Abstract
Histone proteins are subject to modifications, such as acetylation, methylation, phosphorylation, ubiquitination, glycosylation, and ADP ribosylation, some of which are known to play important roles in the regulation of chromatin structure and function. Here we report that histone H4 is modified by small ubiquitin-related modifier (SUMO) family proteins both in vivo and in vitro. H4 binds to the SUMO-conjugating enzyme (E2), UBC9, and can be sumoylated in an E1 (SUMO-activating enzyme)- and E2-dependent manner. We present evidence suggesting that histone sumoylation mediates gene silencing through recruitment of histone deacetylase and heterochromatin protein 1.
Chromatin is composed of nucleosomes in which 146 bp of DNA are wrapped around a core histone octamer. The octamer consists of a (H3–H4)2 tetramer associated with two H2A–H2B dimers. The C-terminal globular domains of core histones bind to each other to form the octamer whereas core histone N-terminal tails protrude from the nucleosome and are accessible to the enzymatic modification machineries that catalyze acetylation, methylation, phosphorylation, glycosylation, and ADP ribosylation (for reviews see refs. 1 and 2). Over the last several years, rapid progress has been made in the field of histone modification, owing largely to the discoveries that histone acetyltransferases and histone deacetylases function as transcriptional coactivators and corepressors, respectively (3). These discoveries were followed by analyses of the mechanism and effects of acetylation and the identification of other histone modifications including phosphorylation, methylation, and ubiquitination as regulators of transcription.
Small ubiquitin-related modifier (SUMO) is the best-characterized member of a growing family of ubiquitin-like proteins involved in posttranslational modifications (for reviews see refs. 4–6). In mammals, there are three members of the SUMO protein family: SUMO-1, SUMO-2 (SMT3a), and SUMO-3 (SMT3b), which are implicated in partly overlapping, yet distinct functions (7, 8). SUMO is covalently attached to other proteins through the activities of an enzyme cascade (E1–E2–E3) similar to that for ubiquitination. There is only one known SUMO-activating enzyme, E1 (a heterodimer of SAE1 and SAE2) and only one known SUMO-conjugating enzyme, E2 (UBC9). There appear to be a number of different SUMO ligases (E3s) in higher eukaryotes such as PIAS family proteins (9–11), RanBP2 (12), and the polycomb protein Pc2 (13). To date, dozens of proteins from different species have been identified as sumoylation substrates. Among these are the Ran-GTPase-activating protein, RanGAP1 (14), PML (15), IκBα (16), p53 (17, 18), and MDM2 (19). Unlike ubiquitination, sumoylation of proteins has not been linked to protein degradation. Proposed functions for sumoylation include regulation of protein–protein interaction and localization, inhibition of ubiquitin-mediated degradation, and enhancement of transcriptional activity.
Histone proteins have been long known to be ubiquitinated (20), a modification that has been more recently linked to transcriptional function (21, 22). SUMO family proteins resemble ubiquitin both in structure and their mechanism of ligation to substrates; yet the functional consequences of sumoylation are quite different from ubiquitination (for reviews see refs. 4, 5, and 23).
Here we investigate the possibility that core histones are sumoylated and demonstrate that histone H4 is modified with SUMO family proteins both in vivo and in vitro. We present evidence suggesting that histone sumoylation mediates transcriptional repression through recruitment of histone deacetylases and heterochromatin protein 1 (HP1). We propose that histone sumoylation is another modification that must be considered to understand chromatin regulation.
Materials and Methods
Cell Culture. The 293 and 293T cells were cultured in DMEM supplemented with 10% calf serum. HeLa cells were cultured in DMEM supplemented with 10% FCS. P493-6 human B cells were cultured in RPMI medium 1640 supplemented with 10% FCS. Calcium phosphate coprecipitation was used for transfection.
Immunoprecipitation and Immunoblotting. For immunoprecipitation under denaturing conditions, the cells were lysed in Ab buffer (20 mM Tris, pH 7.4/50 mM NaCl/0.5% Nonidet P-40/0.5% deoxycholate/0.5% SDS/1 mM EDTA/protease inhibitors). For immunoprecipitation under native conditions, the cells were lysed in TNE buffer (10 mM Tris, pH 7.4/150 mM NaCl/1% Nonidet P-40/1 mM EDTA/protease inhibitors). Immunoprecipitation was performed essentially as described (24). Immunoblotting was done as described (25).
