Abstract
Toxoplasma gondii, an obligate intracellular parasite of the phylum Apicomplexa, can cause severe disease in humans with an immature or suppressed immune system. The outcome of Toxoplasma infection is highly dependent on the strain type, as are many of its in vitro growth properties1. Here we use genetic crosses between type II and III lines to show that strain-specific differences in the modulation of host cell transcription are mediated by a putative protein kinase, ROP16. Upon invasion by the parasite, this polymorphic protein is released from the apical organelles known as rhoptries and injected into the host cell, where it ultimately affects the activation of signal transducer and activator of transcription (STAT) signalling pathways and consequent downstream effects on a key host cytokine, interleukin (IL)-12. Our findings provide a new mechanism for how an intracellular eukaryotic pathogen can interact with its host and reveal important differences in how different Toxoplasma lineages have evolved to exploit this interaction.
Most Toxoplasma gondii isolates that have been identified in Europe and North America belong to three distinct clonal lines2,3, referred to as types I, II and III. The three types differ widely in a number of phenotypes in mice such as virulence, persistence, migratory capacity, attraction of different cell types and induction of cytokine expression1. Recent results indicate that such differences might also exist in human infection4–9. To test the hypothesis that some of these strain-specific differences are a result of how the strains interact with the host cell, we infected human foreskin fibroblasts (HFFs) with each of the three types and used microarray analysis to investigate differences in host gene expression 24 h later. Significance analysis of microarrays10 (SAM) identified 105 human complementary DNAs, representing at least 88 unique genes that were regulated in a strain-specific manner (false discovery rate 15%) (Fig. 1a).
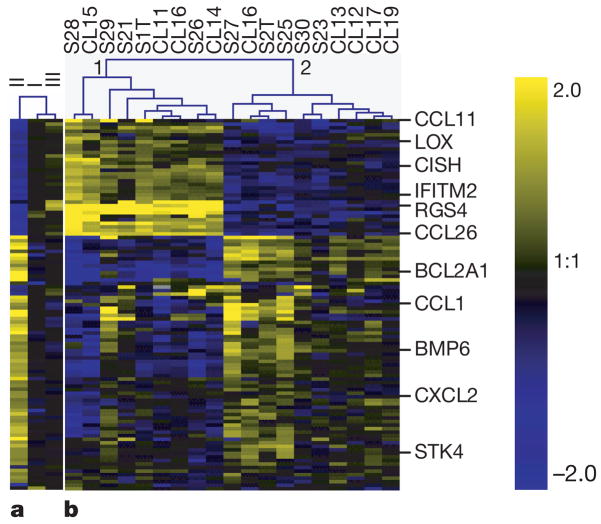
Figure 1. Toxoplasma strain-specific regulation of human gene expression.
a, HFFs were infected with type I, type II or type III Toxoplasma strains. 24 h after infection, expression profiles were obtained using human cDNA arrays. The averaged results (from at least three biological replicates) for median-centred expression levels for cDNAs are displayed using a log2 blue (low) to yellow (high) scale. Values log2 >2 or log2 <−2 were assigned the values 2 and −2, respectively. b, HFFs were infected with 19 F1 progeny derived from crosses (S or CL) between type II and type III strains. Details are as for a. Also displayed is the unsupervised clustering of experiments. For a full-size image and array data see Supplementary Fig. 1 and Supplementary data 1.
If the strain-specific regulation of a host gene has a genetic basis, it should segregate among F1 progeny derived from a cross between two strains that differ in its regulation. We therefore infected HFFs separately with each of 19 F1 progeny derived from crosses between type II and type III parasites and repeated the microarray analyses. The F1 strains formed two distinct clusters and, for a large portion of the genes, the progeny belonging to each cluster modulated human gene expression in either a type II- or a type III-like manner (Fig. 1b).
