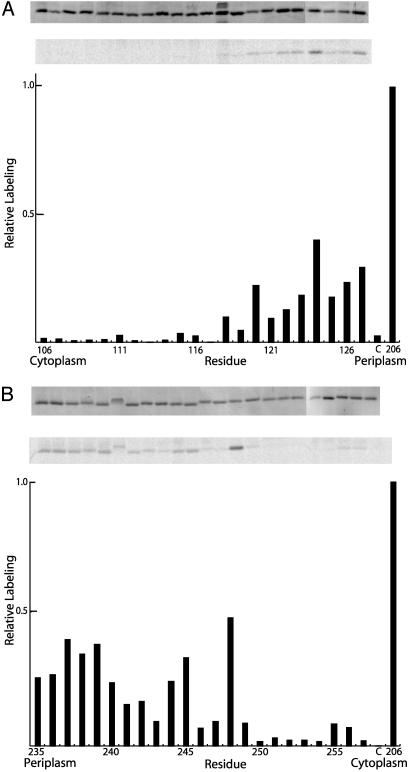
14C-NEM Labeling of Cys in aTMH2 and aTMH5. To check whether NEM reacted with residues in the suggested transmembrane regions without causing inhibition of quenching, inside-out membrane vesicles from the aTMH2 and aTMH5 mutants were treated with 1 mM 14C-NEM for 1 h. The hexahistidine-tagged subunit a was then purified by Ni+2 affinity chromatography, and the amount of label incorporated into subunit a was quantified by PhosphorImage analysis. Of the 45 substitutions tested, only residues 244, 245, and 248 at the center of aTMH5 were labeled moderately relative to the most NEM-sensitive control, Cys-206 in aTMH4.§ Other substitutions showing moderate labeling clustered at the periplasmic sides or in extramembranous loops at the ends of both aTMH2 and aTMH5 (Fig. 3 A and B). Residues at the cytoplasmic sides of the helices showed little or no labeling.
Fig. 3.
Labeling of aTMH2 and aTMH5 with 14C NEM. Membranes from various aTMH2 and aTMH5 mutants were treated at 10 mg/ml in TMG-acetate buffer (pH 7.5) with 1 mM 14C NEM for 1 h at room temperature. The membranes were then solubilized with SDS, and subunit a was purified with nickel-nitrilotriacetic acid-agarose. Samples were subjected to SDS/PAGE, and the radioactivity in the dried gel was quantitated with a PhosphorImager. (A) aTMH2 data. (B) aTMH5 data. (A and B Top) Coomassie stain of dried gel, used to normalize the labeling intensities. (A and B Middle) PhosphorImage of dried gel. (A and B Bottom) Bar graph of labeling intensity normalized to Coomassie staining intensity. In all experiments, aS206C membranes were used as a positive control for labeling, and the labeling intensity for aS206C represents 100% on the bar graph. The C bar on the graphs represent labeling results for the control membranes lacking Cys in F1Fo.
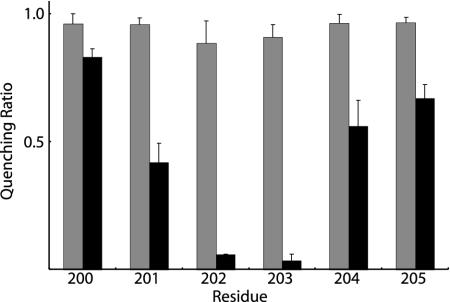
Ag+-Sensitive Residues at the Cytoplasmic End of aTMH4. Cys substitutions in aTMH4 had previously been tested for Ag+ and NEM sensitivity over the span of residues 206–224 (5). The relative Ag+ and NEM sensitivity of residues 200–205 is shown in Fig. 4. Although NEM-insensitive, residues 202 and 203 proved to be extremely sensitive to inhibition by Ag+. Therefore, it appears that the pattern of Ag+-sensitive residues on one helical face of aTMH4 extends from N214C at the center of the membrane to residues S202C, K203C, and S206C at or near the cytoplasmic surface.
Fig. 4.
