Abstract
We designed a series of nine-residue peptides that bound to a defined site on the tumor suppressor p53 and stabilized it against denaturation. To test whether the peptides could act as chaperones and rescue the tumor-suppressing function of oncogenic mutants of p53 in living cells, we treated human tumor cells with the fluorescein-labeled peptide Fl-CDB3 (fluorescent derivative of CDB3). Before treatment, the mutant p53 in the cell was predominantly denatured. Fl-CDB3 was taken up into the cytoplasm and nucleus and induced a substantial up-regulation of wild-type p53 protein and representative mutants. The mutants, His-273 and His-175 p53, adopted the active conformation, with a dramatic decrease in the fraction of denatured protein. In all cases, there was p53-dependent induction of expression of the p53 target genes mdm2, gadd45, and p21, accompanied by p53-dependent partial restoration of apoptosis. Fl-CDB3 sensitized cancer cells that carried wild-type p53 to p53-dependent γ-radiation-induced apoptosis. Although Fl-CDB3 did not elicit a full biological response, it did bind to and rescue p53 in cells and so can serve as a lead for the development of novel drugs for anticancer therapy.
The tumor suppressor p53 protects the cell against cancer by inducing cell cycle arrest and apoptosis in response to oncogenic stress (reviewed in ref. 1). Radiotherapy and much of chemotherapy also function by activating p53. Some 50% of human cancers have missense point mutations in the gene for p53 that inactivate or impair its transcriptional activity (see p53 databases at www.iarc.fr/p53/index.html and http://p53.curie.fr). Most of the mutations in p53 are located in its DNA-binding core domain. Many of the mutations, such as R273H, are of residues that directly interact with DNA, the ”contact mutants” (2). In addition, p53 is an extremely unstable protein, with a melting temperature that is only slightly above body temperature (3, 4). Many other oncogenic mutations are ”structural”; they lower the thermodynamic stability of p53, resulting in its unfolding and inactivation (4). A further source of instability of p53 is that it is degraded by the proteasome after binding to its major negative regulator protein MDM2, a ubiquitin ligase that is induced by p53 in a feedback loop (5). Reactivating mutant p53 is now an important target in the development of novel therapies for cancer (6-8). One promising way to reactivate structural mutants of p53 is to identify small molecules that bind to its native, but not denatured state and thereby raise its melting temperature (9).
Potential p53-stabilizing small-molecule drugs may be found by random screening (10, 11) or rational design (12). An early candidate found by screening the Pfizer drug library, CP-31398 (10), does not bind to the core domain in vitro (13) but functions in vivo by inhibiting ubiquitination of p53 and hence its degradative pathway (14). Another small molecule that rescues p53 function in living cancer cells, PRIMA-1, was discovered by using cell-based screening assays, but its mechanism of action has yet to be established (11). We designed a peptide, CDB3 (REDEDEIEW), by using the structure of the complex between p53 core and p53-binding protein 2 (53BP2 or ASPP) as a starting point. CDB3 binds to and stabilizes the p53 core domain in vitro (12). NMR analysis reveals that CDB3 binds to a site in p53 that partly overlaps with its positively charged binding site for DNA (12). At first sight, CDB3 would be thought to be of little practical use because it would be an inhibitor of p53. However, we envisage a ”chaperone” strategy whereby CDB3 binds to p53 and mutants during biosynthesis, raises it melting temperature to above body temperature so it can fold, and then transfers p53 to its natural binding partners in the cell that would take over the stabilizing function (12, 15). Here, we test the chaperoning strategy in living cells. We chose a fluorescent derivative of CDB3, Fl-CDB3, which is labeled at its N terminus with fluorescein. Fl-CDB3 binds more tightly (12) and may also be tracked by fluorescence microscopy. We chose derivatives of three human cancer cell lines: H1299 (lung adenocarcinoma), which has no intact p53 alleles; H1299-His-175, which contains a vector that expresses p53 with the structural mutation R175H; Saos-2 (osteosarcoma), which also lacks intact p53 alleles, and Saos-2-His-273, which expresses the contact mutant R273H; and HCT116 (colon carcinoma), which produces wild-type p53, and its p53-null counterpart HCT116p53-/-, in which both alleles of p53 were deleted by means of homologous recombination (11, 16). In our study, we addressed the following questions: will cancer cells take up the peptide; if so, will it rescue the active conformation of p53; will the p53 so stabilized still be functional; is there evidence that the peptide binds to p53 in the cells? If the answers are positive, then we have a lead molecule for cancer therapy whose mode of binding to p53 is known.
