Abstract
Using a combination of affinity chromatography and tandem mass spectrometry, we recently identified 8 MHC class II (I-Ab) -bound Chlamydia peptides eluted from dendritic cells (DCs) infected with Chlamydia muridarum. In this study we cloned and purified the source proteins that contained each of these peptides and determined that three of the 8 peptide/protein antigens were immunodominant (PmpG-1, RplF and PmpE/F-2) as identified by IFN-γ ELISPOT assay using splenocytes from C57BL/6 mice recovered from C. muridarum infection. In order to evaluate whether the three immunodominant Chlamydia protein antigens were also able to protect mice against Chlamydia infection in vivo, we adoptively transferred LPS-matured DCs transfected ex vivo with the cationic liposome, DOTAP and individual PmpG-1(25–500aa), RplF or PmpE/F-2(25–575aa) proteins. The results showed that the transfected Chlamydia proteins were efficiently delivered intracellularly into DCs. Mice vaccinated with DCs transfected with individual Chlamydia protein PmpG-125–500, RplF or PmpE/F-225–575 exhibited significant resistance to challenge infection as indicated by reduction in the median Chlamydia inclusion forming units in both the lung and genital tract models. The major outer membrane protein (MOMP) was used as a reference antigen but conferred significant protection only in the genital tract model. Overall, vaccination with DCs transfected with PmpG-125–500 exhibited the greatest degree of protective immunity among the four Chlamydia antigens tested. This study demonstrates that T cell peptide antigens identified by can be successfully exploited as T cell protein based subunit vaccines and that PmpG-125–500 protein may be a suitable vaccine candidate for further evaluation.
Keywords: Antigens, Peptides, Epitopes, Protein, Vaccination, Dendritic Cells, Bacterial
Introduction
Chlamydia trachomatis is the leading cause of sexually transmitted bacterial infection, with over 92 million new infections occurring annually worldwide (1). Although antibiotics are effective against C. trachomatis, the majority of cases are initially asymptomatic and thus are often undetected and untreated. Untreated infection can spread throughout the reproductive tract and cause severe complications in women, such as pelvic inflammatory disease, ectopic pregnancy, and infertility. Additionally, infection with C. trachomatis facilitates the transmission of HIV (2) and might be a cofactor in human papilloma virus (HPV)-induced cervical neoplasia (3). Public health measures to control Chlamydia appear to be failing as case rates have risen over the past decade (4) perhaps due to early antibiotic treatment blunting the development of immunity to C. trachomatis (5). Therefore the development of an effective vaccine remains an urgent public-health priority.
Early vaccination trials in both human and nonhuman primates with whole inactivated C. trachomatis elementary bodies demonstrated that immunity to Chlamydia could be induced but that vaccine efficacy was incomplete and short lived. Moreover, breakthrough C. trachomatis infection in some primate models resulted in more severe disease with worse inflammation post-vaccination (6, 7). These observations were interpreted to suggest that Chlamydia contains both immunoprotective and immunopathological antigens and that an effective Chlamydia vaccine will need to be molecularly defined and delivered in such a manner as to engender long-term protective immune responses. Therefore, contemporary C. trachomatis vaccine research has focused on the production of subunit vaccines that are based on individual protective C. trachomatis proteins, which are administered with adjuvant or other delivery vehicles to enhance immunogenicity (8, 9).
Most subunit vaccine efforts have evaluated the Chlamydia major outer membrance protein (MOMP) because this protein is abundant, highly conserved and elicits T cell responses and neutralizing antibodies (10–12). It has been both surprising and disappointing in primate models that while MOMP vaccines elicit strong immune responses, they confer only marginal protection even to a homologous strain (13). Various other candidate antigens that trigger T cell responses in humans and in mice have therefore been proposed (14–19); however none has been yet evaluated in primate models and thus the search for protective T cell antigens in Chlamydia remains a high research priority.
Studies in animal models and during human infection have established that Chlamydia-specific CD4+ T cells producing gamma interferon (IFN-γ) are critically involved in the clearance of a Chlamydia infection (20–22) and that antibody may play an important role in resistance to reinfection (22, 23); the role of CD8+ T cells appears to be less important (22, 24). Therefore selection of molecularly defined antigens for a subunit vaccine that stimulate CD4+ Th1 cells is central to the current design effort. While development of a vaccine for intracellular pathogens that require protective cell-mediated immunity (CMI) will be more difficult than for pathogens that simply require protective antibodies, protective T cell antigen candidates can be chosen by identifying microbial peptides that bind to MHC molecules.
Using a combination of affinity chromatography and tandem mass spectrometry, we recently identified 8 MHC class II (I-Ab) -bound Chlamydia peptides eluted from C. muridarum infected dendritic cells (DCs) from C57 BL/6 mice. Adoptive transfer of DCs pulsed with a pool of the 8 peptides partially protected mice against challenge infection (25). In the current study, the parent protein containing each peptide sequence was cloned, expressed and purified. Three of the 8 Chlamydia antigens were immunodominant (PmpG-1, RplF and PmpE/F-2) and vaccination with DCs transfected with individual Chlamydia proteins, PmpG-125–500, RplF or PmpE/F-225–575 induced significant protective immunity against lung and genital tract infections.
Materials and Methods
Chlamydia
C. muridarum strain Nigg (the mouse pneumonitis strain) was grown in Hela 229 cells and elementary bodies (EBs) were purified by discontinuous density gradient centrifugation and stored at −80°C as previously described (11). The infectivity of purified EBs was titrated by counting Chlamydia inclusion forming units (IFUs) on Hela cell monolayer with anti-EB mouse polyclonal antibody followed by biotinylated anti-mouse IgG (Jackson ImmunoResearch) and a DAB substrate (Vector Laboratories) (26).
Chlamydia peptides
Eight MHC class II Chlamydia peptides (Table 1) discovered by immunoproteomics (25) were synthesized and purified (Sigma-Aldrich). Peptides were solubilized in DMSO at a concentration of 4 mg/ml and stored at −20°C. One irrelevant sequence known to bind I-Ab and derived from OVA (ISQAVHAAHAEINE) was used as a negative peptide control.
Table 1.