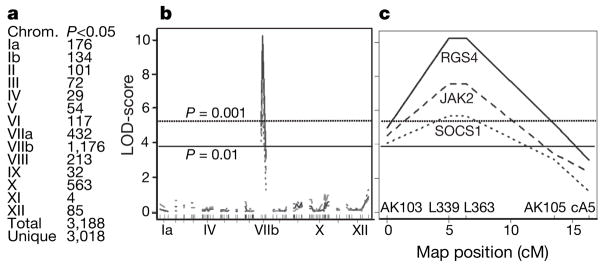
To identify the loci involved in this differential regulation, we performed a genome-wide scan for association of Toxoplasma genetic markers11 and the expression level of each of the 42,000 human cDNAs on the microarray using the package R/qtl (ref. 12). From this, 3,188 cDNAs, could be mapped to a specific Toxoplasma genomic locus (Fig. 2a–c). Interestingly, 1,176 of those cDNAs mapped to chromosome VIIb (Fig. 2b) and, of these, 617 had their highest logarithm of odds (LOD) score around genetic markers L339 and L363 (see Fig. 2c for an example). This indicated that, in the vicinity of these markers, there is at least one polymorphic Toxoplasma gene whose product has a strong effect on gene expression in HFFs. This was also corroborated by the fact that all F1 progeny in cluster 1 of Fig. 1b have type III alleles for markers L339 and L363 while all in cluster 2 have type II alleles.
Figure 2. Genome-wide scans for association of human gene expression with Toxoplasma genetic markers.
a, For each Toxoplasma chromosome, the number of cDNAs that mapped significantly (P <0.05, permutation test) to a genetic marker on that chromosome is shown. b, Plots indicate the log-likelihood association of expression of the human cDNAs for RGS4 (solid line), JAK2 (dashed line) and SOCS1 (dotted line) with markers aligned across the entire Toxoplasma genome in chromosome order. Selected chromosomes are indicated. c, An enlargement of Fig. 2b focusing on chromosome VIIb with the names of genetic markers indicated. LOD-score profiles of all significantly mapped human genes are provided as Supplementary data 2.
Pathway analysis showed that the group of human genes whose strain-specific modulation mapped to Toxoplasma chromosome VIIb was significantly enriched for genes involved in the IL-6, Janus kinase (JAK)/STAT, amyloid processing and IL-4 signalling pathways (Fischer’s exact test for all was P < 5 × 10−3; see Supplementary Fig. 2a–e). A broader, network analysis of molecular relationships between genes and gene products resulted in high scores for three networks whose central transcription factors were STAT3 and STAT5b (network 1), JUN (network 2) and hypoxia-inducible factor (HIF)-1A (network 3) (see Supplementary Fig. 2f–h). Given these results and the fact that the IL-4 and IL-6 signalling pathways culminate in the activation of the transcription factors STAT6 and STAT3, respectively13, we hypothesized that a large part of the strain-specific regulation of host genes is due to differences in how the different Toxoplasma strains intersect the STAT activation pathways, although from the network analysis it is clear that other transcription factors are also probably involved (for example, JUN and HIF1A).
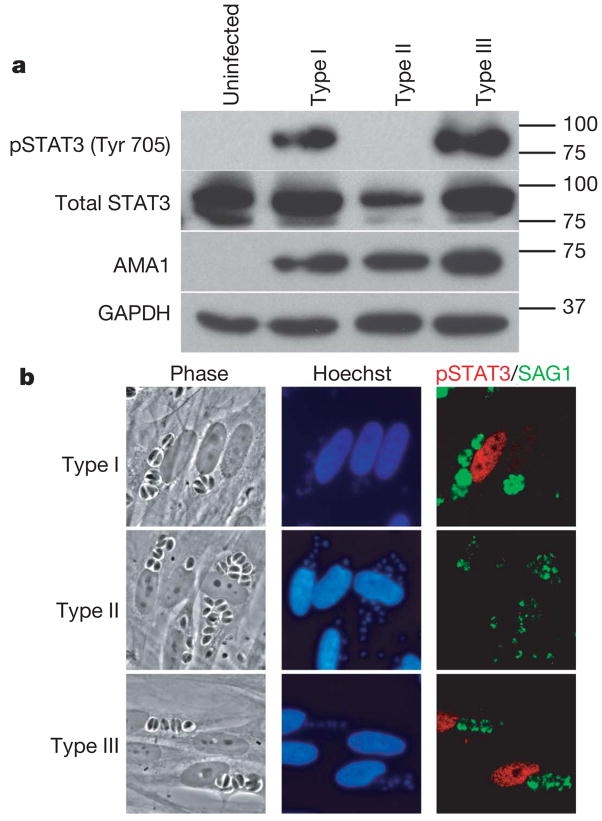
To test whether differences in STAT activation are central to the strain-specific differences, we analysed HFFs infected with type I, II or III parasites using immunofluorescence assays (IFA) and western blotting with antibodies specific for the tyrosine-phosphorylated (activated) forms of STAT3 and STAT6. Both methods revealed that, 18 h after infection, serum-starved HFFs infected with type I or type III strains contain much more activated STAT3 and STAT6 (not shown) than HFFs infected with type II strains (Fig. 3 a, b). At earlier time points (for example, 1 h after infection), HFFs individually infected with any of the three strains showed nuclear localization of tyrosine-phosphorylated STAT3 (data not shown). Somehow, late