Materials and Methods
Peptide Binding in Vitro. The peptides were synthesized by using a Pioneer peptide synthesizer (PerSeptive Biosystems, Framingham, MA) with standard fluorenylmethoxycarbonyl (Fmoc) chemistry. Fluorescein was coupled to the N terminus of the peptides on the Pioneer peptide synthesizer with 4-fold excess of Fluorescein-Osu (Molecular Probes) and 4-fold excess of hydroxybenzotriazole (HoBt). The peptides were purified and characterized as described (12). Human p53 core wild type and mutants (residues 94-312) were cloned, expressed, and purified as described (4). 15N-labeled human p53 core was produced as described (17). Fluorescence anisotropy measurements were performed with fluorescein-labeled CDB3 derivatives in 50 mM Hepes (pH 7.2) and varying concentrations of NaCl to vary ionic strength (Table 1) at 10°C by using a Perkin-Elmer LS-50b luminescence spectrofluorimeter as described (12). Dissociation constants for the peptide-p53 core complex were calculated by fitting the anisotropy titration curves to a simple 1:1 equilibrium model (12).
Table 1. Activity of peptides.
|
Kd, μM
|
Induction of
|
|||||
|---|---|---|---|---|---|---|
| Peptide | I = 0.15 M | I = 0.021 M | PAb1620* epitope induction | p53 | p21 | Apoptosis |
| FI-CDB3 | 100 | 0.6 | +++ | ++ | ++ | + |
| FI-E491/3/5/7A | 1,800 | 136 | ++ | + | + | +/- |
| FI-R490A | 140 | 0.2 | ++ | + | + | +/- |
| FI-polyGlu | 1800 | 0.01 | +++ | + | - | - |
| FI-control | NB | NB | - | - | - | - |
| Biotin-CDB3 | 800 | 12 | ND | + | + | ND |
Peptide sequences are as follows: FI-CDB3, FI-REDEDEIEW; FI-E491/3/5/7A, FI-RADADAIAW; R490A, FI-AEDEDEIEW; FI-polyGlu, FI-(Glu)9; FI-control, FI-RKSKKKITW; Biotin-CDB3, biotin-REDEDEIEW. NB, no binding observed; ND, not done.
Binds to the native conformation of p53
Peptides in Cells. The human Saos-2-His-273 and H1299-His-175 cell lines carry the indicated tetracycline-regulated mutant p53 constructs. The human HCT-116 cell line carries wild-type p53 and in the HCT116p53-/- both p53 alleles were deleted by means of homologous recombination.
For FACS analysis, cells were stained with propidium iodide and analyzed on a FACScan cytometer (BD Biosciences) according to standard procedures. Immunostaining, preparation of cell extracts, ELISA, lacZ staining, and Western blot were performed according to standard procedures. The anti-p53 monoclonal antibodies PAb1620, PAb240, DO1, and PAb1801 were obtained from Calbiochem. The anti-p53 rabbit polyclonal antibody was obtained from Santa Cruz Biotechnology, the anti-MDM2 monoclonal antibody was obtained from NeoMarkers (Fremont, CA), and the anti-p21 monoclonal antibody was obtained from Transduction Laboratories (Lexington, KY). Secondary antibodies (FITC-conjugated horse anti-mouse Ig, Texas red-conjugated goat anti-rabbit Ig) were from Vector Laboratories. All other reagents were from Sigma-Aldrich.