Chlamydia peptides discovered by immunoproteomics and purified source proteins used in this study
| Peptide Sequence | Source protein | Abbreviation | Purified source protein format | |
|---|---|---|---|---|
| KGNEVFVSPAAHIIDRPG | Ribosomal protein L6 | RplF | Full length | N-terminal GST-tag |
| SPGQTNYAAAKAGIIGFS | 3-oxoacyl-(acyl carrier protein) reductase | FabG | Full length | N-terminal GST-tag |
| KLDGVSSPAVQESISE | Anti-anti-factor | Aasf | Full length | N-terminal GST-tag |
| ASPIYVDPAAAGGQPPA | Polymorphic membrane protein G | PmpG-1 | 25–500aa | N-terminal His-tag |
| DLNVTGPKIQTDVD | Hypothetical protein TC0420 | TC0420 | Full length | N-terminal GST-tag |
| IGQEITEPLANTVIA | ATP-dependent Clp protease, proteolytic subunit | ClpP-1 | Full length | N-terminal GST-tag |
| AFHLFASPAANYIHTG | Polymorphic membrane protein F | PmpE/F-2 | 25–575aa | N-terminal His-tag |
| MTTVHAATATQSVVD | Glyceraldehyde 3-phosphate dehydrogenase | Gap | Full length | N-terminal His-tag |
Chlamydia peptide source proteins
The source proteins containing the MHC II binding Chlamydia peptides were cloned, expressed and purified as follows: rplF, fabG, aasf, pmpG-1, TC0420, clp-1, pmpE/F-2 and gap DNA fragments were generated by PCR using genomic DNA isolated from C. muridarum. The PCR products were purified and cloned into either pGEX-6P-3 (GE Healthcare) for rplF, fabG, aasf, TC0420, and clp-1 or pET32a (Novagen) for pmpG-1, pmpE/F-2 and gap after restriction enzyme digestion with BamHI/NotI using standard molecular biology techniques. For pmpG-1, pmpE/F-2, only the first half of the gene (representing amino acids 25 – 500 and 25–575, respectively) was cloned into the vector for expression. The sequences of the sub-cloned genes were confirmed by sequencing with dye-labeled terminators using the ABI PRISM kit (PE Biosystems). Plasmids containing the rplF, fabG, aasf, pmpG-125–500, TC0420, clp-1, pmpE/F-225–575 and gap genes were transformed into the E. coli strain BL21(DE3) (Strategene) where protein expression was carried out by inducing the lac promoter for expression of T7 RNA polymerase using isopropyl-beta-D-thiogalactopyranoside. The expressed RplF, FabG, Aasf, TC0420, and Clp-1 proteins with N-terminal GST-tag were purified from E. coli lysates by affinity chromatography using glutathione sepharose 4 fastflow purification system (GE Healthcare). PmpG-125–500, PmpE/F-225–575 and Gap proteins with N-terminal His-tag were purified by nickel column using the His bind purification system (Qiagen). LPS removal was carried out by adding 0.1% Triton-114 in the wash buffers during purification.
Mice
Female C57BL/6 mice (8 to 10 weeks old) were purchased from Charles River Canada (Saint Constant, Canada). The mice were maintained and used in strict accordance with University of British Columbia guidelines for animal care.
DC generation from bone marrow
DCs were generated following the protocol described by Lutz et al. (27). Briefly, bone marrow cells were prepared from the femora and tibiae of naïve C57BL/6 mice and cultured in DC medium. DC medium is IMDM supplemented with 10% FCS, 0.5 mM 2-ME, 4 mM L-glutamine, 50 μg/ml gentamicin, 5% of culture supernatant of murine GM-CSF transfected plamacytoma X63-Ag8 and 5% of culture supernatant of murine IL-4 transfected plamacytoma X63-Ag8 which contained approximately 10 ng/ml of GM-CSF and 10 ng/ml of IL-4 respectively. The above two cell lines were kindly provided by Dr. F. Melchers, Basilea Institute, Switzerland. Culture medium was changed every three days. After 8-day culture, the cells were harvested for transfection with Chlamydia protein antigens. Approximately 65~70% percent of the cell preparation were DCs as judged by a staining with anti-CD11c monoclonal antibody.
DC transfection with Chlamydia proteins
DCs harvested on day 8 were washed twice in RPMI 1640. Sixty microlitres of the liposomal transfection reagent N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate (DOTAP; Roche) and the Chlamydia proteins PmpG-125–500, RplF, PmpE/F-225–575, MOMP or the negative control protein, GST were mixed with 240 μl RPMI 1640 at room temperature in polystyrene tubes for 20 min (28, 29). The final concentration of PmpG-125–500, PmpE/F-225–575, MOMP or GST protein in the DOTAP/protein mixtures is 0.2 mg/ml and RplF protein is 0.8 mg/ml. DCs (2~3 ×107) in 3 ml RPMI 1640 were added to the DOTAP/protein mixtures. The protein transfected DCs were incubated for 3 h at 37 °C, washed twice, resuspended in DC medium and then cultured overnight in the presence of 0.25 μg/ml LPS for maturation. DCs on day 8 pulsed with live EB (MOI1) for 24 hours was prepared as a positive control. Antigen loaded DCs were used for in vitro immunohistochemical analysis and in vivo immunization.
Immunohistochemistry
The protein transfected DCs were deposited onto Micro Slides using Shandon Cytospin (Thermo Electron Corp.). The DCs on the slides were fixed for 20 min in 4 % paraformaldehyde in PBS. Subsequently, they were permeabilized for 10 min in 0.5 % Triton X-100 in PBS. The cells were blocked for 20 min with PBS containing 1 % horse serum, and incubated with corresponding antigen-specific polyclonal murine serum (1: 200) respectively for 2 h. All anti-Chlamydia protein polyclonal antibodies (PmpG-125–500, RplF, PmpE/F-225–575, or MOMP) were made in our laboratory as follows: Balb/c mice were immunized three times subcutaneously with 10 μg recombinant Chlamydia protein formulated with Incomplete Freunds Adjuvant (Sigma). Two weeks after the final immunization, sera from each group were collected and pooled. All anti-Chlamydia recombinant protein polyclonal antibodies had titers ≥1: 500,000 dilution as determined by ELISA. Biotinylated horse anti-mouse IgG (1: 2000) (Vector Laboratories) was added and then the cells were incubated again for 1 h. Finally, the cells were incubated for 45 min with ABC Reagent (Vector Laboratories) and incubated with peroxidase substrate solution (DAB substrate kit SK-4100; Vector Laboratories) until the desired stain intensity developed. The slides were rinsed in tap water, counterstained with 0.1 % toluidine blue, and again rinsed in tap water. All incubations were performed at room temperature and the slides were washed in PBS three times between incubations.
