Abstract
Endosomes are major sorting stations in the endocytic route that send proteins and lipids to multiple destinations in the cell, including the cell surface, Golgi complex, and lysosomes. They have an intricate architecture of internal membrane structures enclosed by an outer membrane. Recycling proteins remain on the outer membrane, whereas proteins that are destined for degradation in the lysosome are sorted to the interior. Recently, a retrograde pathway was discovered whereby molecules, like MHC class II of the immune system, return from the internal structures to the outer membrane, allowing their further transport to the cell surface for T cell activation. Whether this return involves back fusion of free vesicles with the outer membrane, or occurs via the continuity of the two membrane domains, is an unanswered question. By electron tomography of cryo-immobilized cells we now demonstrate that, in multivesicular endosomes of B-lymphocytes and dendritic cells, the inner membranes are free vesicles. Hence, protein transport from inner to outer membranes cannot occur laterally in the plane of the membrane, but requires fusion between the two membrane domains. This implies the existence of an intracellular machinery that mediates fusion between the exoplasmic leaflets of the membranes involved, which is opposite to regular intracellular fusion between cytoplasmic leaflets. In addition, our 3D reconstructions reveal the presence of clathrin-coated areas at the cytoplasmic face of the outer membrane, known to participate in protein sorting to the endosomal interior. Interestingly, profiles reminiscent of inward budding vesicles were often in close proximity to the coats.
Keywords: clathrin coat, electron tomography, endosomes, MHC class II, multivesicular body
The endocytic pathway is a dynamic system that functions in the cellular housekeeping by internalization, sorting, and breakdown of macromolecules (1). It is composed of primary endocytic vesicles, early endosomes, late endosomes, and lysosomes. Internalized membrane proteins enter early endosomes and can either be recycled to the cell surface or sorted to the lumen of late endosomes and lysosomes for degradation (2, 3). In the electron microscope, endosomes appear as vacuoles containing 50- to 100-nm intralumenal vesicles. The number of these vesicles increases on gradual maturation of early into late endosomes, also referred to as multivesicular endosomes or multivesicular bodies (MVBs). The general role of MVBs, ubiquitously present in all eukaryotic cells, is to recruit proteins on their internal membranes for degradation. In addition, they are known to be the precursors of secretory lysosomes (4).
Recently it has become clear that late endosomes and lysosomes are dynamic organelles from where proteins in the internal membranes can escape and be actively sorted to other destinations in the cell (5–8). In antigen-presenting cells of the immune system, such as B lymphocytes and dendritic cells (DCs), endosomes and lysosomes operate as storage and antigen-loading compartments for MHC class II molecules (9). On antigen loading, MHC class II is transported to the cell surface for recognition by CD4+ T helper cells to initiate an immune response. Crucially, MHC class II is primarily stored in the internal membranes of MVBs. From a recent study by our group it has become clear that the presence of MHC class II at the cell surface of DCs, which is regulated by pathogens and proinflammatory cytokines (10), demands a translocation mechanism from the inner to the outer endosomal membranes (5). The question is whether the translocation of membrane proteins from the endosomal luminal vesicles to the outer membrane occurs through fusion of free vesicles with the outer membrane, or via existing continuities, i.e., that the vesicular profiles are in fact deep invaginations of the outer membrane. The general relevance of this issue is underlined by the fact that other proteins, like the tetraspanin CD63 and mannose 6-phosphate receptors, have been reported to also recycle from the endosomal inner membranes (6, 11).
To resolve the complex architecture of endosomes, high-resolution (≈4 nm) 3D analysis by electron tomography is the method of choice. As model systems, we used B lymphocytes and DCs that both have an elaborate and well characterized endosomal system (12). To preserve the cells in their most natural state and capture fast processes like fission and fusion, they were cryo-immobilized in milliseconds under high pressure (13, 14). The 3D reconstructions of early and late endosomes reveal the high level of compartmentalization of these organelles, at the level of both inner and outer membranes. The internal membrane structures are distinct vesicles, which are not interconnected or continuous with the outer membrane. This strongly argues that protein relocation within the endosome requires fusion between its membrane subcompartments.
Methods and Materials
Cells. The EBV transformed human B cell line RN was maintained as described (12). Normal rat kidney (NRK) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 100 units/ml penicillin, 4.5 g/liter glucose, and 2 mM l-glutamine. DC line D1 was cultured as described (15). Freshy isolated monocytes were differentiated into DCs as described (16). Before an experiment, NRK and D1 cells were seeded in 9-cm culture dishes containing golden grids covered with a formvar film coated with a thin layer of gelatin.
