Abstract
An important goal after structural genomics is to build up the structures of higher-order protein–protein complexes from structures of the individual subunits. Often structures of higher order complexes are difficult to obtain by crystallography. We have used an alternative approach in which the structures of the individual catalytic (C) subunit and RIα regulatory (R) subunit of PKA were first subjected to computational docking, and the top 100,000 solutions were subsequently filtered based on amide hydrogen/deuterium (H/2H) exchange interface protection data. The resulting set of filtered solutions forms an ensemble of structures in which, besides the inhibitor peptide binding site, a flat interface between the C-terminal lobe of the C-subunit and the A- and B-helices of RIα is uniquely identified. This holoenzyme structure satisfies all previous experimental data on the complex and allows prediction of new contacts between the two subunits.
Keywords: matrix-assisted, laser desorption ionization/time-of-flight, protein–protein interactions, PKA holoenzyme (type I)
Protein kinase A (PKA), a central locus for cAMP-mediated signaling in mammalian cells, exists in an inactive state as a tetrameric holoenzyme composed of two regulatory (R) and two catalytic (C) subunits. Binding of cAMP to the R-subunits leads to dissociation of the holoenzyme and unleashing of the active C-subunit. Like many protein kinases, inhibition of the C-subunit by the RIα regulatory subunit occurs by bipartite binding to the C-subunit. The R-subunit has a pseudosubstrate/inhibitor sequence near the N terminus that binds in the active site of the C-subunit. Two cAMP-binding domains (A and B) follow the pseudosubstrate/inhibitor sequence, and it is the A-domain of the RIα isoform that contains the peripheral binding site for inhibition of the C-subunit (1, 2). Although crystal structures are available for the C-subunit in various active and inhibited forms (3), and for the cAMP-bound RIα(113–376) (4), no structural information is available for the holoenzyme complex consisting of the R-subunit and the C-subunit.
Computational docking is a useful approach to build up the structures of larger protein–protein complexes. The program DOT computes the interaction energies of all possible (billions) translational and rotational solutions by using fast Fourier transform algorithms (5, 6). The interaction between the C-subunit and the R-subunit involves two sites, each of which contributes to the overall binding energy, and DOT did not predict a unique docking interface. To filter the results, we first tried using results from complementary mutagenesis experiments, which revealed a contact between Lys-213 in the C-subunit with Glu-143 in the R-subunit (7). This contact alone, however, was insufficient to specify a unique solution (I.T. and L.T.E., unpublished results).
Amide H/2H exchange, followed by proteolytic digestion and mass spectrometry, is a powerful method to map protein–protein and protein–ligand interactions (8–11). Amide H/2H exchange coupled to matrix-assisted laser desorption ionization/time-offlight (MALDI-TOF) mass spectrometry has been used to map the inhibitor peptide ATP and ADP-binding sites within the C-subunit (8, 12).
The results of this study show that filtering the comprehensive docking results with distance constraints derived from the amide H/2H exchange data predicts a single binding interface and a structure of the PKA holoenzyme that is consistent with previous biochemical results. This approach may be particularly useful for kinases and other signaling molecules that have bipartite binding to the active site as well as to a peripheral site.
Materials and Methods
Materials. ATP, cAMP, Mops, and cAMP immobilized on 6% agarose were obtained from Sigma. Deuterium oxide (D2O; 99.9% deuterium) was obtained from Cambridge Isotope Laboratories (Cambridge, MA). Pepsin immobilized on 6% agarose was obtained from Pierce. Trifluoroacetic acid and acetonitrile were obtained from Fisher and were of the highest grade; α-cyano-4-hydroxycinnamic acid (Aldrich) was recrystallized once from ethanol. PD10 columns for buffer exchange were obtained from Amersham Pharmacia Biotech. All other materials were reagent grade from standard commercial sources.
Preparation of RIα(94–244) and C-Subunits. RIα(94–244) was expressed as a polyhistidine-tagged protein and purified by passing through a nickel-nitrilotriacetic acid column and eluted with sodium phosphate buffer (pH 7.0), containing 100 mM imidazole as described (11). The Cα-subunit of murine PKA was expressed in Escherichia coli and purified as described (13). The final buffer was 50 mM Mops (pH 7.0), with 2 mM ATP and 0.2 mM MgCl2, and the protein concentration was 125 μM.
Formation of Holoenzyme. The C-subunit and 1.2 molar excess of cAMP-bound RIα(94–244) or and cAMP-bound full-length RIα were combined in 10 mM Mops (pH 7.0), 50 mM NaCl, 5 mM β-mercaptoethanol with 0.2 mM ATP, 2 mM MgCl2 as described (10). The proteins were concentrated to 172 μM for RIα(94–244)-C holoenzyme and 125 μM for the full-length type I holoenzyme (R2C2) before deuterium exchange.