ELISPOT assay
The IFN-γ ELISPOT assay was performed as described previously (30). Briefly, 96-well MultiScreen-HA filtration plates (Millipore) were coated overnight at 4 °C with 2 μg/ml of murine IFN-γ specific monoclonal antibody (BD PharMingen, Clone R4-6A2). Splenocytes isolated from mice in AIM-V medium were added to the coated plates at 106 cells per well in presence of individual Chlamydia peptide at 2 μg/ml or individual Chlamydia protein at 1 μg/ml. After 20 h incubation at 37 °C and 5% CO2, the plates were washed and then incubated with biotinylated murine IFN-γ specific monoclonal antibodies (BD PharMingen, Clone XMG1.2) at 2 μg/ml. This was followed by incubation with streptavidin–alkaline phosphatase ((BD PharMingen) at a 1:1000 dilution. The spots were visualized with a substrate consisting of 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Sigma–Aldrich).
Adoptive transfer of DCs transfected with Chlamydia protein antigens
Mice were vaccinated three times with a 2-week interval, intravenously (i.v.) into the tail veins with 1 × 106 DCs transfected with Chlamydia protein PmpG-125–500, RplF, PmpE/F-225–575 or MOMP in 200 μl of PBS. DCs pulsed with live EB or GST protein was used as positive or negative controls respectively. Two weeks after the last immunization, six mice of each group were euthanized to isolate splenocytes for IFN-γ ELISPOT assay. The remaining mice were used for Chlamydia infection challenge.
Pulmonary and cervicovaginal challenge and determination of Chlamydia titer
Two weeks after the final immunization, five to ten mice from each group were intranasally (i.n.) challenged with 2000 IFU of C. muridarum. Weight loss was monitored each or every two days. On 10 day after i.n. challenge, the mice were euthanized and the lungs were collected for Chlamydia titration. Single-cell suspensions were prepared by homogenizing the lungs with tissue grinders and coarse tissue debris was removed by centrifugation at 1000 ×g for 10 min at 4°C. The clarified suspensions were serially diluted and immediately inoculated onto HeLa 229 monolayers for titration (31). For genital tract infections, one week after the final immunization, ten mice from each group were injected s.c. with 2.5 mg of medroxyprogesterone acetate (Depo-Provera; Pharmacia and Upjohn). One week after Depo-Provera treatment, the mice were challenged intravaginally with 1500 IFU of C. muridarum. Cervicovaginal washes were taken at day 6 after infection and stored at −80°C for titration on HeLa cells as described previously (31).
Statistical analysis
All data were analyzed with the aid of a software program (GraphPad Prism 3.0). Differences between the means of experimental groups were analyzed using an independent, two-tailed t-test at the level of p < 0.05.
Results
Identification of immunodominant Chlamydia antigens among the 8 MHC class II binding peptides
Using an immunoproteomic approach, we previously identified 8 MHC class II (I-Ab) -bound Chlamydia peptides eluted from DCs infected with C. muridarum (25). For this study we cloned and purified the 8 source proteins that contained the corresponding peptides (Table 1). To determine which individual peptides or proteins are immunodominant in the context of natural infection, we performed IFN-γ ELISPOT assay using splenocytes from C57BL/6 mice recovered from live C. muridarum infection.
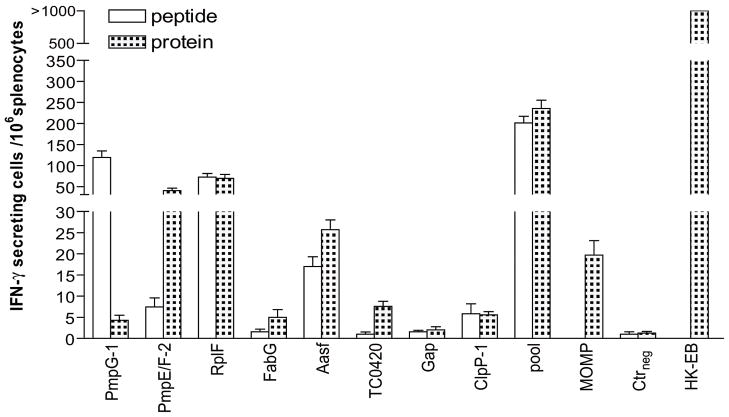
Splenocytes from mice harvested one month after C. muridarum infection were stimulated in vitro for 20 h with 2 μg/ml individual peptide, 1 μg/ml individual protein, pooled peptides or pooled proteins. Irrelevant OVA peptide and GST were used as peptide and protein negative controls respectively and heat killed EB (HK-EB) as positive control. Since MOMP has been long standing candidate in Chlamydia vaccine studies, MOMP was also set up as a reference antigen. As shown in Fig. 1, immune splenocytes exposed to HK-EB developed the highest numbers of IFN-γ secreting cells where more than 1000 IFN-γ secreting cells were detected among 106 splenocytes. In contrast, splenocytes stimulated with the OVA peptide or GST protein as negative controls showed nearly blank background levels indicating that IFN-γ secreting cells detected in the experimental system are Chlamydia antigen-specific. Stimulation by pooled peptides or pooled proteins induced significantly higher numbers of IFN-γ secreting cells than stimulation with individual Chlamydia antigens (p < 0.05).
Figure 1. Recognition of individual Chlamydia peptides eluted from DCs and their source proteins in C57BL/6 mice recovered from live C. muridarum infection identified by IFN-γ ELISPOT assay.