in infections and with type II parasites only, it seems that phospho-STAT3 Tyr 705 levels in the nucleus drop, presumably because tyrosine phosphorylation is repressed or reversed. In all cases, activation of STAT3 and STAT6 depended on parasite invasion, as uninfected neighbouring cells showed little or no signal. Co-infection of HFFs with type II and type III strains showed strong phosphorylation of STAT3 and STAT6, showing that this is an active property of type I and III strains and that type II strains cannot repress this process (not shown).
Figure 3. Strain-specific activation of STAT3.
a, Western blot analysis of strain-dependent phosphorylation of STAT3. Serum-starved HFFs were infected with a type I, type II or type III Toxoplasma strain and 18 h after infection cells were lysed and analysed by immunoblotting. GAPDH (host cell-specific) and AMA1 (parasite-specific) levels are shown as loading controls. b, Immunofluorescence analysis of strain-dependent activation of STAT3. HFFs were infected with type I, II or III parasites (MOI 10) for 18 h, fixed and incubated with antibodies against parasite SAG1 or the phosphorylated forms of STAT3 (Tyr 705).
To confirm and refine the preliminary mapping of the Toxoplasma genomic region involved in activation of STAT3/6, parasites that had recombinations in the region of interest on chromosome VIIb were genotyped and phenotyped for their capacity to activate STAT3/6. These data allowed us to narrow down the locus involved to a region between L363 and AK104, representing a maximum size of 0.56 Mb (see Supplementary Fig. 3). This interval on VIIb contains 78 predicted proteins (http://www.toxodb.org). Assuming that the crucial gene encodes a polymorphic, secreted protein that can substantially alter host cell transcription, we identified ROP16 as the most likely candidate. First, there are 57 single-nucleotide polymorphisms (SNPs; 34 of which are non-synonymous) between the type II allele of ROP16 and the type I and III alleles, which are virtually identical (5 SNPs), consistent with the genetic evidence that the type II allele functions completely differently from those of types I and III (see Supplementary Fig. 4). Second, ROP16 has high homology to serine/threonine protein kinases, which are potent modulators of many cell functions. And, third, ROP16 localizes to the Toxoplasma rhoptries14, a set of apical organelles that can secrete vesicle-like bodies (termed ‘evacuoles’) into the host cell upon invasion15.
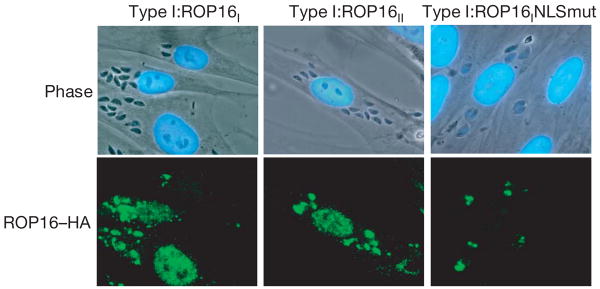
To characterize ROP16 further, a type I strain was engineered to express a carboxy-terminally haemagglutinin (HA)-tagged version of the protein derived from either a type I or type II background; the resulting strains were designated type I:ROP16I and type I:ROP16II, respectively. IFA showed that the tagged ROP16 was correctly targeted to the rhoptries and that within 10 min of invasion it had entered the host cell nucleus; it was still visible at 24 h but the brightest relative staining occurred between 10 min and 4 h (Fig. 4) with an apparent diminution of signal thereafter. Similar results were obtained when the mouse macrophage cell line RAW 264.7 was used as the host cell (not shown). Localization of HA-tagged ROP16 to the host cell nucleus depended on the putative nuclear localization signal (NLS) in ROP16 (RKRKRK at residues 339–344, inclusive): mutation of lysine residues 340, 342 and 344 to methionines (yielding RMRMRM) markedly diminished nuclear localization (Fig. 4).