BSA-Gold Uptake Experiments. To label the endosomal pathway, cells were washed with serum-free medium and incubated for 10 min with a prewarmed 10-nm BSA-gold solution (OD5) in medium containing 0.5% FCS (17, 18). A 10-min time period was used to label the early stages of endosomal maturation, and a 30-min period was used to mark late endosomes (19). After gold uptake, the cells were cryo-immobilized by high-pressure freezing.
High-Pressure Freezing. RN and human DC suspensions were slowly spun down in 1.5-ml Eppendorf tubes for 20 s, after which the supernatant was carefully removed, and the pellet was stored in a 37°C water bath. To high-pressure freeze the cells, one or two golden grids without film were dipped into the pellet, saturating its meshes with cells (20). The grids were sandwiched between the flat sides of standard aluminium cups (ALU Ø 3 × 0.5 mm and an inner cavity of Ø 2 mm × 0.15 mm) used for high-pressure freezing and frozen by using a Leica HPF (Leica Microsystems, Vienna; now M. Wohlwend, Sannwald, Switzerland) (21). The NRK and D1 cells that were growing on grids could be directly sandwiched between the aluminium cups and high-pressure frozen.
Freeze Substitution. High-pressure frozen samples were transferred to the Leica AFS (Leica Microsystems) with precooled substitution fluid (-90°C) of anhydrous acetone, containing 0.5% osmium tetroxide and 0.25% glutaraldehyde (20, 22). Substitution at -90°C lasted for 48 h, and the substitution fluid was refreshed one time. Then the samples were warmed with 1°C per h to -30°C, where they remained for 8 h before transferring them to ice for 1 h (22). Thereafter, the substitution fluid was washed away by anhydrous acetone and the samples with RN and human DCs were pelleted and embedded in Epon. The NRK and D1 cells were embedded with their grids in Epon.
Sample Preparation. Epon sections of 200–300 nm were collected for analysis by electron tomography. Sections were poststained with uranylacetate and lead citrate. Sets of four serial sections were collected on a single slot grid.
Electron Tomography, 3D Reconstruction, and Modeling. MVBs were analyzed by making dual axis tilt series by using a Tecnai20 microscope as described (23). Briefly, digital images were recorded of the structure of interest as it was tilted around two perpendicular axes from -60° to + 60° with 1° increments. With the program package IMOD (24), 3D reconstructions were obtained.
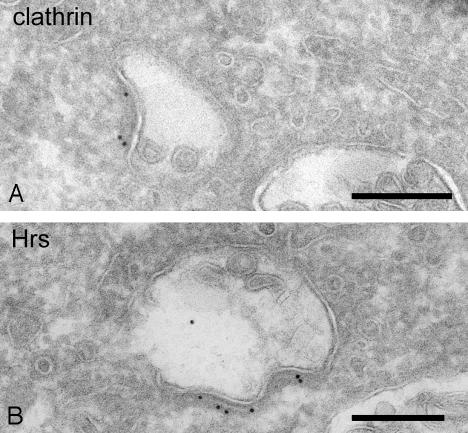
Immunoelectron Microscopy. RN cells were fixed and processed for cryo-immunogold labeling as described (25). Antibodies against clathrin heavy chain (Transduction Laboratories, Lexington, KY) and (hepatocyte growth factor-regulated tyrosine kinase substrate Hrs, a generous gift of S. Urbé, University of Liverpool, Liverpool, U.K.; ref. 26) were used.
Results and Discussion
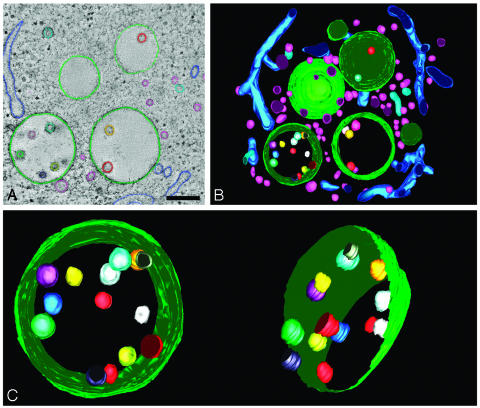
Internal Membrane Structures of MVBs Are Free Vesicles. B-lymphocytes were cryo-immobilized by high pressure freezing and their MVBs analyzed by dual-axis electron tomography. The 3D reconstructions show that internal membrane structures of MVBs are free vesicles (Fig. 1 A–C). Fig. 1B displays the X–Y, X–Z and Y–Z views of the internal vesicles, demonstrating that they have a round shape and are free. The size of the internal vesicles varies from 40 to 100 nm, with an average diameter of 56 nm. The MVB in Figs. 1 and 2 is representative of >20 MVBs that were reconstructed. All of these MVBs were reached by internalized BSA-gold after 30 min of uptake. Fig. 2 A illustrates the marking of membranes to generate a 3D model. The models in Fig. 2 B and C show two different views of the MVB to illustrate that the internal vesicles, each with a different color, are not connected to each other. A movie of the 3D model is shown in Movie 1, which is published as supporting information on the PNAS web site. Our results demonstrate that the MVB is a compartmentalized organelle, consisting of separate inner and outer membrane domains.