Mice were infected with i.n.1000 IFU live C. muridarum. One month later, the splenocytes from recovered mice were harvested and stimulated with in vitro for 20 h with 2 μg/ml individual peptide, 1 μg/ml individual protein, pooled peptides or pooled proteins. One irrelevant OVA peptide and GST were used as peptide and protein negative controls (Ctrneg) respectively and heat killed EB (HK-EB) as positive control. MOMP protein stimulation was also set up as a reference. The results represent the average of duplicate wells and are expressed as the means ± SEM of Chlamydia antigen-induced IFN-γ secreting cells per 106 splenocytes for groups of six mice. These data are representative of three similar experiments.
Immune splenocytes stimulated with individual Chlamydia antigens exhibited markedly different levels of IFN-γ response (Fig. 1). The results demonstrated that IFN-γ responses in immune splenocytes to stimulation of PmpG-1 peptide, PmpE/F-225–575 protein, RplF peptide and protein were strong; Aasf peptide, Aasf protein or MOMP protein were moderate and others were weak. Thus, three of the 8 antigens (PmpG-1, RplF and PmpE/F-2) were determined as immunodominant based on their strong IFN-γ responses by ELISPOT assay to stimulation by either the peptide or parent protein. The above experiments were repeated 3 times and showed very consistent data.
Efficient intracellular uptake of Chlamydia protein by DCs using DOTAP as a delivery system
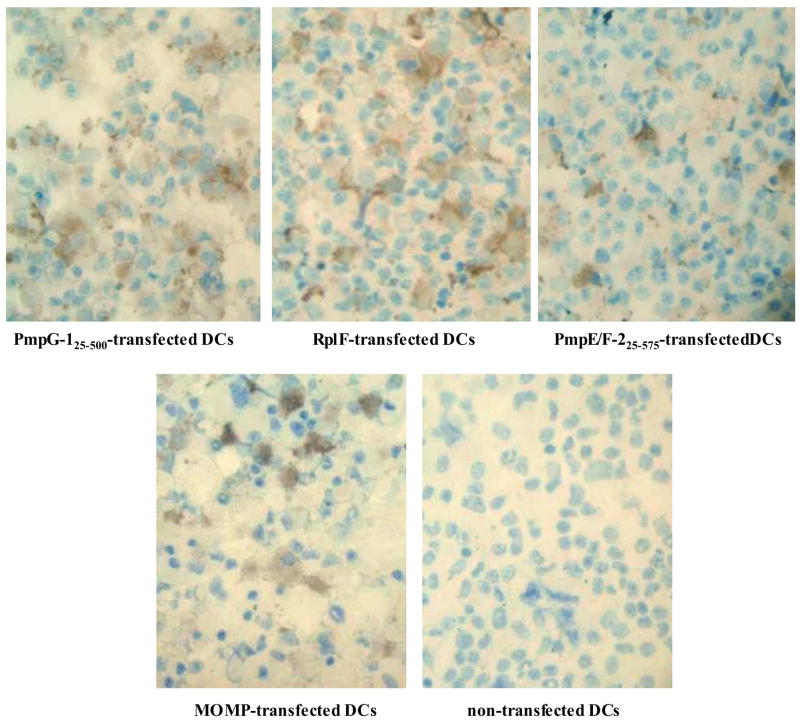
Because protein antigens require endocytotosis and lysosomal processing before peptide loading onto MHC class II molecules can take place, we used the cationic liposome DOTAP to deliver Chlamydia protein intracellularly into DCs. The intracellular uptake of PmpG-125–500, PmpE/F-225–575, RplF or MOMP protein was visualized by immunohistochemistry following transfection. As shown in Fig. 2, strong expression of PmpG-125–500, RplF, PmpE/F-225–575 and MOMP was detected in the cytoplasm of the Chlamydia protein-transfected DCs, whereas no signal was detected in non-transfected DCs. Thus the cationic liposome DOTAP efficiently delivered Chlamydia protein intracellularly into DCs.
Figure 2. Chlamydia protein uptake by DCs detected by immunohistochemistry.
Chlamydia protein uptake in DCs transfected with PmpG-125–500, Rplf, PmpE/F-225–575, MOMP or without protein. The cationic liposome DOTAP was used to deliver Chlamydia protein into the DCs. The presence of Chlamydia protein after transfection was visualized with a protein corresponding polyclonal mouse antibody followed by a biotinylated horse anti-mouse IgG and a DAB substrate.
After DC transfection with Chlamydia proteins, DCs were matured with LPS for 18 h. We evaulated the cell surface antigen expression of the DCs after LPS stimulation and found no phenotypic difference between DCs transfected with different Chlamydia antigens or GST DCs stimulated with LPS expressed enhanced levels of CD40, MHC class II and CD86 compared with unstimulated DCs. (data not shown).
Specific immune responses to Chlamydia antigens following adoptive transfer of DCs transfected with Chlamydia proteins
We next wanted to investigate whether adoptive transfer of transfected DCs induces antigen specific immune responses. Again, a group of DCs transfected with MOMP was set up as a reference control antigen. One group of mice received DCs pulsed with GST protein as a negative control and another group received DCs pulsed with viable C. muridarum EB as a positive control.
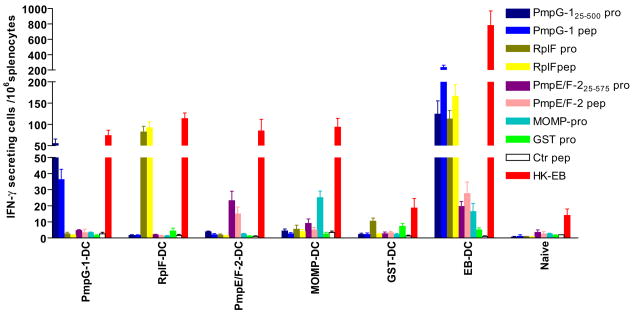
Two weeks following the final adoptive transfer, Chlamydia specific immune responses in vaccinated mice were assessed by enumerating antigen-specific IFN-γ producing cells in splenocytes from each group after exposure to Chlamydia antigens (Fig. 3). The results showed that the groups of DCs transfected with individual Chlamydia proteins developed significant antigen specific IFN-γ responses to corresponding peptides and proteins but not to other non-related Chlamydia antigens. Importantly mice immunized with DCs transfected with individual Chlamydia proteins demonstrated strong specific immune responses to HK EB (p < 0.01). As a positive control, mice that received DCs pulsed with live C. muridarum (EB) developed the strongest IFN-γ responses to HK-EB shown by more than 1000 IFN-γ-secreting cells detected among 106 splenocytes. This group also exhibited strong antigen-specific IFN-γ responses to PmpG-1 peptide/protein and RplF peptide/protein and moderate responses to PmpE/F-2 peptide/protein and MOMP. In contrast, naïve and GST-DC vaccinated splenocytes stimulated with the Chlamydia antigens or HK-EB showed low background levels except for the GST-DC group which exhibited some responses to GST protein and the GST-fusion protein, RplF. IL-4 ELISPOT assays were also performed and showed no or very low Chlamydia antigen specific IL-4 secretion in any groups immunized with DCs transfected with individual Chlamydia protein (data not shown).