Results
Peptide Binding in Vitro. We synthesized a range of variants of Fl-CDB3 to analyze and optimize its binding to p53 to select the best derivative for the experiments in vivo (Table 1). The derivatives also provide important controls for linking activities in vivo and in vitro. Fl-CDB3 is highly negatively charged and binds to a positively charged region of p53. We made mutants that differ in their net charges as well as in key amino acid residues. These include Fl-polyGlu, which is a model of a relatively nonspecific peptide that binds by electrostatic interactions to positively charged sites; Fl-R490A, Fl-E491/3/5/7A, which lacks negatively Glu-charged side chains; and a control peptide, Fl-control (Fl-RKSKKKITW), that is positively charged and should not bind to the positively charged binding site. Binding was assayed by fluorescence anisotropy (12) at various ionic strengths, I. At low ionic strength, the electrostatic interactions from the glutamates or aspartates dominated: Fl-polyGlu bound with Kd = 12 nM, 50 times more tightly than did Fl-CDB3 at I = 21 mM. The positively charged control, Fl-control, had no detectable binding. Binding affinity to p53 core decreased with removal of negatively charged residues. At physiological ionic strength (I = 0.15 M), the parent Fl-CDB3 bound the tightest. Biotinylated CDB3 (biotin-CDB3) bound far more weakly. Fl-CDB3 bound tightly to p53 R273H with dissociation constants of 3.4 μM at I = 21 mM and 200 μM at I = 0.15 M. These and unpublished data on the structure and activity of Fl-CDB3 derivatives show that its specificity of binding results in part from electrostatic interactions and also from specific side chain interactions.
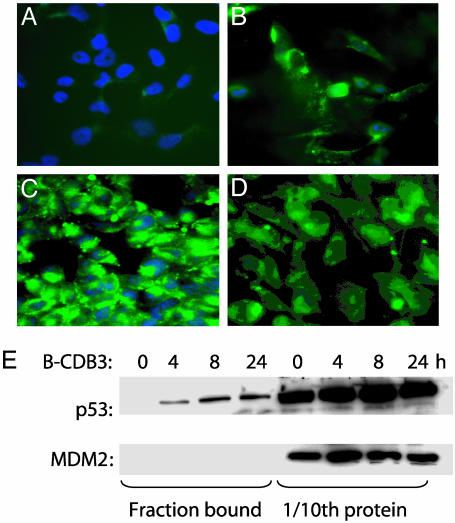
Fl-CDB3 Is Taken into Cancer Cells. We treated the human tumor cell lines with Fl-CDB3, Fl-polyGlu, the control peptide, and other derivatives (Table 1). All of the peptides were able to enter the cells, irrespective of the presence of p53 (Fig. 1). The distribution of peptides was similar, predominantly cytoplasmic, with some nuclear and/or nucleolar localization. A fraction of Fl-CDB3 formed intense clumps in the nucleus.
Fig. 1.
Uptake of Fl-labeled peptides by human tumor cells. (A-D) Fluorescence microscopy analysis of cellular uptake of peptides in H1299-His-175 cells: A, no peptide; B, control peptide; C, Fl-CDB3; D, Fl-polyGlu. (E) In vivo interaction of biotinylated peptides with mutant p53:H1299-His-175 cells incubated with 10 μg/ml biotin-CDB3 for indicated time points. Intracellular peptide was captured from lysates by using avidin-coated Dynabeads (Dynal, Oslo). Bound proteins were analyzed by Western blot. One-tenth of total protein in the cell was analyzed to reference the data.
Biotin-CDB3 Binds p53 in Cells. We next tested whether peptides can bind p53 in the context of cellular proteins. H1299-His-175 cells were treated with biotin-CDB3, biotin-polyGlu, and biotin-control peptides. Mutant p53 from H1299-His-175 cells coprecipitated with biotin-CDB3 and biotin-polyGlu, despite poorer affinity than the Fl derivatives (12), but not with biotin-control (Fig. 1E and data not shown). Thus, the p53-binding peptides selected in vitro can bind to the p53 protein in living cells.