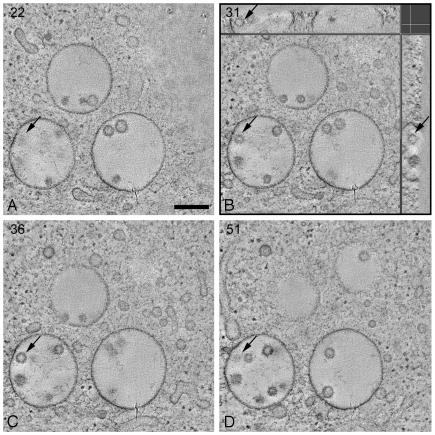
Fig. 1.
MVBs contain free internal vesicles. Shown are tomographic slices (X–Y plane) through the 3D volume of an electron tomographic reconstruction of a cluster of MVBs in a high-pressure frozen freeze-substituted RN cell. The arrows in the different slices point at the same internal vesicle: in A and D, the top and bottom of the vesicle are indicated, respectively; B and C show slices through the vesicle. Numbers specify the slice number through the tomographic volume. (B) In addition to the X–Y plane, tomographic slices through the X–Z plane (Upper) and Y–Z plane (Right) are shown at the level of the indicated internal vesicle. The indicated internal vesicle is contained within the whole 3D volume and is a round structure. Each X–Y slice has a thickness in the z axis of ≈4 nm. The total tomographic volume has a thickness of ≈250 nm. (Bar, 250 nm.)
Fig. 2.
Three-dimensional model of MVB in RN cell. (A) Membranes in consecutive tomographic slices are manually traced to create a 3D model. Internal vesicles in the models are not perfectly round because of the manual tracing of the membranes. (Bar, 250 nm.) (B) Model view of the 3D reconstruction of the MVBs and surrounding area from Fig. 1. The space surrounding the MVBs is filled with tubules and vesicles. Movie 1 shows a movie of the 3D model. (C) Model view of the MVB tilted along two different angles. In the model on the right, the limiting membrane is partially removed to show the internal vesicles. The vesicles were each given a different color to trace them in each different view of the model.
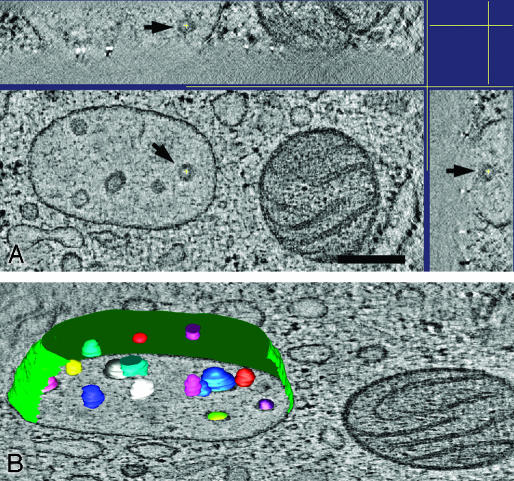
Because MVBs in DCs are dynamic organelles in which proteins, such as MHC class II, are not only stored but can also relocate from the inner to the outer membranes (5), we next analyzed MVBs in this cell type. We used the adherent DC line D1, which is a well characterized long-term culture of isolated mouse spleen DCs that behaves identically to freshly isolated DCs (15, 27) and human blood-derived DCs (Fig. 6 A and B, which is published as supporting information on the PNAS web site). The structure of MVBs in DCs was very similar to the MVBs in B lymphocytes, again showing free internal vesicles, with no connections to the outer membrane (Fig. 3). This finding has the important implication that the relocation of MHC class II from the internal membranes requires fusion with the outer membrane.
Fig. 3.