Computational Docking with dot of C-Subunit and RIα(113-244). Computational docking analyses of the C-subunit with the R-subunit was carried out by using the program DOT. The atomic coordinates used were PDB:1RGS for the regulatory subunit, RIα (4) and PDB:1ATP (3) for the C-subunit. Only residues 113–244 of the RIα subunit were used in the docking, and residues 245–376 were added after the docked solutions were obtained. ATP and the 20-residue peptide PKI(5–24) were retained in the C-subunit structure. The C-subunit was held stationary while the R-subunit was rotated and translated about it. The electrostatic potential grid for the C-subunit was generated by solving the linearized Poisson–Boltzmann equation with the program UHBD (University of Houston Brownian Dynamics) after assigning partial charges according to an AMBER parameter set (14). The potential was evaluated on a 128 × 128 × 128 grid with 1.0-Å spacing, a solvent dielectric of 78.0, a protein dielectric of 3.0, an ionic radius of 1.4 Å, a solvent radius of 1.4 Å, and a solvent ionic strength of 150 mM. The entire calculation took ≈4 hr on a SUN Microsystems Ultra HPC 10000 using 20 processors.
Deuterium Exchange and Mass Spectrometry. Deuterium exchange was initiated by combining the protein solution (10 μl) with deuterated Mops buffer (90 μl) at 25°C. After 0, 0.5, 1, 2, 5, or 10 min, the deuterium exchange reaction was quenched by addition to ice-cold 0.9 ml of 0.1% trifluoroacetic acid (TFA) with 2% TFA added so the final pH was 2.5. Deuterium exchange at time t = 0 was determined by adding the protein solution in H2O (10 μl) to a mixture of ice-cold 0.9 ml 0.1% TFA and Mops buffer in D2O (90 μl). A portion of the quenched reaction (0.1 ml) was mixed with 50 μl of pepsin bead slurry (previously washed two times in 1 ml of cold 0.1% TFA). The mixture was incubated on ice with occasional mixing for 5 min, centrifuged for 15 s at 12,000 × g at 4°C, divided in aliquots, frozen in liquid N2, and stored at -80°C until analyzed.
Frozen samples were quickly defrosted to 0°C, mixed with matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid in 1:1:1 acetonitrile, ethanol, 0.1% TFA, final pH 2.5, 0°C), and 1 μl was spotted on a chilled MALDI target. The target was quickly dried and analyzed as described (15). The back exchange that occurred during the analysis was 46% as determined by carrying out control experiments where each of the protein samples was deuterated for 24 h at 25°C. After MALDI-TOF mass spectrometry, spectra were calibrated by using peptides of m/z = 1,088.6582 and 1,628.8886. The average mass of a peptide was calculated by determining the centroid of its isotopic envelope as described (15). The difference between the centroids of each deuterated vs. nondeuterated peptide gave the number of deuterons incorporated. Side chain deuteration, which occurs in MALDI H/2H exchange, was corrected for before back exchange correction as described (10). Kinetic plots of deuteration fit best to a single exponential model accounting for deuterons exchanging at a rapid rate (mainly solvent accessible amides). The fit was implemented in KALEIDAGRAPH 3.0 (Synergy Software, Reading, PA). Errors in the fits were ≈10%, and similar errors were obtained from individual time points measured in triplicate.
Design of Distance Constraint Filters by Using Amide Exchange Protection Data. Distance constraint filters were designed on the basis of amide H/2H exchange data. The 100,000 solutions with the best energies were filtered (16). “Qualitative” backbone filters specified that at least 1 Cα atom on any residue from each protected region on one subunit was required to be within 10 Å of any Cα atoms on the opposite subunit. A “quantitative” filter was also designed based on the observation that some fragments had more than one backbone amide with decreased solvent accessibility. The filter specified a minimum number of different Cα atoms required to be within a certain distance of Cα atoms on the opposite subunit. Others have shown that the intersection of solutions filtered with Cα constraints and heavy atom constraints results in the removal of some docking artifacts (17). We therefore also designed a “heavy atom” filter that specified a minimum number of non-hydrogen atoms to be within 7 Å of any non-hydrogen atom in the partner subunit. This filter used the same quantitative data already described. A distance constraint of 7 Å resulted in 23 solutions being retained. Intersection of the quantitative Cα-filtered solutions with those from the quantitative heavy atom filter retained 15 final solutions.