Figure 4. ROP16 is secreted from the parasite and localizes to the host cell nucleus.
Type I parasites expressing HA-tagged type I ROP16 (TypeI:ROP16I, left), type II ROP16 (Type I:ROP16II, middle) or type I ROP16 with a mutated NLS (Type I:ROP16INLSmut, right) were added (MOI 10) to HFFs and cells were fixed after 4 h. ROP16 was visualized using anti-HA antibodies followed by incubation with secondary (anti-rat Alexa 488) antibodies.
In both HFFs and macrophages, the intensity of ROP16 staining in the nucleus depended on the number of invasion events (as determined by the number of parasitophorous vacuoles), not the total number of parasites present, which increases as a function of time (Fig. 4 and data not shown). Combined with the fact that ROP16 reaches the nucleus within 10 min of initial contact, these data indicate that ROP16 is introduced during the invasion process and not in a slow, steady release from intracellular parasites. Recently, we have seen that a second rhoptry protein, in this case a protein phosphatase, is injected into host cells with similar kinetics to ROP16. It also possesses an NLS and reaches the host nucleus with similar kinetics16.
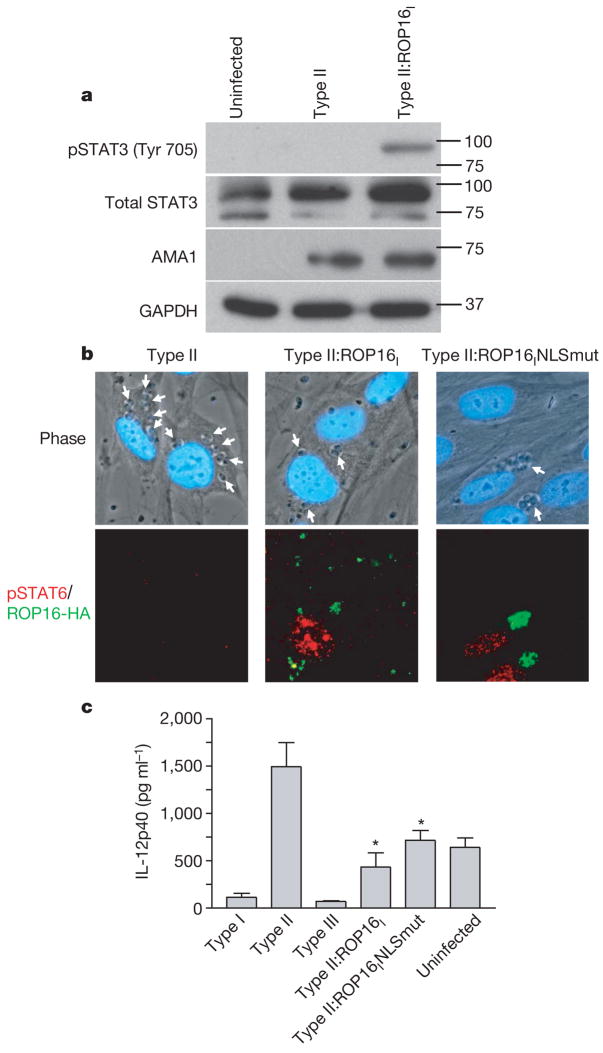
To test whether ROP16 is involved in the strain-specific activation of STAT3/6, we engineered type II strains to express an additional, type I allele of ROP16 (designated type II:ROP16I). Western blot and IFA revealed that, as before, HFFs infected with the parental type II strain showed little if any STAT3/6 activation 18 h after infection, whereas those infected with the type II:ROP16I showed strong activation in both assays (Fig. 5a, b). These experiments show that ROP16 has an important role in maintaining the activation of STAT3 and STAT6 in infected HFFs and indicate that the type I allele is probably dominant over the type II version.