The following Abs were used: anti-FLAG M2 mAb (Sigma), anti-hemagglutinin (HA) 16B12 Ab (Covance, Princeton), anti-FLAG polyclonal Ab (Santa Cruz Biotechnology), anti-SUMO-1 Ab (Zymed), anti-acetylated histone H4 and H3 Abs (06–866 and 06–599, Upstate Biotechnology, Lake Placid, NY), and anti-HP1-γ Ab (Chemicon).
Cell Fractionation. Soluble (S) and chromatin-enriched (C) fractions were prepared as described (26).
In Vitro Sumoylation Reaction. In vitro-translated, 35S-Met-labeled histone H4 was incubated with purified E1 (SAE1 0.15 μg/SAE2 0.45 μg), purified E2 (UBC9 0.5 μg), and GST-SUMO-1 (0.5 μg) (where indicated) in sumoylation buffer (50 mM Tris, pH 7.4/5 mM ATP/10 mM MgCl2/0.2 mM DTT) at 30°C for 3 h.
GST Pull-Down Assay and Luciferase Assay. In vitro-translated, 35S-Met-labeled protein was incubated with 5 μg of purified GST fusion protein in 1× PBS containing 0.4% Nonidet P-40 for 1 h at 4°C and washed three times, and bound proteins were analyzed by SDS/PAGE. The 4xGAL14D luciferase and 14D luciferase reporters were as described (27). Luciferase assay was performed as described (28).
Chromatin Immunoprecipitation. Chromatin immunoprecipitation under denaturing conditions was performed as described (29). Chromatin immunoprecipitation under nondenaturing conditions was performed as described (30). The region amplified comprised the 330-bp sequence at the 5′ end of the luciferase cDNA, located 100 bp downstream of the GAL4 sites. The DNA sequence of the 5′ primer was 5′-GACGCCAAAAACATAAAGAAAGGCC-3′and of the 3′ primer was 5′-TTCACGTTCATTATAAATGTCGTTC-3′. PCRs were repeated by using varying cycle numbers and different amounts of templates to ensure that results were within the linear range of PCR.
Results
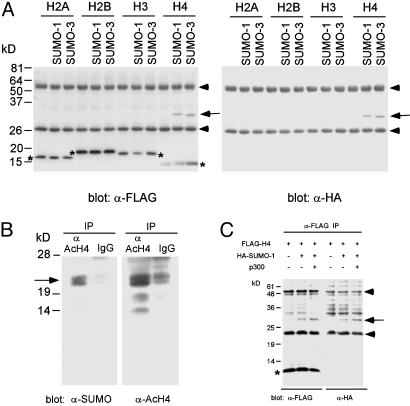
To investigate the possible sumoylation of core histones, we have cloned the four major core histone genes, added a FLAG tag to their N termini, and examined their sumoylation after transfection in 293T cells. FLAG-tagged histone H2A, H2B, H3, or H4 was transfected alone or together with HA-tagged SUMO-1 or SUMO-3, and the cell lysates were immunoprecipitated with anti-FLAG Ab under denaturing conditions. As shown in Fig. 1A, modification of histone H4 with SUMO-1 or SUMO-3 (≈15-kDa size shift) was readily detectable under these conditions. Incorporation of the SUMO moiety into H4 was confirmed by anti-FLAG (-H4) immunoprecipitation followed by anti-HA (-SUMO) immunoblotting (Fig. 1 A Right). The sumoylation of H4 detected by this procedure was not affected by known modulators of sumoylation such as UV, H2O2, heat shock, or nocodazole treatment (data not shown). Upon longer exposure, weak sumoylation of H2A, H2B, and H3 was also observed (data not shown). To examine whether endogenous histone H4 is sumoylated, we prepared chromatin fractions from human B cells and performed antiacetylated H4 immunoprecipitation followed by anti-SUMO-1 immunoblotting. As shown in Fig. 1B, sumoylation (and acetylation) of endogenous H4 was confirmed by this procedure. Coexpression of histone acetyltransferase p300 enhanced the sumoylation of FLAG-H4 (Fig. 1C), suggesting that acetylation of histone(s) may facilitate the sumoylation of H4 and that the use of antiacetylated H4 Ab may have increased the sensitivity of detection of H4 sumoylation.
Fig. 1.