Results and Discussion
Computation of All Possible Docked Structures of the PKA Holoenzyme Complex. The program DOT (6) was used to compute the potential energy of all possible rotations (6° steps) and translations (1-Å steps) of the R-subunit [RIα(113–244) (4)], and the C-subunit (3). The PKI(5–24) peptide was left in the C-subunit active site during docking to mimic residues 94–99 of the R-subunit, which correspond to the pseudosubstrate/inhibitor sequence, and were not observed in the crystal structure of the R-subunit presumably because they were disordered. After the docking was completed, the R-subunit B-domain was added by using the coordinates of RIα(113–376), and none of the docked structures showed interaction of the cAMP-B-domain of RIα with the C-subunit (4). The top 100,000 solutions with the lowest energy values calculated by DOT of a total of 117 billion results were initially identified. The binding energies of the top 100,000 complexes saved ranged from -21.5 kcal/mol to -10.9 kcal/mol. These structures showed the R-subunit distributed all around the C-subunit with no preferred binding modes (16).
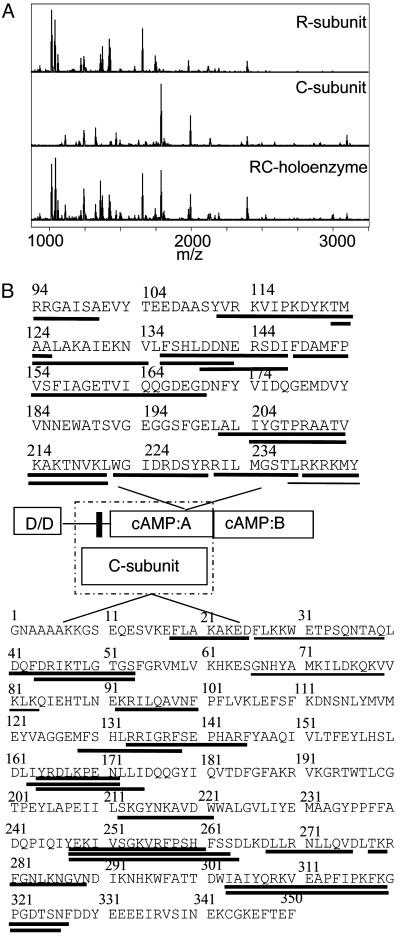
Solvent Accessibility Changes Identify the R–C Interface. We have previously shown that, in general, amides that exchange within 10 min at pH 7.0 and 25°C correspond to surface residues and are useful probes of changes that occur on protein–protein binding (8, 9). Amides within the protein core exchange much more slowly and generally report on protein folding and/or conformational changes. Pepsin digestion after the H/2H exchange reaction is quenched to 0°C and a pH of 2.5, allowing localization of the exchanged amides to short sequences of the protein (18). Both the R- and C-subunits were digested, and the MALDI-TOF mass spectra contained pepsin digest peptides from both proteins (Fig. 1A).
Fig. 1.
(A) MALDI-TOF mass spectrum of the peptic digest from RIα (Top), from the C-subunit (Middle), and from the RIα2–C2 holoenzyme (Bottom). (B) Sequence of the RIα(94–244) and C-subunit showing the fragments that were observed in the mass spectrum of the peptic digest. Lines underneath the sequence are fragments for which quantifiable data were obtained. A schematic is provided showing the domains of each protein, and the dashed box encloses those domains known to participate in the R–C interaction. In the regulatory subunit, D/D refers to the dimerization/docking domain, and the black bar shows the pseudosubstrate/inhibitor sequence.
In previous work, 12 pepsin digest peptides covering 70% of the R-subunit were analyzed, and these data are re-presented in Table 1. The amide exchange data from the R-subunit was complex in the sense that regions of the protein became either more or less accessible on binding of cAMP or the C-subunit. Comparison of the data from cAMP-free R-subunit with the cAMP-bound R-subunit indicated that there is no gross conformational change between these states (10). The pseudosubstrate/inhibitor sequence and a helical subdomain (αA and αB) of the cAMP-A-domain became protected from exchange on C-subunit binding (10). This subset of peptides exchanged rapidly in the absence of the C-subunit, were on the surface of the structure of the R-subunit, and were protected from exchange in the presence of the C-subunit. All three of these criteria are considered essential for identification of a protein–protein interface from amide exchange data (9). Experiments were performed by using both the full-length RIα and the fragment containing only the A-domain RIα(94–244). No additional protection was seen in the B-domain, confirming that the interaction involves only the cAMP-A-domain (data not shown).