NEM and Ag+ sensitivity of residues at the cytoplasmic end of aTMH4. Membranes were treated as described in the legend to Fig. 1. The results are presented as the ratios of quenching in the presence of NEM or Ag+ to the quenching in the absence of either reagent. The gray bars represent the quenching ratio ± NEM treatment, and the black bars represent the quenching ratio ± Ag+ treatment. Each bar represents the average ratio from n = 3 determinations ± SD.
Discussion
Here we report experiments defining aqueous accessible residues in the second and fifth TMHs in subunit a of the rotary ATP synthase. Previously, we had reported on the aqueous accessibility of residues in aTMH4 (5). Several second-site revertants support an arrangement of TMHs with aTMH2 and aTMH5 in contact with aTMH4 (4, 6, 10, 15, 16). Such an arrangement is now supported by crosslinking between the helical pairs of aTMH2/TMH4, aTMH2/TMH5, and aTMH4/TMH5 (B. Schwem and R.H.F., unpublished work). The results presented here support the idea that all three helices form parts of the aqueous access pathway to Asp-61 of subunit c. Very few of the residues reported here are sensitive to inhibition by NEM. This may be due to the restricted size of the pathway through which protons reach cAsp-61. The small size of the Ag+ ion, relative to the bulkier NEM, may make it an ideal probe of the aqueous access pathways in subunit a.
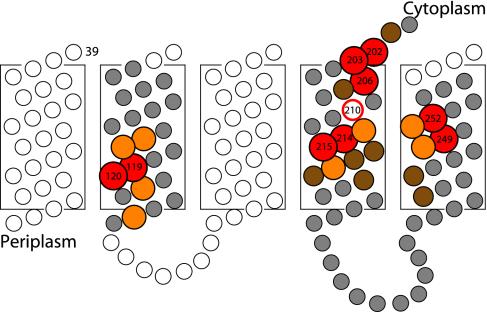
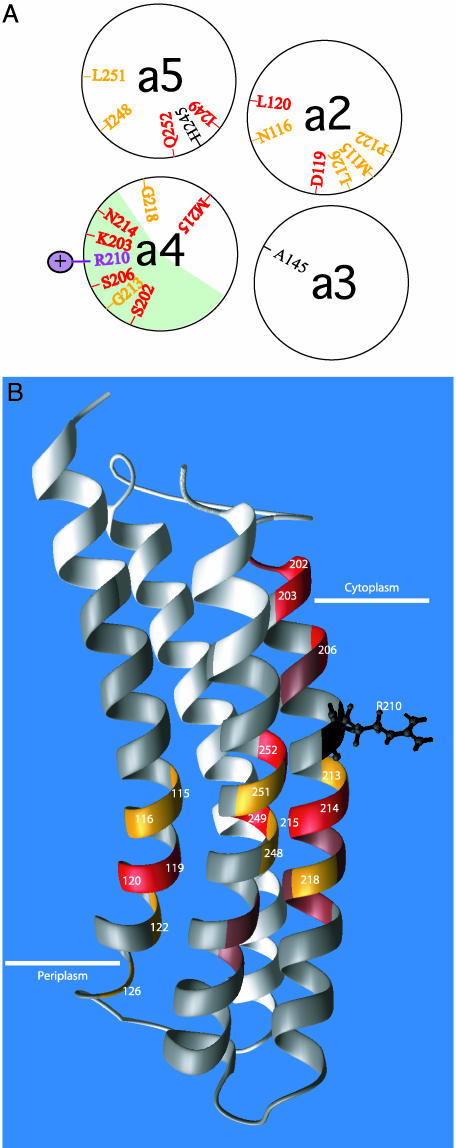
The positions of Ag+-sensitive residues in TMH2, -4, and -5 are shown in a 2D topological representation of subunit a in Fig. 5. aTMH2 appears to provide an aqueous pathway leading from the periplasm to the center of the membrane, whereas Ag+-sensitive residues in aTMH4 extend from the center of the membrane to the cytoplasmic surface. The most Ag+-sensitive residues in aTMH5 cluster at the center of the membrane and extend toward the periplasmic surface. When TMH2, -4, and -5 are brought into juxtaposition, they may form an aqueous cavity at the center of the membrane, which would locate primarily to the center of the modeled four-helix bundle (Fig. 6A). A 3D representation of the four-helix bundle is shown in Fig. 6B. From inspection of Fig. 6A, it is apparent that there are two distinct regions of silver sensitivity in subunit a. The first region is the face of aTMH4 that crosslinks with cTMH2. This is also the most NEM-sensitive region, NEM reactivity extending from residue 214 at the center of the membrane to residue 206 at the cytoplasmic surface. Other regions of Ag+-sensitive residues cluster toward the interior of the four-helix bundle modeled for subunit a. The clustering of Ag+-sensitive residues at the center of the four-helix bundle raises the question of how protons in such an aqueous accessible pocket would reach Asp-61 of subunit c. It therefore seems likely that movement and repositioning of these helices may be necessary during the course of the proton transport cycle, as discussed below.