Figure 3. Specific immune responses to Chlamydia antigens following adoptive transfer of DCs transfected with Chlamydia proteins detected by IFN-γ ELISPOT assay.
Mice were vaccinated three times with DCs transfected with Chlamydia protein PmpG-125–500, RplF, PmpE/F-225–575 or MOMP and matured overnight with LPS. DCs pulsed with live C. muridarum or GST protein was used as positive or negative control respectively. Two weeks after the last immunization, the splenocytes of each group were harvested for IFN-γ ELISPOT assay. The results are expressed as the means ± SEM of Chlamydia antigen-induced IFN-γ secreting cells per 106 splenocytes for groups of six mice. These data are representative of two similar experiments.
Protection against Chlamydia infection following adoptive transfer of DCs transfected ex vivo with the three immunodominant Chlamydia protein antigens
To evaluate whether the Chlamydia protein antigens were able to protect mice against Chlamydia pulmonary and genital tract infection, we undertook adoptive transfer studies using LPS-matured DCs transfected ex vivo with PmpG-125–500, RplF, PmpE/F-225–575 or MOMP. Mice received DCs transfected with GST or pulsed with viable C. muridarum were set up as negative and positive controls respectively. Two weeks following the final adoptive transfer, mice were challenged intranasally or vaginally with C. muridarum.
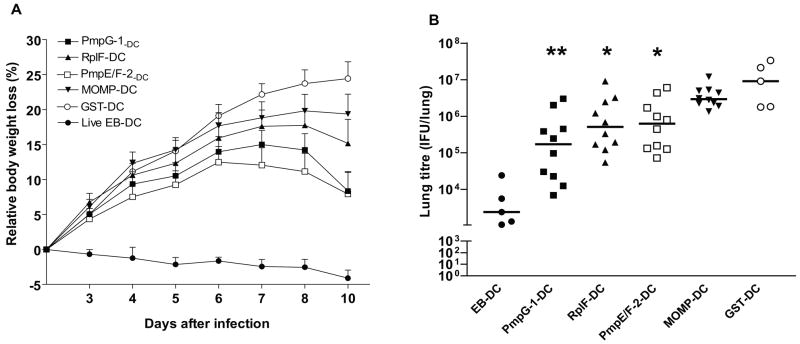
After the intranasal challenge, protection was measured by body weight loss and bacterial load in the lungs. As shown in Fig. 4A, mice adoptively immunized with live EB-pulsed DC demonstrated excellent protection against infection as indicated by no body weight loss. In contrast, mice immunized with GST-pulsed DC exhibited the largest weight loss. The mean body weight loss on day 10 post infection reached 24.4±2.4% in the negative control group (p < 0.001 vs. positive control). Mice vaccinated with individual Chlamydia antigen-transfected DCs showed varying levels of protection as indicated by different degrees of body weight loss during the 10-day period. The mean relative body weight loss at day 10 in groups of PmpE/F-2-DC, PmpG-1-DC, RplF-DC, or MOMP was 7.9±3.1%, 8.1±2.7%, 15.2±3.4%, and 19.4±2.8% respectively. During the late of period following infection, the recovery from lost body weight appeared most rapid in PmpG-1-DC group among the four immunized Chlamydia antigen-transfected DC groups.
Figure 4. Resistance to Chlamydia Pulmonary Infection following adoptive transfer of DCs transfected with Chlamydia proteins.
Adoptive transfer of DCs was described in Materials and Methods. Two weeks after the last immunization, mice were challenged intranasally with 2000 IFU live C. muridarum. Weight loss was monitored each or every two days after challenge (A). Ten days after intranasal challenge, the lungs were collected and bacterial titers were measured on HeLa 229 cells (B). *, p < 0.05; **, p < 0.01 vs. GST-DC group. These data are representative of two similar experiments.
Ten days after the intranasal challenge, lungs were harvested and Chlamydia inclusion forming units were determined by plating serial dilutions of homogenized lungs onto HeLa 229 cells. When compared to the negative control group, the median Chlamydia titers decreased 1.8 orders of magnitude (log10) in mice vaccinated with PmpG-1-DC (p < 0.01) and decreased 1.2 and 1.1 orders of magnitude in mice vaccinated with RplF-DC (p < 0.05) and PmpE/F-2-DC (p < 0.05) respectively. There was no statistically significant difference in lung Chlamydia titers between mice vaccinated with MOMP-DC and the negative control group (Fig. 4B).