Table 1. Summary of H/D exchange data or RIα(94—244).
| Deuteration after 10 min
|
||||
|---|---|---|---|---|
| Sequence of PKA RIα(94—244) (m/z)* | Number of amides | cAMP-Free RIα | Holoenzyme RIα(94—244)-C | cAMP-bound RIα |
| 94—101 (946.51) | 8 | 6.68±0.07 | 4.77±0.08 | 5.53±0.11 |
| 111—125 (1,783.00) | 13 | 11.49±0.39 | 10.73±0.18 | 12.29±0.24 |
| 122—136 (1,619.93) | 14 | 12.17±0.24 | 9.75±0.12 | 10.06±0.08 |
| 222—229 (1,011.46) | 7 | 1.56±0.19 | 0.91±0.03 | 1.55±0.05 |
| 230—238 (1,046.61) | 8 | 4.23±0.24 | 3.86±0.17 | 4.40±0.13 |
| 239—244 (881.51) | 5 | 2.48±0.25 | 2.6±0.06 | 2.77±0.09 |
| 136—143 (976.40) | 7 | 3.06±0.17 | 2.60±0.09 | 2.87±0.07 |
| 140—148 (1,110.47) | 8 | 2.87±0.13 | 2.45±0.08 | 3.70±0.05 |
| 136—148 (1,594.73) | 12 | 5.31±0.41 | 3.88±0.14 | 5.81±0.17 |
| 148—170 (2,473.12) | 21 | 8.16±0.36 | 6.23±0.36 | 8.34±0.12 |
| 202—221 (2,115.27) | 18 | 7.55±0.18 | 6.72±0.03 | 4.28±0.05 |
| 204—221 (1,931.15) | 16 | 4.92±0.18 | 5.83±0.09 | 2.11±0.02 |
Data from Anand et al. (10)
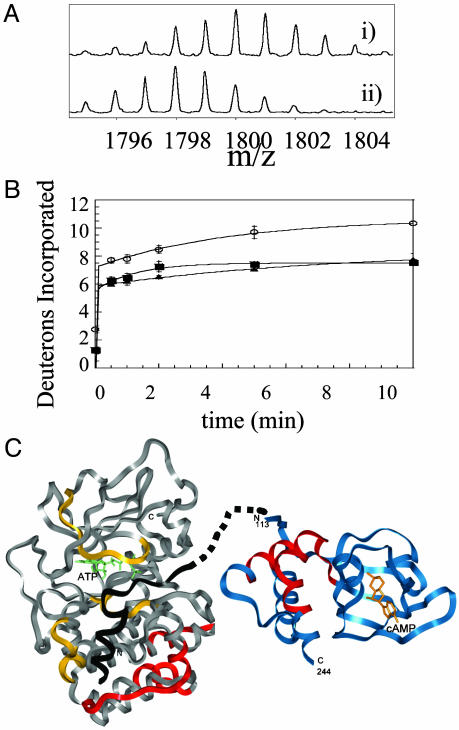
The changes in surface solvent accessibility of the Mg-ATP-containing C-subunit were probed in monomeric complex with RIα(94–244), and in dimeric complex with full-length RIα. Seventeen peptides from the C-subunit could be analyzed from all of the samples, and these covered 40% of the sequence of the C-subunit (Fig. 1B). As was previously observed, the active site of the C-subunit becomes protected on binding of ATP and the inhibitor peptide within the R-subunit (8). Protection from exchange in the glycine-rich loop that closes over the ATP and pseudosubstrate/inhibitor sequence was reflected in decreased deuteration of residues 44–54. The catalytic loop that contacts ATP (residues 164–172) was also protected. Finally, protection of the pseudosubstrate-binding site (residues 133–145) was also observed. New regions of the C-subunit that were protected on complexation with the R-subunit that could not be attributed to binding of Mg-ATP or the pseudosubstrate sequence were all within the C-terminal lobe of the kinase and included residues 212–221, residues 247–261 (Fig. 2), and residues 278–289. These segments of the C-subunit define a contiguous surface for R-subunit interaction.
Fig. 2.
(A) Expansion of the MALDI-TOF mass spectra to show one of the 17 fragments (m/z = 1,793.97, residues 247–261) from the analysis of the C-subunit that experienced slowed exchange in the RIα2–C2 holoenzyme complex. (Ai) The isotopic envelope for the fragment from the free C-subunit bound to MgATP after 10 min of deuteration. (Aii) The isotopic envelope for the same fragment from the RIα2–C2 holoenzyme after 10 min of deuteration. (B) Plot of deuterium incorporation into the amide positions of the region of C-subunit for the peptide fragment; m/z = 1,793.97 from C-subunit +MgATP (○), RIα2–C2 holoenzyme (▪), and RIα(94–244)-C holoenzyme (▴). (C) The structure of the C-subunit is shown in gray; the residues protected by the R-subunit are shown in red; the inhibitor peptide PKI(5–24), which mimics the pseudosubstrate, is shown in black; and the residues protected by it are shown in yellow (8). The structure of RIα(113–244) is shown in blue with the residues protected by the C-subunit in red. The pseudosubstrate/inhibitor sequence is shown connected by dots to the N terminus of the structured part of RIα.