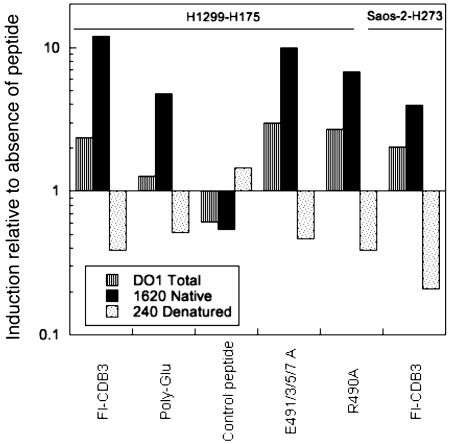
Conformational Rescue of p53. We assessed the conformation of p53 in cells by ELISA, using the wild-type-specific conformational antibody PAb1620 and, in contrast, PAb240 that detects unfolded protein. On treatment of H1299-His-175 and Saos-2-His-273 cells with 10 μg/ml Fl-CDB3, we observed a remarkable increase in the fraction of both His-273 and His-175 p53 mutants (Fig. 2) that bound to native-directed PAb1620. Accordingly, there was a large decrease in the fraction recognized by PAb240, which binds the denatured conformation. Thus, Fl-CDB3 can restore the wild-type conformation to mutant p53 both in vitro and in living cells. Fl-E491/3/5/7A and R490A, which bind p53 in vitro, rescued the wild-type conformation of R175H mutant almost as efficiently as Fl-CDB3. It is noteworthy that treatment of H1299-His-175 cells with Fl-polyGlu also induced the PAb1620 epitope, supporting the idea that the binding of any ligand to the p53 core domain can stabilize its wild-type conformation (9). In contrast, the control peptide that did not bind p53 in vitro did not induce changes in p53 conformation either (Fig. 2).
Fig. 2.
Restoration of the wild-type conformation to mutant p53 by p53-binding peptides. Fl-CDB3 and indicated peptides were incubated with Saos-2-His 273 or H1299-His-175 cells for 24 h. The conformation of p53 in cell lysates was evaluated by ELISA analysis with monoclonal ant-p53 antibodies PAB1620, which recognizes properly folded p53, and PAb240 for the detection of unfolded protein, and DO1 for the total p53. Shown are the ratios of change on a log scale.
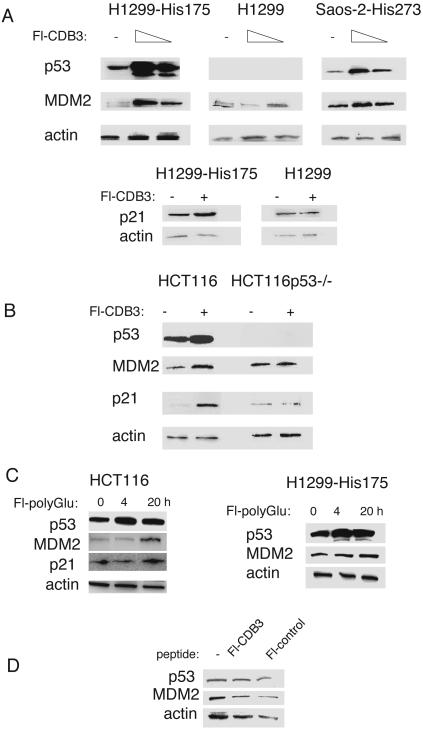
Up-Regulation of p53. Importantly and unexpectedly, treatment of cells with both Fl-CDB3 and Fl-polyGlu resulted in a substantial increase in total p53 levels as detected by ELISA by the conformationally insensitive antibody DO1 (Fig. 2). Western blot analysis confirmed the induction of mutant p53 levels in H1299-His-175 and Saos-2-His-273 cells by treatment with Fl-CDB3, biotin-CDB3, and Fl-polyGlu, but not by the control peptide (Fig. 3 and data not shown). In addition, wild-type p53 in HCT116 cells was also induced by Fl-CDB3 (Fig. 3B).
Fig. 3.
Restoration of transcriptional transactivation function to p53 in cells. (A) Induction of target genes by Fl-CDB3 is mutant p53-dependent. Mutant p53-carrying cells H1299-His-175 and Saos-2-His-273 and their p53-null counterparts were treated with increasing concentrations of Fl-CDB3 (1 and 10 μg/ml) for 24 h, and the levels of p53 and its target genes mdm2 and p21 were assessed by Western blot. Treatment with Fl-CDB3 resulted in a substantial increase in the expression of MDM2 and p21, along with induction of p53 levels. No induction of p21 or MDM2 was observed in p53-null cells. (B) Activation of wild-type p53 in HCT116 cells. Induction of p53 and its target genes was observed on treatment of colon carcinoma HCT116 cells with Fl-CDB3. Up-regulation of p21 and MDM2 was mediated by p53, because no changes in p21 or MDM2 expression was detected in p53-null cells. (C) p53 level induced by p53-binding peptide Fl-polyGlu. (D) Fl-CDB3 did not induce p53 protein-carrying substitutions of residues 22 and 23 and did not cause the induction of MDM2.