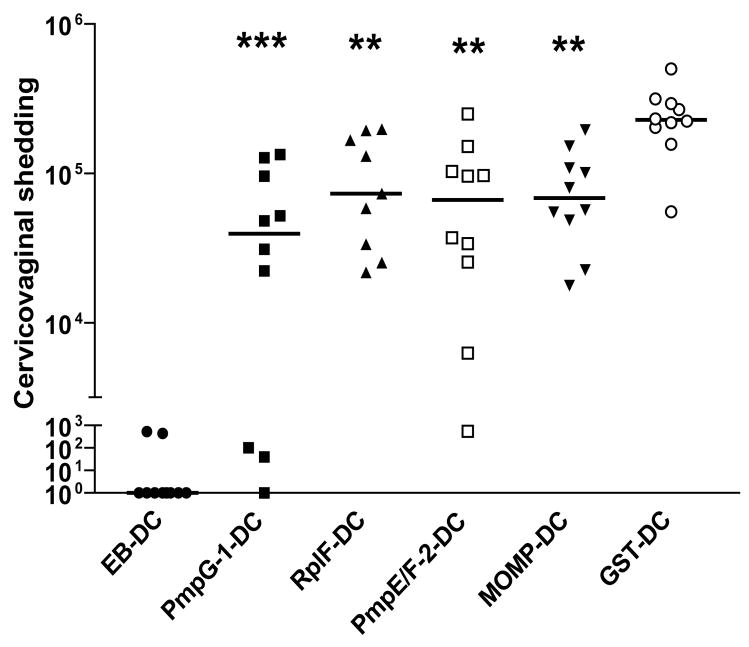
Protection against intravaginal infection was assessed by isolation of Chlamydia from cervicovaginal wash and determination of the number of IFU recovered from each experimental group at day 6 post-infection (Fig. 5). The results showed that the cervicovaginal shedding of C. muridarum in mice immunized with any of the four Chlamydia protein-transfected DCs was significantly lower than that of mice who received GST-transfected DCs (p < 0.001 in PmpG-1 group; p < 0.01 in RplF group; p <0.01 in PmpE/F-2 group; p < 0.01 in MOMP group).
Figure 5. Resistance to Chlamydia genital tract infection following adoptive transfer of DCs transfected with Chlamydia proteins.
Adoptive transfer of DCs was described in Materials and Methods. One week after the final immunization, the mice from each group were injected with Depo-Provera. One week after Depo-Provera treatment, the mice were infected intravaginally with 1500 IFU live C. muridarum. Cervicovaginal washes were taken at day 6 after infection and bacterial titer were measured on HeLa 229 cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 vs. GST-DC group. These data are representative of two similar experiments.
Taken together, mice vaccinated with DCs transfected with Chlamydia protein PmpG-125–500, RplF or PmpE/F-225–575 exhibited significant resistance to challenge infection as indicated by log10 reduction in the median Chlamydia titer in comparison with the negative control group in both lung model and genital tract model. MOMP, as a reference antigen, conferred significant protection but only in the genital tract model. Overall the data demonstrated that vaccination with DCs transfected with PmpG-125–500 developed the greatest degree of protective immunity among the four Chlamydia antigens evaluated.
Discussion
Chlamydia immunity is currently understood as T cell-mediated based on CD4 Th1 T cells producing IFN-γ. A major impediment to developing a vaccine against this intracellular pathogen lies in identifying relevant T cell antigens able to stimulate protective immunity (8). Our previous study using an immunoproteomic approach successfully identified 8 MHC class II-bound Chlamydia peptides directly eluted from murine dendritic cells infected with Chlamydia. A pool of the 8 Chlamydia MHC II binding peptides partially protected mice against Chlamydia infection when peptide loaded DCs were adoptively transferred to mice prior to challenge infection (25). In this study we demonstrated that three of the 8 antigens (PmpG-1, RplF and PmpE/F-2) were immunodominant as identified by IFN-γ ELISPOT assay using splenocytes from mice recovered from natural infection. Furthermore, mice vaccinated with DCs transfected with individual Chlamydia protein PmpG-125–500, RplF or PmpE/F-225–575 elicited IFN-γ immune responses and exhibited significant resistance to challenge infection as indicated by log10 reduction in the median Chlamydia titer in both the lung and genital tract models. Vaccination of DCs transfected with MOMP as a reference antigen demonstrated significant protective immunity in the genital tract model but not in the lung model. Among the four evaluated immunogens, PmpG-125–500 was the most promising vaccine candidate.
To our knowledge, this is the first published report to show that PmpG-125–500, PmpE/F-225–575 or RplF are able to engender protective immunity against challenge with C. muridarum. PmpG-1 and PmpE/F-2 belong to the polymorphic membrane protein (Pmp) family unique to Chlamydiales (32). There are 9 pmp genes (pmpA to pmpI) identified for C. trachomatis (33, 34). The pmp genes are predicted to be localized in the outer membrane, and all Pmps have been detected by proteomics as membrane constituents (35). The C. trachomatis Pmps are abundant and account for 3.15% of the organism’s protein coding capacity and are immunogenic for humans (18, 36). By protein structure analysis, they are predicted to be autotransporters that mediate the translocation of the N terminus to the bacterial surface (35). They contain multiple GGAI motifs, which have been associated in other organisms with adhesion to the host cell (37). It is likely that they play an important role in Chlamydia structural, functional or antigenic polymorphism. Chlamydia genome analysis (38) showed that the PmpE/F-2 homolog from C. trachomatis (PmpF) contains a disproportionate number of single-nucleotide polymorphisms, many of which are located at predicted sites of T cell epitopes that bind to human MHC (HLA) class I and II alleles indicating Chlamydia PmpF could be a particularly important immune target of T cells. A recent study based on comparative expression profiling of the C. trachomatis pmp gene family demonstrated that pmpF also had the highest mRNA expression level among the nine pmp genes of tested C. trachomatis strains (39). There is less immunobiological information on PmpG of C. trachomatis, however PmpG has been identified as recognized by CD4 T cells during C. pneumoniae infection in mice (40). RplF is the ribosomal protein L6 that has homology to the N terminus of a putative membrane protein adhesion of 18 kDa possessing Hela-binding ability (41). Virtually nothing is known regarding immune recognition of RplF or the other Chlamydia peptides/proteins, Gap, FabG, Aasf, TC0420 and ClpP-1 evaluated in this study. Since PmpG-125–500, RplF and PmpE/F-225–575 induced significant levels of protection against Chlamydia challenge infection, they are a high priority for further detailed immunobiological investigation including their potential role in human immunity to Chlamydia infection.