Sumoylation of histone H4 in vivo.(A) Modification of histone H4 by SUMO-1 and SUMO-3. FLAG-tagged histone H2A, H2B, H3, or H4 was cotransfected with HA-tagged SUMO-1 or SUMO-3 (where indicated) into 293T cells. Two days later, anti-FLAG immunoprecipitates (under denaturing conditions) were analyzed by anti-FLAG (Left) or anti-HA (Right) immunoblotting. The asterisks indicate the positions of unmodified histones. The arrow indicates the position of sumoylated H4. The addition of the FLAG tag increases the apparent molecular mass of H4 from 10 to 14 kDa. The apparent molecular mass of the SUMO monomer is ≈15 kDa. Arrowheads indicate Ig chains. The presence of the SUMO moiety in the modified H4 was confirmed by anti-HA immunoblotting (Right). (B) Sumoylation of endogenous histone H4 in human B cells. A chromatin fraction derived from human P493-6 B cells (1 × 108 cells; see Materials and Methods) was immunoprecipitated under denaturing conditions with antiacetylated histone H4 Ab or a control Ab, and the immuno-precipitate was analyzed by anti-SUMO-1 immunoblotting (Left) or antiacetylated histone H4 immunoblotting (Right). The position of sumoylated, acetylated histone H4 is indicated by the arrow. The identity of ≈15- and 18-kDa bands detected by antiacetylated H4 immunoblotting is unknown. The sumoylation of histone H4 appeared to enhance the efficiency of electrotransfer. Most of the nonsumoylated histone H4 remained in the gel after transfer under the conditions used. (C) Enhancement of H4 sumoylation by histone acetyltransferase p300. FLAG-tagged histone H4 was transfected alone or with HA-tagged SUMO-1 or p300 (where indicated) into 293T cells. Two days later, anti-FLAG immunoprecipitates (under denaturing conditions) were analyzed by anti-FLAG (Left) or anti-HA (Right) immunoblotting. The asterisk indicates the positions of unmodified histones. The arrow indicates the position of sumoylated H4. Arrowheads indicate Ig chains. The presence of the SUMO moiety in the modified H4 was confirmed by anti-HA immunoblotting (Right).
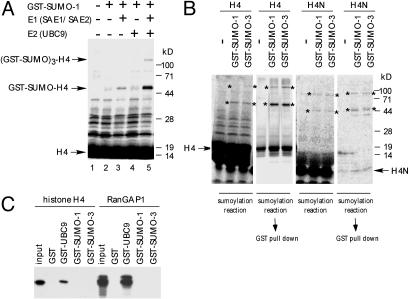
To analyze the mechanism of H4 sumoylation, we tested whether H4 can be sumoylated in vitro. As shown in Fig. 2A, lanes 2–5, in vitro-translated H4 was sumoylated in an E1 (SAE1/SAE2)- and E2 (UBC9)-dependent manner. The weaker sumoylation of H4 observed in the absence of exogenous UBC9 (Fig. 2 A, lane 3) is most likely caused by endogenous UBC9 present in the reticulocyte lysate. The high molecular weight band present in Fig. 2 A, lane 5, may represent H4 sumoylated at three sites.
Fig. 2.
Sumoylation of histone H4 in vitro. (A) E1- and E2-dependent sumoylation of histone H4. In vitro-translated, 35S-Met-labeled histone H4 was incubated with purified E1 (SAE1 0.15 μg/SAE2 0.45 μg), purified E2 (UBC9 0.5 μg), and GST-SUMO-1 (0.5 μg) (where indicated) at 30°C for 3 h. The positions of unmodified H4 and H4 modified with GST-SUMO-1 are indicated by arrows. (B) Sumoylation of the N-terminal tail of histone H4. In vitro-translated, 35S-Met-labeled full-length histone H4 or the N-terminal tail of H4 (H4N, amino acid 1–26) was subjected to the sumoylation reaction with GST-SUMO-1, or GST-SUMO-3, and E1 and E2 as in A. The positions of modified H4 or H4N are indicated by asterisks. An aliquot of the sumoylation reaction mixture was bound to glutathione agarose beads to assess the association of GST-SUMO proteins with H4. (C) Binding of histone H4 to UBC9. In vitro-translated, 35S-Met-labeled histone H4 was incubated with GST, GST-UBC9, GST-SUMO-1, or GST-SUMO-3 attached to glutathione agarose beads. The bound proteins were analyzed by SDS/PAGE (Left). As a positive control, the interaction of RanGAP1 with GST-UBC9 was also examined (Right). The input lanes represent 10% of the input to the binding reactions.
The core histone proteins possess C-terminal globular domains involved in histone octamer formation and N-terminal tail domains that are subject to various modifications. To determine whether the N-terminal tail of histone H4 can be modified with SUMO proteins, we examined the sumoylation of in vitro-translated H4 N-terminal tail. As shown in Fig. 2B, the H4 N-terminal tail, as well as full-length H4 is modified with GST-SUMO-1 or GST-SUMO-3, which was further confirmed by precipitation with glutathione agarose beads. The high molecular weight bands observed in the presence of GST-SUMO were also precipitated with glutathione agarose (Fig. 2B), suggesting that these represent H4 and H4N modified with GST-SUMO at multiple sites.