Restoration of Transcriptional Transactivation Activity. We next tested whether Fl-CDB3 could restore the transcriptional transactivation function to mutant p53 in living cells by examining the induction of two classical p53 target genes, p21 and mdm2. Treatment with Fl-CDB3 of H1299-His-175 and Saos-2-His-273 cells resulted in a significant induction of both MDM2 and p21 in a dose-dependent manner (Fig. 3A). Importantly, treatment of the p53-null isogenic lines Saos-2 and H1299 in the absence of mutant p53 expression did not cause any induction of MDM2 or p21 (Fig. 3A and data not shown). Thus, the effect of peptide on MDM2 and p21 expression is p53-dependent. In addition, Fl-CDB3-induced MDM2 and p21 in HCT116 carrying endogenous wild-type p53 (Fig. 3B) and the GADD45 protein, which is involved in DNA repair and G2 arrest, was induced in H1299-His-175 cells (data not shown). However, we did not see growth arrest. No significant changes of MDM2 and p21 protein levels on Fl-CDB3 treatment were observed in the isogenic line HCT116p53-/- that does not express p53 (Fig. 3B), supporting the notion that Fl-CDB3 exerts its effects on gene expression in a p53-dependent manner. Similar results were obtained with biotin-CDB3 (data not shown).
To test further whether the up-regulation of p53 target genes by Fl-CDB3 depends on the transcriptional activity of p53, we used p53-null SKOV cells stably transfected with His-175 mutant p53 carrying substitutions of Leu-22/Trp-23 for Gln/Ser in the transactivation domain that make p53 unable to induce transcription and abrogate binding to MDM2 (18). We did not observe induction of MDM2 after Fl-CDB3 treatment in these cells, implying that the transcriptional activity of p53 is required for the induction of MDM2 by Fl-CDB3 (Fig. 3D). Further evidence is that treatment of carcinoma A431 cells that carry the endogenous His-273 mutant p53 and a stably transfected p53-responsive lacZ reporter with Fl-CDB3 resulted in the appearance of lacZ-positive cells: incubation of cells for 8 h with either 20 μg/ml Fl-CDB3 or Fl-polyGlu gave a 20-fold increase in lacZ-positive cells, whereas control peptide did not induce lacZ expression significantly above background level (data not shown).
Treatment with Fl-polyGlu did not induce p21 in H1299-His-175 and HCT116 cells (Fig. 3C). PolyGlu is a very nonspecific molecule for mimicking electrostatic interactions in DNA binding and so may bind promiscuously and inhibit other DNA binding reactions in the cell. Treatment with the control peptide did not affect the levels of p53 or MDM2 (data not shown). Interestingly, we did not observe the induction of expression of the apoptosis-inducing gene PUMA on treatment with Fl-CDB3 (data not shown).
Partial Restoration of p53-Dependent Apoptosis. To determine whether Fl-CDB3 can rescue the apoptosis-inducing function of p53, we analyzed cell survival after treatment with peptide by using FACS analysis. Analysis of the DNA content profile of Saos-2 and Saos-2-His-273 cells treated with 10 μg/ml Fl-CDB3 for 24 h showed that Fl-CDB3 caused a mild increase (19%) in the fraction of cells with a sub-G1 DNA content in the presence of mutant p53, indicating DNA fragmentation and cell death. We analyzed the biological response of human tumor cell lines with different p53 statuses (p53-null, wild-type p53, and mutant p53) to Fl-CDB3 and its derivatives. Comparisons between the isogenic lines that differ only in p53 status, i.e., Saos-2 and Saos-2-His-273 and H1299 and H1299-His-175, showed that Fl-CDB3 induced apoptosis in cell lines carrying mutant p53 but not in their p53-null counterparts.