Since IFN-γ has been found to play a major role in mediating control of Chlamydia infection, we measured IFN-γ responses to Chlamydia antigens to evaluate antigenicity of the identified antigens in mice recovered from live C. muridarum infection and in mice vaccinated with the corresponding protein antigen. Following adoptive transfer of live EB-pulsed DCs, mice demonstrated strong IFN-γ responses to both peptides and proteins of PmpG-1, RplF and moderate IFN-γ responses to both peptides and proteins of PmpE/F-2, whereas mice who recovered from live EB infection showed comparable levels of IFN-γ responses to both peptide and protein of RplF, PmpG-1 peptide and PmpE/F-225–575 protein but none or low IFN-γ responses to PmpG-125–500 protein and PmpE/F-2 peptide (compare Fig.1 with Fig.3). These results may be explained in part by differential MHC class II loadng from peptide versus protein sources. Additionally it may be that PmpG-125–500 and PmpE/F-225–575 contain multiple immunoprotective peptides and DCs pulsed ex vivo with live C. muridarum present a greater variety of Chlamydia peptides than do DCs in vivo which sample Chlamydia antigens via infected mucosal tissues during natural infection. Importantly the results following vaccination with DCs transfected with PmpG-125–500 or PmpE/F-225–575 protein showed that both EBs and corresponding peptide and protein induced IFN-γ responses (Fig. 3).
Because an optimal Th1 polarized adjuvant that efficiently delivers Chlamydia antigens is as yet undefined, we used the cationic liposome DOTAP as a tool to deliver Chlamydia proteins directly into DCs and subsequently matured DCs with LPS overnight before vaccination. Our study demonstrated that liposome DOTAP remarkably increased the uptake of PmpG-125–500, RplF, Pmp E/F225–575 and MOMP protein by DCs compared to the uptake without DOTAP (data not shown). Although we did not evaluate the protective effect against Chlamydia challenge between groups of mice immunized with and without DOTAP in this study, our previous work showed that MOMP-pulsed DCs without DOTAP induced very poor protection (unpublished data), whereas MOMP-pulsed DC using DOTAP conferred significant protection against genital tract infection. One recent study reported that delivery of ovalbumin to DCs using cationic liposomes resulted in more than 100-fold increase in the efficiency of MHC class II antigen presentation (42). LPS-matured DCs exhibit enhanced levels of MHC class II, CD86 and CD40 expression. The LPS treatment, in particular, dramatically increased the expression of CD40, which functions in the adaptive immune response as a trigger for efficient T-cell activation (43).
Further studies will be conducted to determine whether the same Chlamydia protein antigens engender cross-species and cross-serovar immunity and provide protection in different MHC genetic backgrounds. A successful Chlamydia vaccine will likely need to be composed of multiple purified recombinant proteins and provide broad coverage in an outbred population and cross protect against multiple variants of C. trachomatis. Additionally immunogenicity for vaccine antigens will need to be optimized using compatible adjuvants in order to improve the partial protection that has been found with the current immunization protocol. This study provides a direct and useful strategy for discovery of T cell-based subunit molecular vaccines against Chlamydia as well potentially for other intracellular pathogens such as tuberculosis, malaria and HIV.
Footnotes
This work was supported by grants from Canadian Institutes of Health Research and National Institute of Health (Grant No. R01AI076483).
Abbreviations used in this paper: CMI, cell-mediated immune response; DC, dendritic cell; BMDC, bone marrow-derived DC; IFU, inclusion-forming units; EB, elementary body; HK-EB, heat-killed EB; DOTAP, N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate
References
- 1.Starnbach MN, Roan NR. Conquering sexually transmitted diseases. Nat Rev Immunol. 2008;8:313–317. doi: 10.1038/nri2272. [DOI] [PubMed] [Google Scholar]
- 2.Plummer FA, Simonsen JN, Cameron DW, Ndinya-Achola JO, Kreiss JK, Gakinya MN, Waiyaki P, Cheang M, Piot P, Ronald AR, et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 3.Anttila T, Saikku P, Koskela P, Bloigu A, Dillner J, Ikaheimo I, Jellum E, Lehtinen M, Lenner P, Hakulinen T, Narvanen A, Pukkala E, Thoresen S, Youngman L, Paavonen J. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. Jama. 2001;285:47–51. doi: 10.1001/jama.285.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J Infect Dis. 2005;192:1836–1844. doi: 10.1086/497341. [DOI] [PubMed] [Google Scholar]
- 5.Su H, Morrison R, Messer R, Whitmire W, Hughes S, Caldwell HD. The effect of doxycycline treatment on the development of protective immunity in a murine model of chlamydial genital infection. J Infect Dis. 1999;180:1252–1258. doi: 10.1086/315046. [DOI] [PubMed] [Google Scholar]
- 6.Wang SP, Grayston JT, Alexander ER. Trachoma vaccine studies in monkeys. Am J Ophthalmol. 1967;63(Suppl):1615–1630. doi: 10.1016/0002-9394(67)94155-4. [DOI] [PubMed] [Google Scholar]
- 7.Grayston JT, Wang SP. The potential for vaccine against infection of the genital tract with Chlamydia trachomatis. Sex Transm Dis. 1978;5:73–77. doi: 10.1097/00007435-197804000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 9.Stanberry LR, Rosenthal SL. Progress in vaccines for sexually transmitted diseases. Infect Dis Clin North Am. 2005;19:477–490. xi. doi: 10.1016/j.idc.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Brunham RC, Peeling RW. Chlamydia trachomatis antigens: role in immunity and pathogenesis. Infect Agents Dis. 1994;3:218–233. [PubMed] [Google Scholar]
- 11.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen J, Jensen KT, Follmann F, Agger EM, Theisen M, Andersen P. Liposome delivery of Chlamydia muridarum major outer membrane protein primes a Th1 response that protects against genital chlamydial infection in a mouse model. J Infect Dis. 2008;198:758–767. doi: 10.1086/590670. [DOI] [PubMed] [Google Scholar]
- 13.Kari L, Whitmire WM, Crane DD, Reveneau N, Carlson JH, Peterson EM, Pal S, de la Maza LM, Caldwell HD. Chlamydia trachomatis native-MOMP induces partial protection in non-human primates. Fourth Meeting of the European Society for Chlamydia Research; Aarhus, Denmark. 1–4 July 2008.2008. [Google Scholar]