Distance Constraint Filters from Amide H/2H Exchange. Protection of amide exchange on complex formation was primarily localized to three segments in each protein (Tables 1 and 2 and Fig. 2C). Within the R-subunit, residues 136–148, 222–229, and 230–238 contained amides that were protected. Within the C-subunit, residues 212–221, 247–261, and 278–289 contained amides that were protected. We have previously shown that the segment from residues 18–26 becomes less mobile on R-subunit binding, so this segment was not included in the docking as it was likely that the small decrease in amide exchange was due to helix tightening rather than interface protection (19). Each of these segments contained at least three residues that were surface exposed, and the number of amides that were protected in each case was at least one.
Table 2. Summary of H/D exchange data for the PKA catalytic subunit.
| Deuteration after 10 min
|
||||
|---|---|---|---|---|
| Sequence of PKA C-subunit (m/z) | Number of amides | C-subunit (+Mg2+, ATP) | Holoenzyme RIα(94—244)-C | Holoenzyme (RIα)2C2 |
| 18—26 (1,068.57) | 8 | 7.16 | 6.22±0.09 | ND |
| 44—54 (1,194.65) | 10 | 6.84±0.11 | 3.54±0.29 | 3.64±0.06 |
| 43—54 (1,341.72) | 11 | 7.26±0.19 | 4.11±0.01 | ND |
| 41—54 (1,584.8) | 13 | 9.11±0.45 | 6.15±0.18 | 6.01±0.02 |
| 66—83 (2,113.2323)* | 17 | 1.69±0.01 | 1.55±0.04 | ND |
| 92—100 (1,088.66) | 8 | 3.15±0.28 | 3.16±0.10 | 3.11±0.16 |
| 133—145 (1,628.89) | 11 | 5.75±0.01 | 3.24±0.19 | 4.83±0.05 |
| 164—172 (1,147.61) | 7 | 2.58±0.10 | 1.21±0.01 | 1.28±0.02 |
| 163—172 (1,260.69) | 8 | 2.94±0.06 | 1.11±0.14 | 1.13±0.08 |
| 212—221 (1,167.58) | 9 | 6.18±0.25 | 3.37±0.00 | 3.64±0.06 |
| 247—261 (1,793.97) | 13 | 10.32±0.15 | 7.74±0.42 | 7.53±0.02 |
| 247—264 (2,084.22) | 16 | 12.13±0.01 | 9.37±0.15 | 8.49±0.06 |
| 247—265 (2,197.33) | 17 | 12.01±0.02 | 9.58±0.05 | 8.46±0.12 |
| 268—276 (1,083.65) | 8 | 2.82±0.11 | 2.78±0.15 | ND |
| 278—289 (1,347.75) | 11 | 6.09±0.01 | 5.25±0.03 | 4.99±0.05 |
| 303—326 (2,676.45)† | 20 | 14.12±0.45 | 14.13±0.17 | 13.10±0.13 |
| 303—327 (2,823.52) | 21 | 15.00±0.43 | 14.88 | 13.77±0.25 |
ND, Not determined.
This row of data is from a partial centroid because of peak overlap and does not represent the absolute number of amides exchanging although relative differences can be seen
One additional amide is protected in C-subunit upon binding the full-length R-subunit, and this result is due to the presence of residues upstream of the pseudosubstrate sequence (30)
For all peptides in both the R- and C-subunits, only one to three amides within each peptide segment were protected by the binding partner in the complex. To use this type of experimental data to filter the docked protein complexes for consistency with experiment, we first tried a “qualitative” approach wherein at least one Cα from the segment showing protection was required to be within 10 Å of the binding partner, as a fairly “loose” filter (20). By using this approach, 96 solutions survived the filter. Table 3 shows that there was a progressive decrease in the number of surviving structures with each additional constraint. The filters were, as expected, commutative, and the order in which they were applied did not change the results.