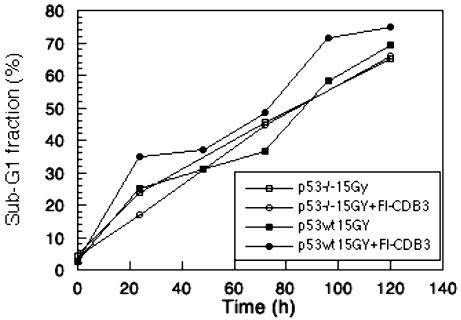
Sensitization to γ-Radiation. As assessed by FACS analysis, Fl-CDB3 did not induce apoptosis in wild-type p53 carrying HCT116 cells. However, Fl-CDB3 sensitized HCT116 cells, but not their p53-null counterpart HCT116 p53-/- cells, to apoptosis induced by γ-irradiation (Fig. 4).
Fig. 4.
Fl-CDB3-sensitized tumor cells carrying wild-type p53 to ionizing radiation-induced apoptosis. Wild-type p53-positive HCT116 or p53-negative HCT116p53-/- cells were incubated with 10 μg/ml Fl-CDB3 for 8 h and then irradiated with 15 Gy. The fraction of sub-G1 cells was determined as above at indicated time points. The sub-G1 fraction was increased by 20% in HCT116 cells but was not increased in p53-negative HCT116p53-/- cells.
Discussion: Validity of the Chaperoning Strategy
The major goal of the study was to test whether the peptide Fl-CDB3 would be active in living cells. The peptide Fl-CDB3 and its designed derivatives are imported by cancer cell lines. Fl-CDB3 induces the up-regulation of wild-type p53, a representative destabilized structural mutant, R175H, and a representative contact mutant, R273H. The up-regulated proteins are all in the native conformation and only a small fraction are in the denatured state, as evidenced from antibody-binding studies. Wild-type and mutant proteins induced by the peptide are transcriptionally active, as shown by the up-regulation of expression of p53 target genes p21, gadd45, and mdm2, as well as a p53-dependent lacZ reporter gene.
One of the classic dilemmas in designing a drug from experiments in vitro is whether it binds to the same target in cells and is active in the desired manner. The absence of effect of p53-binding peptides on gene expression or cell growth in p53-negative cells provides strong evidence that the effects observed in p53-positive cells are exerted through p53. Indeed, our results show that biotin-CDB3 binds p53 in vivo. Importantly, the activities in cells roughly parallel the binding affinities in vitro (Table 1). Induction of p53 target genes by Fl-CDB3 requires the transactivation function of p53: Fl-CDB3 did not induce MDM2 and p21 when added to cells expressing His-175 p53 that also had the 22/23 double mutation that abolishes its transcriptional activity. Taken together, our data provide solid support to the idea that Fl-CDB3 functions as a p53-specific molecular chaperone.
The up-regulation of p53 and its mutants is a bonus because a massive increase in the concentration of, for example, R273H would compensate for its lower activity. This may be the basis for the rescue of the contact mutant R273H by Fl-CDB3. R273H has a residual DNA-binding activity: we have measured its affinity to the gadd45 promoter and found it to be 50 μM at I = 0.15 M. Under these conditions, R273H binds Fl-CDB3 with Kd = 200 μM (data not shown). In the cell, the concentration of R273H is too low to bind DNA efficiently, but after its upregulation by Fl-CDB3 its concentration may be high enough to bind its target DNA.
We do not yet know the mechanism of the up-regulation of p53, which could be at the level of transcription or degradation by the proteasome. Further work is also required to understand why there is only partial restoration of apoptosis and no observed growth arrest. Perhaps Fl-CDB3 inhibits the interaction of p53 with the 53BP2 protein, from whose structure Fl-CDB3 was derived. Interestingly, the p53-reactivating compound PRIMA-1 (11) does not up-regulate p53. Because we have failed to detect any binding of it to the core domain (unpublished data), PRIMA-1 clearly has a mode of action different from Fl-CDB3.
It was highly unexpected that a negatively charged peptide such as Fl-CDB3 would be taken up by cells, because most known cell-penetrating peptides are positively charged. It was also not known whether Fl-CDB3 would be sufficiently stable in the cellular environment or have sufficient and specific affinity to p53 to be effective. In the light of the present results, Fl-CDB3 could be a lead compound for a chaperoning agent for rescuing p53 in vivo. Because radiotherapy and much of chemotherapy function via the p53 pathways, we envisage that Fl-CDB3 or an optimized derivative could be infused before radiotherapy or chemotherapy, be taken up by cancer cells, and activate them for more effective treatment.