- 14.Eko FO, He Q, Brown T, McMillan L, Ifere GO, Ananaba GA, Lyn D, Lubitz W, Kellar KL, Black CM, Igietseme JU. A novel recombinant multisubunit vaccine against Chlamydia. J Immunol. 2004;173:3375–3382. doi: 10.4049/jimmunol.173.5.3375. [DOI] [PubMed] [Google Scholar]
- 15.Fling SP, Sutherland RA, Steele LN, Hess B, D’Orazio SE, Maisonneuve J, Lampe MF, Probst P, Starnbach MN. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2001;98:1160–1165. doi: 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starnbach MN, Loomis WP, Ovendale P, Regan D, Hess B, Alderson MR, Fling SP. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–4749. doi: 10.4049/jimmunol.171.9.4742. [DOI] [PubMed] [Google Scholar]
- 17.Deane KH, Jecock RM, Pearce JH, Gaston JS. Identification and characterization of a DR4-restricted T cell epitope within chlamydia heat shock protein 60. Clin Exp Immunol. 1997;109:439–445. doi: 10.1046/j.1365-2249.1997.4711371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodall JC, Yeo G, Huang M, Raggiaschi R, Gaston JS. Identification of Chlamydia trachomatis antigens recognized by human CD4+ T lymphocytes by screening an expression library. Eur J Immunol. 2001;31:1513–1522. doi: 10.1002/1521-4141(200105)31:5<1513::AID-IMMU1513>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol. 2008;180:3375–3382. doi: 10.4049/jimmunol.180.5.3375. [DOI] [PubMed] [Google Scholar]
- 20.Su H, Caldwell HD. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect Immun. 1995;63:3302–3308. doi: 10.1128/iai.63.9.3302-3308.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Fan Y, Brunham RC, Yang X. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur J Immunol. 1999;29:3782–3792. doi: 10.1002/(SICI)1521-4141(199911)29:11<3782::AID-IMMU3782>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Morrison SG, Su H, Caldwell HD, Morrison RP. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun. 2000;68:6979–6987. doi: 10.1128/iai.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison SG, Morrison RP. Resolution of secondary Chlamydia trachomatis genital tract infection in immune mice with depletion of both CD4+ and CD8+ T cells. Infect Immun. 2001;69:2643–2649. doi: 10.1128/IAI.69.4.2643-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry LL, Feilzer K, Hughes S, Caldwell HD. Clearance of Chlamydia trachomatis from the murine genital mucosa does not require perforin-mediated cytolysis or Fas-mediated apoptosis. Infect Immun. 1999;67:1379–1385. doi: 10.1128/iai.67.3.1379-1385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, Gabel BR, Yu H, Foster LJ, Brunham RC. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol. 2008;180:2459–2465. doi: 10.4049/jimmunol.180.4.2459. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, HayGlass KT, Brunham RC. Genetically determined differences in IL-10 and IFN-gamma responses correlate with clearance of Chlamydia trachomatis mouse pneumonitis infection. J Immunol. 1996;156:4338–4344. [PubMed] [Google Scholar]
- 27.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Chiriva-Internati M, Grizzi F, Salati E, Roman JJ, Lim S, Hermonat PL. Rapid induction of cytotoxic T-cell response against cervical cancer cells by human papillomavirus type 16 E6 antigen gene delivery into human dendritic cells by an adeno-associated virus vector. Cancer Gene Ther. 2001;8:948–957. doi: 10.1038/sj.cgt.7700391. [DOI] [PubMed] [Google Scholar]
- 29.Nonn M, Schinz M, Zumbach K, Pawlita M, Schneider A, Durst M, Kaufmann AM. Dendritic cell-based tumor vaccine for cervical cancer I: in vitro stimulation with recombinant protein-pulsed dendritic cells induces specific T cells to HPV16 E7 or HPV18 E7. J Cancer Res Clin Oncol. 2003;129:511–520. doi: 10.1007/s00432-003-0462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioannou XP, Griebel P, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. The immunogenicity and protective efficacy of bovine herpesvirus 1 glycoprotein D plus Emulsigen are increased by formulation with CpG oligodeoxynucleotides. J Virol. 2002;76:9002–9010. doi: 10.1128/JVI.76.18.9002-9010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilenki L, Wang S, Yang J, Fan Y, Joyee AG, Yang X. NK T cell activation promotes Chlamydia trachomatis infection in vivo. J Immunol. 2005;175:3197–3206. doi: 10.4049/jimmunol.175.5.3197. [DOI] [PubMed] [Google Scholar]
- 32.Vandahl BB, Birkelund S, Christiansen G. Genome and proteome analysis of Chlamydia. Proteomics. 2004;4:2831–2842. doi: 10.1002/pmic.200400940. [DOI] [PubMed] [Google Scholar]
- 33.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 35.Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes JP, Hsia RC, Mead S, Borrego MJ, Dean D. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect. 2005;7:410–420. doi: 10.1016/j.micinf.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Grimwood J, Stephens RS. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb Comp Genomics. 1999;4:187–201. doi: 10.1089/omi.1.1999.4.187. [DOI] [PubMed] [Google Scholar]
- 38.Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunes A, Gomes JP, Mead S, Florindo C, Correia H, Borrego MJ, Dean D. Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PLoS ONE. 2007;2:e878. doi: 10.1371/journal.pone.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mygind T, Vandahl B, Pedersen AS, Christiansen G, Hollsberg P, Birkelund S. Identification of an in vivo CD4+ T cell-mediated response to polymorphic membrane proteins of Chlamydia pneumoniae during experimental infection. FEMS Immunol Med Microbiol. 2004;40:129–137. doi: 10.1016/S0928-8244(03)00300-6. [DOI] [PubMed] [Google Scholar]
- 41.Gray GJ, Kaul R, Roy KL, Wenman WM. Isolation and molecular characterization of the ribosomal protein L6 homolog from Chlamydia trachomatis. J Bacteriol. 1991;173:1663–1669. doi: 10.1128/jb.173.5.1663-1669.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korsholm KS, Agger EM, Foged C, Christensen D, Dietrich J, Andersen CS, Geisler C, Andersen P. The adjuvant mechanism of cationic dimethyldioctadecylammonium liposomes. Immunology. 2007;121:216–226. doi: 10.1111/j.1365-2567.2007.02560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Sullivan B, Thomas R. CD40 and dendritic cell function. Crit Rev Immunol. 2003;23:83–107. doi: 10.1615/critrevimmunol.v23.i12.50. [DOI] [PubMed] [Google Scholar]