We also examined the effect of a PIAS protein (31), a recently identified E3 for sumoylation (9–11) and observed no effect on H4 sumoylation in vitro or in vivo (data not shown). Because some sumoylation substrates have been demonstrated to directly bind UBC9 (32, 33), we performed an in vitro-binding assay to assess this possibility for histone H4. As shown in Fig. 2C, in vitro-translated H4 bound to GST-UBC9, but not to GST, GST-SUMO-1, or GST-SUMO-3. As a positive control, the binding of RanGAP1 to GST-UBC9 was also examined (Fig. 2C Right).
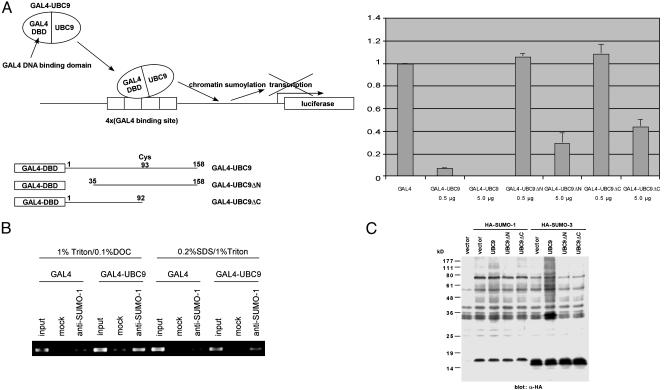
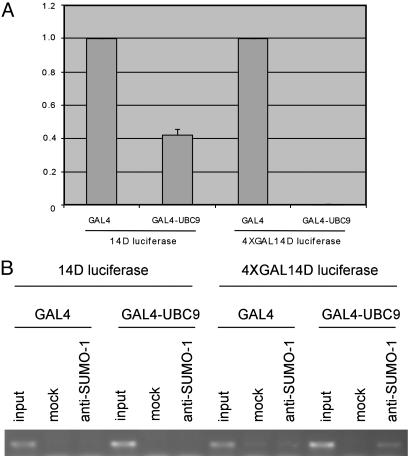
As an approach to understanding the function of histone sumoylation, we examined the transcriptional consequences of recruiting UBC9 to a promoter. We fused UBC9 to a GAL4 DNA-binding domain and determined its effect on a GAL4-dependent reporter gene. As shown in Fig. 3A, GAL4-UBC9 strongly repressed transcription, which is consistent with the notion that sumoylation of chromatin component(s) mediates transcriptional repression. To determine whether sumoylation of chromatin is associated with transcriptional repression by GAL4-UBC9, we performed anti-SUMO-1 chromatin immuno-precipitation of the GAL4 reporter plasmid cotransfected with a vector expressing GAL4-UBC9 or GAL4 alone. As shown in Fig. 3B, GAL4-UBC9 induced chromatin sumoylation proximal to the GAL4-binding sites. Chromatin sumoylation by GAL4-UBC9 was detected under both nondenaturing (Fig. 3B Left) (30) and denaturing (Fig. 3B Right) (29) chromatin immunoprecipitation conditions. Deletion of the N-terminal E1-binding domain (UBC9ΔN) (34) or the C-terminal domain including the active site Cys-93 (UBC9ΔC) strongly attenuated transcriptional repression by GAL4-UBC9 (Fig. 3A). In contrast to intact UBC9, these UBC9 deletion mutants could not induce the sumoylation of cellular proteins (Fig. 3C). These results suggest that sumoylation of chromatin component(s) by GAL4-UBC9 mediates transcriptional repression. Moreover, the ability of GAL4-UBC9 to induce strong repression (>200-fold) depends on GAL4-binding sites because a reporter lacking such sites is repressed by GAL4-UBC9 only ≈2-fold (Fig. 4A). As shown in Fig. 4B, chromatin sumoylation by GAL4-UBC9 also depended on the presence of GAL4-binding sites.
Fig. 3.