Figure 5. ROP16 mediates strain-specific activation of STAT3/6 and consequent downstream effects on IL-12.
a, Type II parasites expressing the type I ROP16 gene induce phosphorylation of STAT3. HFFs were infected with type II or type II:ROP16I and lysates were prepared for immunoblot analysis as described in the legend to Fig. 3a. b, Type II parasites expressing the normal type I ROP16 gene or the type I ROP16 NLS mutant gene induce translocation of phosphorylated STAT6 to the host cell nucleus. White arrows indicate parasites. c, Type II parasites expressing the type I ROP16 gene or the type I ROP16 NLS mutant gene inhibit the production of IL-12p40 from infected macrophages. Error bars represent s.e.m. of three replicates. Data shown are from one representative experiment out of three. Asterisks denote significantly less IL-12p40 production relative to type II parasites (Student’s t-test).
Tyrosine phosphorylation of STAT3 and STAT6 generally occurs in the cytoplasm of a cell. It is therefore surprising that ROP16 seems to be concentrated in the host cell nucleus. To investigate whether nuclear localization is essential for its function, we engineered a type II strain to express a type I allele of ROP16 carrying the NLS disruption described above (RKRKRK to RMRMRM). Infection with the resulting strain, designated type II:ROP16INLSmut, showed a comparable level of STAT6 activation as infection with the type II:ROP16I strain (Fig. 5b). These results indicate that ROP16 can probably execute its function in the cytosol (perhaps as it transits to the nucleus) although we cannot exclude the possibility that the small amount of ROP16 that still goes to the nucleus in the NLS mutant (not shown) is responsible for its function in cells infected with this strain.
To investigate how ROP16 mediates its effect on STAT3/6, we used a battery of antibodies that recognize the activated (phosphorylated) forms of proteins upstream of the STAT3/6 signalling pathways. We found no strain differences in the activation of extracellular signal regulated kinase (ERK)1/2, P38, Jun amino-terminal kinase (JNK), AKT, JAK2 and tyrosine kinase (TYK)2. Combined with the fact that all three strains activate STAT3/6 early (see above and Supplementary Fig. 5), these results indicate that the pathway that leads to the initial activation of these key transcription factors is similar between strains but that HFFs infected with type I/III strains are actively prevented from subsequent downregulation.
Our results provide a potential molecular basis for at least one of the strain-dependent differences in how a host cell responds to Toxoplasma infection: type II but not type I or III strains induce mouse macrophages to produce high levels of IL-12 (both p40 and p70; ref. 17) and constitutive activation of STAT3 by type I parasites prevents lipopolysaccharide-triggered production of IL-12p40 by macrophages18. To determine whether ROP16 is involved in these differences, we examined STAT3 phosphorylation and IL-12p40 production by murine RAW264.7 macrophages infected with our engineered parasites and stimulated with interferon (IFN)-γ and LPS. The amount of phosphorylated STAT3 in macrophages infected with the different Toxoplasma strains paralleled the results described above for HFFs (see Supplementary Fig. 6). We confirmed that infection of macrophages with type II, but not with the type I or III strains, results in very high secretion of IL-12p40 and further showed that the type II:ROP16I and type II:ROP16INLSmut strains elicit significantly less IL-12p40 than parental type II parasites (Fig. 5c), as predicted by our results showing increased activation of STAT3 by these strains. Such effects could be an important part of the different disease outcomes seen with the type I, II and III strains. The facts that the type II strain induced more IL-12p40 than IFN-γ and LPS alone, and that the type II:ROP16I strain did not bring the IL-12p40 down to the levels of type I and III strains, indicate that in addition to ROP16, other molecules might be involved in the strain-specific regulation of IL-12p40.
To come full circle and show that because of or in addition to its role in strain-specific activation of STAT3/6, ROP16 is involved in the strain-specific differences originally observed in the human cDNA microarray experiments, we compared the gene expression profiles of HFFs infected with type II and type II:ROP16I strains. Of 13,382 cDNAs that met the quality control threshold, 127 were previously shown (Fig. 2a) to map to chromosome VIIb with an average difference greater than twofold between the original F1 progeny based on which ROP16 allele they carried. Of these 127 cDNAs, 29 (out of 242 total) were differentially regulated (average of at least twofold difference in the two arrays) when comparing the type II and type II:ROP16I strains. This is significantly more genes than expected by chance (hypergeometric distribution, P <10−15), confirming a role for ROP16 in this strain-specific regulation of host gene expression. Furthermore, all 29 genes changed in the expected direction (See Supplementary data 3).