Table 3. Number of docked structures remaining after filtering based on 10-Å Cα—Cα distance.
| Fragment | 10-Å Cα—Cα filter* | 10-Å Cα—Cα filter (no. of amides protected)† | 7-Å heavy atom filter (no. of amides protected)‡ |
|---|---|---|---|
| 212—221 (C) | 17,466 | 9,075 (3) | 11,026 (3) |
| 247—261 (C) | 6,015 | 1,609 (3) | 1,981 (3) |
| 278—289 (C) | 2,856 | 1,087 (1) | 1,414 (1) |
| 136—148 (R) | 615 | 158 (2) | 356 (2) |
| 222—229 (R) | 103 | 23 (1) | 61 (1) |
| 230—238 (R) | 96 | 23 (1) | 60 (1) |
The filter required that at least one Cα from the fragment be within 10 Å of any Cα in the binding partner
The filter required that at least the number of Cα in parentheses from the fragment be within 10 Å of any Cα in the binding partner
The filter required that at least the number of heavy atoms in parentheses from different residues in the fragment be within 7 Å of any heavy atom in the binding partner
The results from the qualitative approach did not yield a single interface between the two proteins, so a quantitative approach was implemented. Within the C-subunit, the peptides corresponding to residues 212–221 and 247–261 each showed differences of three amides between complexed and free. Within the R-subunit, the peptide corresponding to residues 136–148 showed a difference of two amides. These results were then used to further filter the results so that, if two amides were protected, two Cα atoms were required to be within 10 Å, etc. This quantitative filter resulted in 23 solutions (Table 3).
Because protection from amide exchange at protein–protein interfaces may require only side-chain contact and not necessarily backbone contact, filters were also devised that required the same quantitative number of heavy atoms from different residues on one protein to be within 7 Å of any heavy atom on the partner protein (21–23). This “heavy atom” filter resulted in 60 solutions (Table 3). The intersection of the set of structures from the quantitative Cα and heavy atom filters yielded 15 structures that defined a unique interface between the C-terminal lobe of the C-subunit and the A- and B-helices of the R-subunit. There was an axis of rotation through this flat interface.
Predicted New Interactions Between Residues at the Interface. Examination of the 15 DOT solutions that survived the distance constraint filters revealed that they all satisfied the interaction between Glu-143 (from RIα) and Lys-213 (from C-subunit) required by the experimental result that mutations at Lys-213 of the C-subunit complemented those at Glu-143 in RIα (1, 7). They also all satisfied a distance constraint connecting the pseudosubstrate/inhibitor sequence (mimicked by the PKI(5–24) and the N terminus of the structure of RIα (residue 113). Finally, residues 92–100 in the C-subunit and residues 122–136, 204–221, and 239–244 in RIα that did not show any protection on complex formation were not at the interface in the final model.
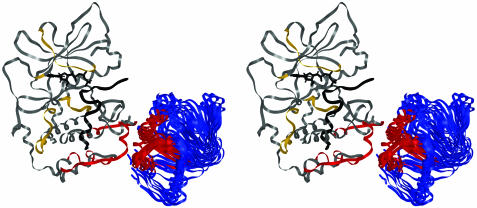
The 15 final structures were clustered according to center of mass distances and difference in orientation between all 210 pairs of configurations for the R-subunit (Table 4, which is published as supporting information on the PNAS web site). There are three obvious clusters that differ internally by <5 Å and 20°, but between clusters differ by at least 14 Å and 50°. All clusters of solutions predict a common R–C interface and differ in their rotational orientations. The solution that was most often found, represented by the cluster containing eight solutions, also contains the lowest energy solution from the filtered DOT solutions. All of the structures except structure 13, the most weakly associated member of the cluster, had several contacts [as defined by the Critical Assessment of PRedicted Interactions (CAPRI) contact scoring scheme] between the activation loop and the R-subunit (24). This cluster was, therefore, taken as the most likely model for the holoenzyme complex (Fig. 3).
Fig. 3.
Stereo image of the top cluster of eight solutions that represent the final docked structure. The structure of the C-subunit is colored as in Fig. 2. The regions of RIα(113–244) that showed decreased solvent accessibility exclusively on binding the C-subunit are colored red.
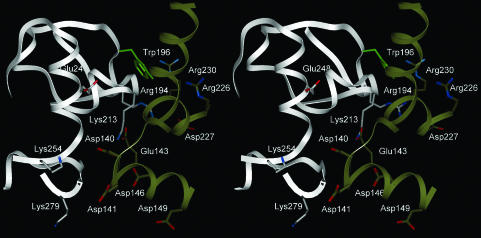
The model predicts that the C-subunit activation loop (residues 189–199) contacts RIα. It therefore predicts the basis for the observation that phosphorylation of Thr-197 is essential for RIα binding (G. H. Iyer and S.S.T., unpublished results). Key contacts between Trp-196 (C) and Arg-230 (R) and between Arg-194 (C) and Asp-227 (R) are predicted by the model (Fig. 4). Previous studies highlighted the importance of a conserved Trp at position 196 in binding both isoforms of the R-subunit (25), and subsequent studies showed that mutation of this residue to Ala and Arg greatly decreased binding to RIα with the Trp-196 → Arg mutation being more disruptive for holoenzyme formation (26). Our model predicts that the Trp-196 → Arg mutation not only disrupts a favorable interaction with Arg-230 of RIα but also introduces a charge repulsion. The model also predicts a complementary registry of charged residues in which one side of the interface has Glu-248 from the C-subunit approaching Arg-226, -230, and -231 from the R-subunit whereas the other side of the interface has Lys-213 and Arg-194 from the C-subunit approaching Asp-140, -141, -146, and -149 and Glu-143 from the R-subunit.