Chromatin sumoylation is associated with transcriptional repression by GAL4-UBC9. (A) Transcriptional repression by GAL4-UBC9. HeLa cells were cotransfected with 2 μg of 4XGAL14D luciferase reporter and indicated amount of GAL4, GAL4-UBC9 (full-length, amino acid 1–158), GAL4-UBC9ΔN (35–158), or GAL4-UBC9ΔC(1–92). After transfection (48 h), the luciferase activities were determined. The data represent the average of two independent experiments. Comparable expression of GAL4, GAL4-UBC9, GAL4-UBC9ΔN, and GAL4-UBC9ΔC was confirmed by anti-GAL4 immunoblotting (data not shown). (B) Chromatin sumoylation by GAL4-UBC9. HeLa cells were cotransfected with 4XGAL14D luciferase reporter and GAL4 or GAL4-UBC9. Two days later, sumoylation of chromatin near the GAL4 site was examined by anti-SUMO-1 chromatin immunoprecipitation. Mock lanes represent the chromatin immunoprecipitation without the Ab. Chromatin sumoylation by GAL4-UBC9 was detected by chromatin immunoprecipitation under both nondenaturing (Left) and denaturing (Right) conditions. (C) SUMO-conjugating activities of UBC9 deletion mutants. The 293T cells were cotransfected with HA-SUMO-1 or HA-SUMO-3 together with a vector, UBC9, UBC9DN, or UBC9DC. Whole-cell lysate (30 μg) was analyzed by anti-HA immunoblotting for sumoylation of cellular proteins.
Fig. 4.
Transcriptional repression and chromatin sumoylation by GAL4-UBC9 depend on the GAL4-binding sites. (A) Transcriptional repression by GAL4-UBC9 depends on the GAL4-binding sites. HeLa cells were cotransfected with 2 μg of 14D luciferase reporter (without GAL4-binding sites) or 4XGAL14D luciferase reporter (with GAL4-binding sites) and 5 μg of GAL4 or GAL4-UBC9. Luciferase activities were determined 48 h after transfection. The data shown represent the average of two independent experiments. (B) Chromatin sumoylation by GAL4-UBC9 depends on the presence of GAL4-binding sites. HeLa cells were cotransfected with 14D luciferase reporter (without GAL4-binding sites) or 4XGAL14D luciferase reporter (with GAL4-binding sites) and GAL4 or GAL4-UBC9. Chromatin sumoylation by GAL4-UBC9 was detected by chromatin immunoprecipitation under nondenaturing condition as in Fig. 3B.
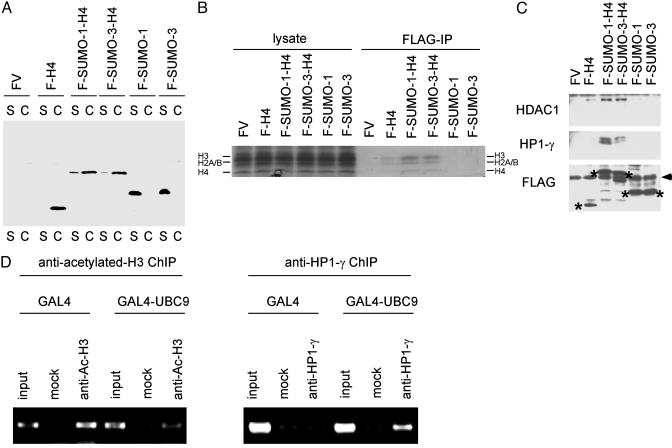
To examine the possible interaction of sumoylated histone H4 with transcriptional repression/gene silencing components, we fused SUMO-1 or SUMO-3 to the N terminus of H4. This process generates a form of H4 that is constitutively sumoylated at its N terminus (see Fig. 2B) without the complication of indirect effects associated with overexpression of SUMO or SUMO-conjugating enzymes. SUMO-H4 fusion constructs were transfected into 293 cells, and the cells were fractionated into soluble (S) and chromatin (C) fractions (26, 35). As shown in Fig. 5A, SUMO-H4 fusion proteins were detected predominantly in chromatin fractions. The transfected SUMO-H4 fusion proteins were immunoprecipitated under native conditions and were shown to be associated with core histones (Fig. 5B). We were unable to detect the association of transfected SUMO-1 or SUMO-3 with endogenous H4. This finding is possibly caused by the high rate of SUMO incorporation into RanGAP1 compared to the lower efficiency of incorporation into H4. These results indicate that SUMO-H4 fusion proteins are functional in the sense that they associate with other core histones as well as chromatin. Because we observed a connection between transcriptional repression and chromatin sumoylation (Fig. 3), we decided to examine the possible interaction of SUMO-H4 fusion proteins with histone deacetylase (HDAC) and HP1, two well studied proteins that mediate transcriptional repression/gene silencing. SUMO-H4 fusion constructs were transfected into 293 cells, and the expressed SUMO-H4 fusion proteins were immunoprecipitated under native conditions. As shown in Fig. 5C, SUMO-H4 fusion proteins associate with endogenous HDAC1 and HP1-γ. Furthermore, by using chromatin immunoprecipitation, we confirmed that GAL4-UBC9 induces histone deacetylation and recruitment of HP1-γ proximal to GAL4-binding sites (Fig. 5D). These results suggest that GAL4-UBC9 represses transcription by sumoylating histone H4 and recruiting HDAC or HP1 to sumoylated H4.