The phenomenology of ROP16 introduction is analogous to type III and type IV secretion systems in prokaryotes19 but there are no indications for the existence of such systems outside bacteria. One of the earliest events in Toxoplasma invasion is a break in the host cell membrane20 and this might represent the moment when injection of ROP16 occurs, although we have been unable to detect ROP16 free in the cytosol or even associated with evacuoles15 (data not shown). Our results do not reveal the primary target of ROP16. It probably does not directly phosphorylate STAT3/6 but instead is likely to intersect pathways that lead to or maintain such activation. This could involve something akin to what has been reported21 for v-Abl, an oncogenic tyrosine kinase that causes constitutive activation of Jaks and STATs by disrupting suppressor of cytokine signalling (SOCS)-1 function (SOCS-1 normally targets Jaks and STATs for proteasomal degradation). Infection with type I and III strains results in a higher level of SOCS-1, SOCS-2 and SOCS-3 mRNA than infection with type II (see Supplementary data 2) and this difference maps to ROP16 (Fig. 2). This result is consistent with the fact that STAT3 is a positive regulator of SOCS (that is, the sustained activation of STAT3 results in their upregulation) and indicates that the mechanism involved in the type I and III-specific maintenance of activated STAT3 involves a block in SOCS function or some other downstream effect rather than a decrease in SOCS expression.
Rhoptries are a defining property of all Apicomplexa and some of the proteins found in the Toxoplasma rhoptries have homologues in Plasmodium. Thus, the ability to co-opt host cell gene expression for the parasite’s own purposes might be found in other Apicomplexa that also are otherwise trapped within a membrane-limited vacuole (for example, Plasmodium species as they grow within hepatocytes). This represents a previously undescribed mechanism for interaction between eukaryotic pathogens and their hosts.
METHODS
Details of parasite strains, genetic crosses and microarray analysis can be found in Supplementary Information.
Immunofluorescence analysis
Parasites were allowed to invade confluent HFF monolayers on coverslips for 0.17, 0.5, 1, 4, 6, 8, 18 or 24 h. The cells were then fixed, blocked and permeabilized. Coverslips were incubated with 3F10 (anti-HA) antibody, antibodies specific for the tyrosine phosphorylated forms of STAT3 and STAT6, or a mouse monoclonal antibody against surface antigen (SAG)-1 (DG52)22. Fluorescent secondary antibodies and Hoechst dye were used for antigen and DNA visualization, respectively.
Western blots
Parasites (MOI 10) were added to HFFs and infection was allowed to proceed for 18 h. Western blots were performed as described23 using antibodies specific for total STAT3 (Cell Signaling Technologies), GAPDH (Calbiochem), AMA1 (mAb B3.9024) or for the phosphorylated forms of STAT3 (phospho-Tyr705) and STAT6 (phospho-Tyr641) (both from Cell Signaling Technologies).
IL-12 enzyme-linked immunosorbent assay (ELISAs)
RAW264.7 macrophages were activated with IFN-γ (20 U ml−1) and LPS (100 ng ml−1) and 1 h later parasites were added at a multiplicity of infection (MOI) of 10. After 24 h of infection, supernatants were collected and IL-12p40 ELISAs were performed as per manufacturer’s recommendations (BD Biosciences).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
This work was supported by grants from the NIH (to J.C.B., J.P.B. and M.W.W.), the Ellison Medical Foundation (to J.C.B.), the USDS (to M.W.W.) and the California Universitywide AIDS Research Program (to J.P.J.S. and S.C.). We thank K. Broman for help with R/qtl and the Stanford Functional Genomics Facility for the human cDNA microarrays used for this study.