Fig. 4.
Stereo image of a close-up of the R–C subunit interface from the lowest energy filtered dot solution. The C-subunit is shown in gray, and RIα is in gold. Glu-143 (R) is in contact with Lys-213 (C). The side chain of Trp-196 (C) is in green and is predicted to contact Arg-230/Arg-231 (R). Arg-194 (C) is predicted to contact Asp-227 (R). Lys-254 (C) and Glu-248 (C) are also in the interface.
A General Approach to Structural Genomics of Higher Order Complexes. Although protein–protein docking methods attempt to correctly predict the structure of a protein–protein complex from the structures of the individual interacting proteins, the overall change in energy between free and docked proteins is often relatively small, and side-chain positioning, as well as conformational changes, can significantly alter the measured binding energy. This finding is particularly relevant when the structures were not crystallized as a complex, and when the binding site is of relatively weak affinity.
One solution to the problem that several groups have attempted is to filter the docking results with experimental data (17). Recent attempts to use experimental data in docking have met with limited success (27). Attempts to filter the PKA holoenzyme docked solutions with the complementary mutagenesis constraint, which would be analogous to a protein–protein crosslink, generated a very different model that was inconsistent with other experimental results (I.T., V. Kotlovyi, and L.T.E., unpublished results). The complementary mutagenesis constraint was also combined with small-angle x-ray scattering information to yield yet a different model (28). Whereas both models predict that the intersubunit interface can be localized to a small surface on the helical subdomain of cAMP:A domain, neither found Trp-196 at the interface, nor were they consistent with the role of the activation loop in holoenzyme formation.
Our results show that amide H/2H exchange mass spectrometric interface mapping can provide the experimental data needed to filter docking results (8, 9, 29). First, the absence of large changes in amide exchange indicates that no major conformational change has occurred, an implicit assumption when using individual structures for molecular docking. Second, amide exchange gives comprehensive information about the surface of each protein that must be in contact. Of the original 100,000 low-energy solutions from DOT, only 15 remained after application of two distance filters designed on the basis of amide H/2H exchange data, and these structures specified a single protein–protein interaction that varied only in rotation about the flat plane of the interface. The majority of the solutions formed a single cluster of the PKA holoenzyme complex that was consistent with all known biochemical data and from which new predictions could be made. It is our hope that the combination of amide H/2H exchange mass spectrometry and computational docking will become a high throughput method to rapidly obtain accurate structural models of protein–protein complexes.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grants MCB9808286 (to E.A.K.) and DBI-9911196 (to L.F.T.E.), the Howard Hughes Medical Institute (S.S.T.), and National Institutes of Health Grant GM19301 (to S.S.T.). D.L. was the recipient of a Graduate Assistance in Areas of National Need fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MALDI-TOF, matrix-assisted laser desorption ionization/time-of-flight; R, regulatory; C, catalytic; TFA, trifluoroacetic acid.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1PVK).
References
- 1.Gibson, R. M., Ji-Buechler, Y. & Taylor, S. S. (1997) J. Biol. Chem. 272, 16343-16350. [DOI] [PubMed] [Google Scholar]
- 2.Huang, L. J. & Taylor, S. S. (1998) J. Biol. Chem. 273, 26739-26746. [DOI] [PubMed] [Google Scholar]
- 3.Zheng, J., Knighton, D. R., Ten Eyck, L. F., Karlsson, R., Xuong, N., Taylor, S. S. & Sowadski, J. M. (1993) Biochemistry 32, 2154-2161. [DOI] [PubMed] [Google Scholar]
- 4.Su, Y., Dostmann, W. R. G., Herberg, F. W., Durick, K., Xuong, N.-h., Ten Eyck, L. F., Taylor, S. S. & Varughese, K. I. (1995) Science 269, 807-819. [DOI] [PubMed] [Google Scholar]
- 5.Ten Eyck, L. F., Mandell, J. G., Roberts, V. A. & Pique, M. E. (1995) in Proceedings of the 1995 ACM/IEEE Supercomputing Conference (IEEE Comput. Soc. Press, Los Alamitos, CA).