Fig. 5.
Histone sumoylation is associated with recruitment of HDAC and HP1. (A) Incorporation of SUMO-H4 fusion proteins into chromatin. The 293 cells were transfected with FLAG-vector (FV), FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and the cells were fractionated into soluble (S) and chromatin (C) fractions as described (26). The location of each FLAG-tagged protein was determined by anti-FLAG immunoblotting. (B) Association of SUMO-H4 fusion proteins with core histones. The 293 cells were transfected with FLAG vector, FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and 2 days later cell lysates were immunoprecipitated with anti-FLAG Ab under nondenaturing conditions. The immunoprecipitates were analyzed by SDS/PAGE and Coomassie staining for the presence of core histones (H4, H2A/B, and H3). (C) Interaction of SUMO-H4 fusion proteins with HDAC1 and HP1-γ. The 293 cells were transfected with FLAG vector, FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and 2 days later cell lysates were immunoprecipitated with anti-FLAG Ab (under native conditions) and analyzed by anti-HDAC1 (Top) or anti-HP1-γ (Middle) immunoblotting. The expression level of each FLAG-tagged protein (shown by *) was examined by anti-FLAG immunoblotting (Bottom). The position of Ig light chain is shown by the arrowhead (Bottom). (D) Histone deacetylation and recruitment of HP1-γ induced by GAL4-UBC9. HeLa cells were cotransfected with 4XGAL14D luciferase reporter and GAL4 or GAL4-UBC9. Two days later, histone acetylation status and recruitment of HP1-γ near the GAL4 sites were examined by antiacetylated histone H3 (Left) and anti-HP1-γ (Right) chromatin immunoprecipitation under nondenaturing conditions.
Discussion
Although a large number of proteins with varied activities and functions have been reported to be posttranslationally modified by SUMO moieties (4, 5), to our knowledge there have been no reports of sumoylation of general chromatin components. Here we provide several lines of evidence that the core histone H4 is modified by sumoylation both in vivo and in vitro and that this modification is likely to be linked to transcriptional repression. We have demonstrated that, of the four major core histones, only histone H4 was efficiently modified by both SUMO-1 or SUMO-3 after transfection (Fig. 1A) whereas a relatively lower degree of sumoylation was observed for H2A, H2B, and H3. Furthermore endogenous H4 was found to be covalently bound to SUMO after immunoprecipitation (Fig. 1B). Because our Ab recognized only acetylated H4, and because coexpression of the histone acetyltransferase p300 appeared to promote sumoylation of H4 (Fig. 1C), we believe that acetylation provides at least a partial signal for histone sumoylation.
The sumoylation of histone H4 can be carried out in vitro and depends on the known sumoylation pathway enzymes, specifically the E1 SUMO-activating enzyme (SAE1 and SAE2) and UBC9, the only known SUMO-conjugating enzyme (Fig. 2 A). We have found that histone H4 can associate with UBC9 (Fig. 2C) perhaps explaining why in vitro sumoylation of H4 apparently does not require a separate SUMO ligase. Indeed, the known SUMO ligase, PIAS (9–11) had no effect on H4 sumoylation in vivo or in vitro. These data are consistent with previous reports showing that, unlike ubiquitination, an E3 for sumoylation is not absolutely required, and some sumoylation substrates directly bind to E2 (UBC9) (32, 33). We have also shown that the N-terminal tail of H4 is a target for linkage of multiple SUMO moieties (Fig. 2B). This region is the same as that targeted for most of the known histone posttranslational modifications. Unfortunately, however, we have not yet been able to obtain sufficient amounts of sumoylated endogenous H4 to definitively determine the residues linked to SUMO. Possibly the low levels of endogenous sumoylated H4 are indicative of the transient nature of histone sumoylation, similar to that proposed for histone ubiquitination (S. Berger, personal communication).