Footnotes
Author Contributions J.P.J.S. and S.C. contributed equally to this work. J.P.J.S. performed the microarrays and pathway analyses. S.C. and J.P.J.S. performed the experiments in Fig. 3. S.C. performed the experiments in Fig. 4 and Fig 5. J.P.B., M.E.J. and M.W.W. performed the genetic crosses that produced the progeny D3X1 and JD4. J.P.B. genotyped D3X1 and JD4. J.P.J.S., S.C., J.P.B. and J.C.B. wrote the paper. All authors discussed the results and commented on the manuscript.
Author Information The microarray data have been deposited in ArrayExpress with the accession number E-MEXP-783. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Saeij JP, Boyle JP, Boothroyd JC. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 2005;21:476–481. doi: 10.1016/j.pt.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- 3.Darde ML, Bouteille B, Pestre-Alexandre M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiological implications. J Parasitol. 1992;78:786–794. [PubMed] [Google Scholar]
- 4.Grigg ME, Ganatra J, Boothroyd JC, Margolis TP. Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis. 2001;184:633–639. doi: 10.1086/322800. [DOI] [PubMed] [Google Scholar]
- 5.Howe DK, Honore S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35:1411–1414. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuentes I, Rubio JM, Ramirez C, Alvar J. Genotypic characterization of Toxoplasma gondii strains associated with human toxoplasmosis in Spain: direct analysis from clinical samples. J Clin Microbiol. 2001;39:1566–1570. doi: 10.1128/JCM.39.4.1566-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan A, et al. Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains. J Clin Microbiol. 2005;43:5881–5887. doi: 10.1128/JCM.43.12.5881-5887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darde ML, Villena I, Pinon JM, Beguinot I. Severe toxoplasmosis caused by a Toxoplasma gondii strain with a new isoenzyme type acquired in French Guyana. J Clin Microbiol. 1998;36:324. doi: 10.1128/jcm.36.1.324-324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carme B, et al. Severe acquired toxoplasmosis in immunocompetent adult patients in French Guiana. J Clin Microbiol. 2002;40:4037–4044. doi: 10.1128/JCM.40.11.4037-4044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan A, et al. Composite genome map and recombination parameters derived from three archetypal lineages of Toxoplasma gondii. Nucleic Acids Res. 2005;33:2980–2992. doi: 10.1093/nar/gki604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 13.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 14.Bradley PJ, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 15.Hakansson S, Charron AJ, Sibley LD. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 2001;20:3132–3144. doi: 10.1093/emboj/20.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert LA, Ravindran S, Turetzky JM, Boothroyd JC, Bradley PJ. Toxoplasma gondii targets a protein phosphatase 2C to the nucleus of infected host cells. Eukaryot Cell. doi: 10.1128/EC/00309-06. published online 3 November 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robben PM, et al. Production of IL-12 by macrophages infected with Toxoplasma gondii depends on the parasite genotype. J Immunol. 2004;172:3686–3694. doi: 10.4049/jimmunol.172.6.3686. [DOI] [PubMed] [Google Scholar]
- 18.Butcher BA, et al. IL-10-independent STAT3 activation by Toxoplasma gondii mediates suppression of IL-12 and TNF-alpha in host macrophages. J Immunol. 2005;174:3148–3152. doi: 10.4049/jimmunol.174.6.3148. [DOI] [PubMed] [Google Scholar]
- 19.Remaut H, Waksman G. Structural biology of bacterial pathogenesis. Curr Opin Struct Biol. 2004;14:161–170. doi: 10.1016/j.sbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Suss-Toby E, Zimmerberg J, Ward GE. Toxoplasma invasion: The parasitophorous vacuole is formed from host cell pasma membrane and pinches off via a fission pore. Proc Natl Acad Sci USA. 1996;93:8413–8418. doi: 10.1073/pnas.93.16.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limnander A, et al. v-Abl signaling disrupts SOCS-1 function in transformed pre-B cells. Mol Cell. 2004;15:329–341. doi: 10.1016/j.molcel.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 22.Burg JL, Perelman D, Kasper LH, Ware PL, Boothroyd JC. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- 23.Chan SM, et al. Protein microarrays for multiplex analysis of signal transduction pathways. Nature Med. 2004;10:1390–1396. doi: 10.1038/nm1139. [DOI] [PubMed] [Google Scholar]
- 24.Mital J, et al. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol Biol Cell. 2005;16:4341–4349. doi: 10.1091/mbc.E05-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.