Fig. 5.
Ag+-sensitive residues represented in a 2D topological model. Residues that are most sensitive to modification by Ag+ are shown in red (>90% inhibition), orange (76–90% inhibition), and brown (50–75% inhibition). Less-sensitive and insensitive residues are shown in gray, and untested residues are white.
Fig. 6.
Two distinct regions of Ag+ sensitivity in subunit a.(A) Cross-sectional view of TMH2–5 of subunit a depicted as α-helical wheels. The most Ag+-sensitive residues are shown in red (>90% inhibition) and orange (76–90% inhibition). The position of the aArg-210 side chain is also indicated. The light green area on aTMH4 represents the helical face that crosslinks to TMH2 of subunit c.(B) Placement of Ag+-sensitive residues in 3D model of subunit a by Rastogi and Girvin (25) (Protein Data Bank entry 1C17). Ag+-sensitive residues are indicated as described in Fig. 5, i.e., red (>90% inhibition), orange (76–90% inhibition), and brown (50–75% inhibition). Insensitive residues are shown in gray, and untested residues are white.
We have previously proposed a model for rotary function in which the concerted swiveling of helices within the four-helix bundle of subunit a may provide a mechanism of gating alternate access of protons to cAsp-61, and in turn be linked to the mechanical movements driving c-ring rotation (4). In the model, the H+-transport cycle is initiated by the swiveling of TMH2 of subunit c, bringing cAsp-61 and aArg-210 in juxtaposition and simultaneously aligning the helical faces of cTMH2 and aTMH4 in the position required for crosslinking (9). After deprotonation of cAsp-61 and exit of the proton to the cytoplasm via the aS206C NEM-sensitive channel, the helices of subunit a are proposed to swivel in a concerted fashion to move aArg-210 away from cAsp-61 to facilitate its reprotonation, and simultaneously gate movement of protons from the periplasmic channel centered in the four-helix bundle. After protonation of cAsp-61, the helices are proposed to swivel against each other to return to their original position, and in a manner akin to meshed gears, drive the stepwise rotation of the c-rotor. The mapping of Ag+-sensitive residues (presumed to reflect aqueous access) to the interior of the proposed four-helix bundle lends support to a model of this type and immediately predicts that aqueous exposure of different regions of the protein should change during the H+-transport cycle. We expect that this prediction can be tested in future experiments.
Supplementary Material
Acknowledgments
This study was supported by U.S. Public Health Service Grant GM23105 from the National Institutes of Health. C.M.A. was supported in part by a traineeship awarded through the Biotechnology Training Program of the National Institutes of Health (Grant 5T32 GM08349) to the University of Wisconsin.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ACMA, 9-amino-6-chloro-2-methoxyacridine; NEM, N-ethylmaleimide; TMH, transmembrane helix.
Footnotes
NIH IMAGE was developed at the National Institutes of Health and is available at http://rsb.info.gov/nih-image.
We previously reported that the aH245C/D119H mutant membranes, which were made partially functional by the D119H second-site suppressor, were inhibited by ≈50% by 40 μMAg+ and nearly completely by 80 μMAg+. The membranes used here were expressed from a plasmid carrying the complete unc operon, vs. the partial operon plasmid used previously, and expressed with an F1 containing no Cys. These differences likely account for the quite robust activity and slightly reduced Ag+ sensitivity observed here.