We have also taken several approaches to define the consequences of histone H4 sumoylation for transcription. We show that fusion of the GAL4 DNA-binding domain to UBC9 mediates repression of a reporter gene containing GAL4-binding sites (Figs. 3A and 4A). This repression depends on both an intact E1-binding site and the catalytic domain of UBC9. Importantly, strong repression by GAL4-UBC9 corresponded to the appearance of sumoylated chromatin, presumably at H4 within nucleosomes, in the vicinity of the GAL4-binding sites (Figs. 3B and 4B). Consistent with the observed repression, we also detected decreased acetylation of histone H3 and a sharp increase in binding by HP1-γ. Interestingly, UBC9 has been previously shown to interact with a number of DNA-binding transcription factors [such as c-Jun, p53, androgen receptor, and TEL (4)] and some of these transcription factors may use UBC9 as a corepressor. We think it is plausible that recruitment of UBC9 by these transcription factors leads to sumoylation of histone H4 in chromatin, which in turn promotes deacetylation and HP1-γ binding. Indeed, in an experimental approach similar to that used to examine the function of ubiquitination and sumoylation of transcription factors (36, 37), we fused SUMO to the N terminus of histone H4 (SUMO-H4) and found that it cofractionated with chromatin and core histones and was associated with both endogenous HDAC1 and HP1-γ (Fig. 5 A–C).
Although we show that UBC9 can sumoylate histone H4 and induce HDAC1 and HP1-γ recruitment and repression, it is likely that UBC9 also sumoylates chromatin proteins other than histone H4. Both the PIAS and Pc2 proteins have been shown to be associated with chromatin (38) and thus may function as SUMO ligases for transcriptional components exclusive of H4. A number of chromatin proteins such as p53, proliferating cell nuclear antigen, DNA topoisomerases, and HDAC4 are known to be sumoylated, and it is likely that sumoylation of chromatin proteins are involved in the regulation of chromatin.
Based on the results presented here, we propose histone sumoylation as a component of the group of modifications that appear to govern chromatin structure and function to mediate transcriptional repression and gene silencing (1, 39). In Drosophila polytene chromosomes, the SUMO moiety was detected in many euchromatic sites and the chromocenter (40), suggesting that histone (chromatin) sumoylation plays a role in both euchromatic transcriptional repression and heterochromatic gene silencing. Future work should clarify the precise roles of histone/chromatin sumoylation in the regulation of chromatin function.
Acknowledgments
We thank Ronald Hay for SAE1 and SAE2 cDNAs, Jorma Palvimo for PIAS cDNA, Edward Seto for anti-HDAC1 Ab, and Dirk Eick for P493-6 cells. We thank Sara Hook, Mark Groudine, Steve Henikoff, and Dan Gottschling for critical readings of the manuscript and Shelley Berger for communication of unpublished information. This work was supported by National Institutes of Health Grant R01CA57138 (to R.N.E.) and the Interdisciplinary Research Training Fellowship from the Fred Hutchinson Cancer Research Center (to Y.S.). R.N.E. is a Research Professor of the American Cancer Society.
Abbreviations: SUMO, small ubiquitin-related modifier; HDAC, histone deacetylase; HP1, heterochromatin protein 1; HA, hemagglutinin.
See Commentary on page 13118.
References
- 1.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L. (2001) Oncogene 20, 3007-3013. [DOI] [PubMed] [Google Scholar]
- 3.Kuo, M. H. & Allis, C. D. (1998) BioEssays 20, 615-626. [DOI] [PubMed] [Google Scholar]
- 4.Melchior, F. (2000) Annu. Rev. Cell Dev. Biol. 16, 591-626. [DOI] [PubMed] [Google Scholar]
- 5.Hay, R. T. (2001) Trends Biochem. Sci. 26, 332-333. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, E. S. & Gupta, A. A. (2001) Cell 106, 735-744. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh, H. & Hinchey, J. (2000) J. Biol. Chem. 275, 6252-6258. [DOI] [PubMed] [Google Scholar]
- 8.Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H. & Hay, R. T. (2001) J. Biol. Chem. 276, 35368-35374. [DOI] [PubMed] [Google Scholar]