Acknowledgments
We thank Michael Fritsche (Institute for Biomedical Research, Frankfurt) for Saos-2-His-273 cells, P. Chumakov (Engelhardt Institute of Molecular Biology, Moscow) for H1299-His-175 and A431-LacZ cells, and Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore) for HCT-116 cells and PUMA antibodies. This work was supported by project grants from the Swedish Cancer Society (Cancerfonden), the Swedish Medical Research Council, the Swedish Royal Academy of Sciences, the Cancer Society of Stockholm, the Gustav V Jubilee Foundation, Cancer Research UK, and the Medical Research Council. A.F. was supported by Long-Term Fellowship LT00056/2000-M from the Human Frontier Science Program.
Abbreviations: biotin, biotinylated; CDB3, core domain binding peptide 3; Fl, fluorescein-labeled derivative; polyGlu, polyglutamic acid.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 2.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 3.Bullock, A. N., Henckel, J. & Fersht, A. R. (2000) Oncogene 19, 1245-1256. [DOI] [PubMed] [Google Scholar]
- 4.Bullock, A. N., Henckel, J., DeDecker, B. S., Johnson, C. M., Nikolova, P. V., Proctor, M. R., Lane, D. P. & Fersht, A. R. (1997) Proc. Natl. Acad. Sci. USA 94, 14338-14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vousden, K. H. (2002) Biochim. Biophys. Acta 1602, 47-59. [DOI] [PubMed] [Google Scholar]
- 6.Selivanova, G. (2001) Curr. Opin. Investig. Drugs 2, 1136-1141. [PubMed] [Google Scholar]
- 7.Lane, D. P. & Lain, S. (2002) Trends Mol. Med. 8, S38-S42. [DOI] [PubMed] [Google Scholar]
- 8.Lane, D. P. & Hupp, T. R. (2003) Drug Discov. Today 8, 347-355. [DOI] [PubMed] [Google Scholar]
- 9.Bullock, A. N. & Fersht, A. R. (2001) Nat. Rev. Cancer 1, 68-76. [DOI] [PubMed] [Google Scholar]
- 10.Foster, B. A., Coffey, H. A., Morin, M. J. & Rastinejad, F. (1999) Science 286, 2507-2510. [DOI] [PubMed] [Google Scholar]
- 11.Bykov, V. J., Issaeva, N., Shilov, A., Hultcrantz, M., Pugacheva, E., Chumakov, P., Bergman, J., Wiman, K. G. & Selivanova, G. (2002) Nat. Med. 8, 282-288. [DOI] [PubMed] [Google Scholar]
- 12.Friedler, A., Hansson, L. O., Veprintsev, D. B., Freund, S. M., Rippin, T. M., Nikolova, P. V., Proctor, M. R., Rudiger, S. & Fersht, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rippin, T. M., Bykov, V. J., Freund, S. M., Selivanova, G., Wiman, K. G. & Fersht, A. R. (2002) Oncogene 21, 2119-2129. [DOI] [PubMed] [Google Scholar]
- 14.Wang, W., Takimoto, R., Rastinejad, F. & El-Deiry, W. S. (2003) Mol. Cell. Biol. 23, 2171-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedler, A., Veprintsev, D. B., Hansson, L. O. & Fersht, A. R. (2003) J. Biol. Chem. 278, 24108-24112. [DOI] [PubMed] [Google Scholar]
- 16.Bunz, F., Hwang, P. M., Torrance, C., Waldman, T., Zhang, Y., Dillehay, L., Williams, J., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1999) J. Clin. Invest. 104, 263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong, K. B., DeDecker, B. S., Freund, S. M., Proctor, M. R., Bycroft, M. & Fersht, A. R. (1999) Proc. Natl. Acad. Sci. USA 96, 8438-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez, G. S., Nister, M., Stommel, J. M., Beeche, M., Barcarse, E. A., Zhang, X. Q., O'Gorman, S. & Wahl, G. M. (2000) Nat. Genet. 26, 37-43. [DOI] [PubMed] [Google Scholar]