The 14C-NEM labeling of aI248C shown in Fig. 3B appears more extensive than would be predicted from the 38% inhibition shown in Fig. 2B and Table 1. The discrepancy may be due to differences in the conditions used for labeling with 14C-NEM vs. that for inhibition of ATP-driven quenching (see Experimental Procedures).
References
- 1.Senior, A. E. (1988) Physiol. Rev. 68, 177-231. [DOI] [PubMed] [Google Scholar]
- 2.Jiang, W., Hermolin, J. & Fillingame, R. H. (2001) Proc. Natl. Acad. Sci. USA 98, 4966-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cain, B. D. (2000) J. Bioenerg. Biomembrane 32, 365-371. [DOI] [PubMed] [Google Scholar]
- 4.Fillingame, R., Angevine, C. & Dmitriev, O. (2002) Biochim. Biophys. Acta 1555, 29-36. [DOI] [PubMed] [Google Scholar]
- 5.Angevine, C. M. & Fillingame, R. H. (2003) J. Biol. Chem. 278, 6066-6074. [DOI] [PubMed] [Google Scholar]
- 6.Valiyaveetil, F. I. & Fillingame, R. H. (1998) J. Biol. Chem. 273, 16241-16247. [DOI] [PubMed] [Google Scholar]
- 7.Long, J. C., Wang, S. & Vik, S. B. (1998) J. Biol. Chem. 273, 16235-16240. [DOI] [PubMed] [Google Scholar]
- 8.Wada, T., Long, J. C., Zhang, D. & Vik, S. B. (1999) J. Biol. Chem. 274, 17353-17357. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, W. & Fillingame, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 6607-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatch, L. P., Cox, G. B. & Howitt, S. M. (1995) J. Biol. Chem. 270, 29407-29412. [DOI] [PubMed] [Google Scholar]
- 11.Valiyaveetil, F. I. & Fillingame, R. H. (1997) J. Biol. Chem. 272, 32635-32641. [DOI] [PubMed] [Google Scholar]
- 12.Fillingame, R. H. (1990) in The Bacteria, ed. Krulwich, T. A. (Academic, New York), Vol. XII, pp. 345-391. [Google Scholar]
- 13.Dunn, S. D., McLachlin, D. T. & Revington, M. (2000) Biochim. Biophys. Acta 1458, 356-363. [DOI] [PubMed] [Google Scholar]
- 14.Jones, P. C., Hermolin, J., Jiang, W. & Fillingame, R. H. (2000) J. Biol. Chem. 275, 31340-31346. [DOI] [PubMed] [Google Scholar]
- 15.Cain, B. D. & Simoni, R. D. (1988) J. Biol. Chem. 263, 6606-6612. [PubMed] [Google Scholar]
- 16.Hartzog, P. E. & Cain, B. D. (1994) J. Biol. Chem. 269, 32313-32317. [PubMed] [Google Scholar]
- 17.Tamura, N., Konishi, S., Iwaki, S., Kimura-Someya, T., Nada, S. & Yamaguchi, A. (2001) J. Biol. Chem. 276, 20330-20339. [DOI] [PubMed] [Google Scholar]
- 18.Mordoch, S. S., Granot, D., Lebendiker, M. & Schuldiner, S. (1999) J. Biol. Chem. 274, 19480-19486. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Q. & Miller, C. (1995) Science 268, 304-307. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., Xu, Q., Cortes, D. M., Perozo, E., Laskey, A. & Karlin, A. (2002) Proc. Natl. Acad. Sci. USA 99, 11605-11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo, P. H., Ketchum, C. J. & Nakamoto, R. K. (1998) FEBS Lett. 426, 217-220. [DOI] [PubMed] [Google Scholar]
- 22.Barik, S. (1996) Methods Mol. Biol. 57, 203-215. [DOI] [PubMed] [Google Scholar]
- 23.Gay, N. J. & Walker, J. E. (1981) Nucleic Acids Res. 9, 2187-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schagger, H. & von Jagow, G. (1987) Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- 25.Rastogi, V. K. & Girvin, M. E. (1999) Nature 402, 263-268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.