- 9.Kahyo, T., Nishida, T. & Yasuda, H. (2001) Mol. Cell 8, 713-718. [DOI] [PubMed] [Google Scholar]
- 10.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, Y., Kahyo, T., Toh, E. A., Yasuda, H. & Kikuchi, Y. (2001) J. Biol. Chem. 276, 48973-48977. [DOI] [PubMed] [Google Scholar]
- 12.Pichler, A., Gast, A., Seeler, J. S., Dejean, A. & Melchior, F. (2002) Cell 108, 109-120. [DOI] [PubMed] [Google Scholar]
- 13.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127-137. [DOI] [PubMed] [Google Scholar]
- 14.Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997) Cell 88, 97-107. [DOI] [PubMed] [Google Scholar]
- 15.Muller, S., Matunis, M. J. & Dejean, A. (1998) EMBO J. 17, 61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desterro, J. M., Rodriguez, M. S. & Hay, R. T. (1998) Mol. Cell 2, 233-239. [DOI] [PubMed] [Google Scholar]
- 17.Gostissa, M., Hengstermann, A., Fogal, V., Sandy, P., Schwarz, S. E., Scheffner, M. & Del Sal, G. (1999) EMBO J. 18, 6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez, M. S., Desterro, J. M., Lain, S., Midgley, C. A., Lane, D. P. & Hay, R. T. (1999) EMBO J. 18, 6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buschmann, T., Fuchs, S. Y., Lee, C. G., Pan, Z. Q. & Ronai, Z. (2000) Cell 101, 753-762. [DOI] [PubMed] [Google Scholar]
- 20.Goldknopf, I. L., Taylor, C. W., Baum, R. M., Yeoman, L. C., Olson, M. O., Prestayko, A. W. & Busch, H. (1975) J. Biol. Chem. 250, 7182-7187. [PubMed] [Google Scholar]
- 21.Pham, A. D. & Sauer, F. (2000) Science 289, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 22.Sun, Z. W. & Allis, C. D. (2002) Nature 418, 104-108. [DOI] [PubMed] [Google Scholar]
- 23.Muller, S., Hoege, C., Pyrowolakis, G. & Jentsch, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202-210. [DOI] [PubMed] [Google Scholar]
- 24.Luscher, B. & Eisenman, R. N. (1988) Mol. Cell. Biol. 8, 2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiio, Y., Donohoe, S., Yi, E. C., Goodlett, D. R., Aebersold, R. & Eisenman, R. N. (2002) EMBO J. 21, 5088-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wysocka, J., Reilly, P. T. & Herr, W. (2001) Mol. Cell. Biol. 21, 3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayer, D. E., Laherty, C. D., Lawrence, Q. A., Armstrong, A. P. & Eisenman, R. N. (1996) Mol. Cell. Biol. 16, 5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasier, A. R. & Fortin, J. J., eds. (1995) Current Protocols in Molecular Biology (Wiley, New York), Vol. 3.
- 29.Wu, W. S., Vallian, S., Seto, E., Yang, W. M., Edmondson, D., Roth, S. & Chang, K. S. (2001) Mol. Cell. Biol. 21, 2259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, Y., Rayman, J. B. & Dynlacht, B. D. (2000) Genes Dev. 14, 804-816. [PMC free article] [PubMed] [Google Scholar]
- 31.Moilanen, A. M., Karvonen, U., Poukka, H., Yan, W., Toppari, J., Janne, O. A. & Palvimo, J. J. (1999) J. Biol. Chem. 274, 3700-3704. [DOI] [PubMed] [Google Scholar]
- 32.Lee, G. W., Melchior, F., Matunis, M. J., Mahajan, R., Tian, Q. & Anderson, P. (1998) J. Biol. Chem. 273, 6503-6507. [DOI] [PubMed] [Google Scholar]
- 33.Kim, Y. H., Choi, C. Y. & Kim, Y. (1999) Proc. Natl. Acad. Sci. USA 96, 12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bencsath, K. P., Podgorski, M. S., Pagala, V. R., Slaughter, C. A. & Schulman, B. A. (2002) J. Biol. Chem. 277, 47938-47945. [DOI] [PubMed] [Google Scholar]
- 35.Mendez, J. & Stillman, B. (2000) Mol. Cell. Biol. 20, 8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salghetti, S. E., Caudy, A. A., Chenoweth, J. G. & Tansey, W. P. (2001) Science 293, 1651-1653. [DOI] [PubMed] [Google Scholar]
- 37.Ross, S., Best, J. L., Zon, L. I. & Gill, G. (2002) Mol. Cell 10, 831-842. [DOI] [PubMed] [Google Scholar]
- 38.Hari, K. L., Cook, K. R. & Karpen, G. H. (2001) Genes Dev. 15, 1334-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber, S. L. & Bernstein, B. E. (2002) Cell 111, 771-778. [DOI] [PubMed] [Google Scholar]
- 40.Lehembre, F., Badenhorst, P., Muller, S., Travers, A., Schweisguth, F. & Dejean, A. (2000) Mol. Cell. Biol. 20, 1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]