- 6.Mandell, J. G., Roberts, V. A., Pique, M. E., Kotlovyi, V., Mitchell, J. C., Nelson, E., Tsigelny, I. & Ten Eyck, L. F. (2001) Protein Eng. 14, 105-113. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, R. M., Ji-Buechler, Y. & Taylor, S. S. (1997) Protein Sci. 6, 1825-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandell, J. G., Falick, A. M. & Komives, E. A. (1998) Proc. Natl. Acad. Sci. USA 95, 14705-14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandell, J. G., Baerga-Ortiz, A., Akashi, S., Takio, K. & Komives, E. A. (2001) J. Mol. Biol. 306, 575-589. [DOI] [PubMed] [Google Scholar]
- 10.Anand, G. S., Hughes, C. A., Jones, J. M., Taylor, S. S. & Komives, E. A. (2002) J. Mol. Biol. 323, 377-386. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, C. A., Mandell, J. G., Anand, G. S., Stock, A. M. & Komives, E. A. (2001) J. Mol. Biol. 307, 967-976. [DOI] [PubMed] [Google Scholar]
- 12.Andersen, M. D., Shaffer, J., Jennings, P. A. & Adams, M. R. (2001) J. Biol. Chem. 276, 14204-14211. [DOI] [PubMed] [Google Scholar]
- 13.Herberg, F. W., Bell, S. M. & Taylor, S. S. (1993) Protein Eng. 6, 771-777. [DOI] [PubMed] [Google Scholar]
- 14.Davis, M. E., Madura, J. D., Sines, J., Luty, B. A., Allison, S. A. & McCammon, J. A. (1991) Methods Enzymol. 202, 473-497. [DOI] [PubMed] [Google Scholar]
- 15.Mandell, J. G., Falick, A. M. & Komives, E. A. (1998) Anal. Chem. 70, 3987-3995. [DOI] [PubMed] [Google Scholar]
- 16.Law, D. S., Ten Eyck, L. F., Katzenelson, O., Tsigelny, I., Roberts, V. A., Pique, M. E. & Mitchell, J. C. (2003) Proteins 52, 33-40. [DOI] [PubMed] [Google Scholar]
- 17.Gabb, H. A., Jackson, R. M. & Sternberg, M. J. (1997) J. Mol. Biol. 272, 106-120. [DOI] [PubMed] [Google Scholar]
- 18.Rosa, J. J. & Richards, F. M. (1979) J. Mol. Biol. 133, 399-416. [DOI] [PubMed] [Google Scholar]
- 19.Gangal, M., Clifford, T., Deich, J., Cheng, X., Taylor, S. S. & Johnson, D. A. (1999) Proc. Natl. Acad. Sci. USA 96, 12394-12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández-Recio, J., Totrov, M. & Abagyan, R. (2003) Proteins 52, 113-117. [DOI] [PubMed] [Google Scholar]
- 21.Chen, R., Tong, W., Mintseris, J., Li, L. & Weng, Z. (2003) Proteins 52, 68-73. [DOI] [PubMed] [Google Scholar]
- 22.Smith, G. R. & Sternberg, M. J. E. (2003) Proteins 52, 74-79. [DOI] [PubMed] [Google Scholar]
- 23.Schneidman-Duhovny, D., Inbar, Y., Polak, V., Shatsky, M., Halperin, I., Benyamini, H., Barzilai, A., Dror, O., Haspel, N., Nussinov, R. & Wolfson, H. J. (2003) Proteins 52, 107-112. [DOI] [PubMed] [Google Scholar]
- 24.Mendez, R., Leplae, R., De Maria, L. & Wodak, S. J. (2003) Proteins 52, 51-67. [DOI] [PubMed] [Google Scholar]
- 25.Orellana, S. A. & McKnight, G. S. (1992) Proc. Natl. Acad. Sci. USA 89, 4726-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson, R. M. & Taylor, S. S. (1997) J. Biol. Chem. 272, 31998-2005. [DOI] [PubMed] [Google Scholar]
- 27.Janin, J., Henrick, K., Moult, J., Ten Eyck, L., Sternberg, M. J. E., Vajda, S., Vakser, I. & Wodak, S. J. (2003) Proteins 52, 2-9. [DOI] [PubMed] [Google Scholar]
- 28.Tung, C. S., Walsh, D. A. & Trewhella, J. (2002) J. Biol. Chem. 277, 12423-12431. [DOI] [PubMed] [Google Scholar]
- 29.Baerga-Ortiz, A., Hughes, C. A., Mandell, J. G. & Komives, E. A. (2002) Protein Sci. 11, 1300-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng, X., Phelps, C. & Taylor, S. S. (2001) J. Biol. Chem. 276, 4102-4108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.