Dendritic cell MVB in 3D. (A) Tomographic slice of an electron tomographic reconstruction of a MVB in a high-pressure frozen dendritic cell that shows multiple free internal vesicles. In X–Z(Upper) and Y–Z plane (Right) is shown that the vesicle, indicated by an arrow, is contained within the 3D volume. (Bar, 250 nm.) (B) 3D model of MVB displayed on a tomographic slice showing that the intralumenal membranes are free vesicles.
Literature is contradictory concerning the MVB ultrastructure (28–31). The internal membranes of endosomes in Hep2 cells have been described as free vesicles (29), in agreement with our results, whereas those in NRK cells as deep invaginations of the outer membrane (28). However, neither of these studies was conclusive because (i) lack of resolution in the z direction, (ii) loss of material with serial sectioning, and (iii) use of chemical fixation techniques, which take seconds to even minutes to stop all cellular processes and may cause changes of the cell ultrastructure and disrupt membrane continuities. In the present study we avoid these problems by ultimate preservation of ultrastructure combined with high-resolution electron tomography and find, also in NRK cells (Fig. 6 C and D), no continuities between internal vesicles and the outer membrane.
An additional strong argument in favor of free internal vesicles is the ability of hematopoietic cells to release the luminal membranes of MVBs in the extracellular milieu (12, 32, 33), from which they are collected as free vesicles, so-called exosomes. In case of antigen-presenting cells, exosomes are enriched in MHC class II molecules, which is in agreement with the storage of these proteins in the internal vesicles of MVBs.
Clathrin Coats and Inward Budding Sites on the Outer Membrane. The formation of internal vesicles and their protein composition depends on multiple sorting mechanisms (34, 35). These start in early endosomes and specifically target proteins like MHC class II to the luminal membranes, resulting in different protein compositions of internal and outer membranes.
Recently, a novel type of clathrin coat was identified on endosomes (26, 35, 36), which has been postulated to play a role in protein retention and sorting toward the internal vesicles, in concert with the ESCRT (endosomal sorting complex required for transport) I, II, and III complexes. Our 3D reconstructions of early endosomes, marked by internalization of BSA-gold for 10 min, clearly revealed this clathrin coat as an extended oval-shaped area with a diameter ranging from 80 to 500 nm (Fig. 4 A and B, area between arrowheads). Electron tomography on two consecutive thick sections shows that one endosomal vacuole can contain multiple coated areas (Fig. 4C, yellow). Immunoelectron microscopy showed that the observed coat contains clathrin and Hrs (Fig. 5). As shown previously, Hrs is specifically enriched in this type of clathrin coat and may bind ubiquitinated proteins that are sorted to the internal vesicles of endosomes (reviewed in ref. 35). Ubiquitination is a known signal to target receptors, like the epidermal growth factor receptor and growth hormone receptor, both enriched in the clathrin coat (26), to the internal membranes (34). Whether all proteins that are enriched on the internal membranes, like the tetraspan family members CD9 and CD63, are sorted to the endosomal lumen in this way has yet to be revealed (37). In addition, a role for lipids in endosomal sorting is expected because certain lipids, such as cholesterol, lysobismonoacylglycerol phosphate, and phosphatidylinositol 3,4,5-trisphosphate, are particularly enriched in the internal vesicles (19, 38, 39) and inhibition of the phosphatidylinositol 3-kinase interferes with the formation of MVBs (40).
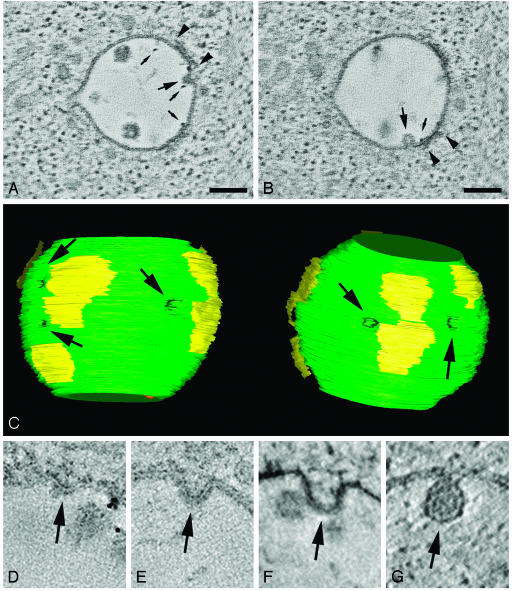
Fig. 4.
Internal vesicle formation in endosomes in RN cells. (A and B) Tomographic slices showing that inward budding profiles of the outer membrane (large arrows) are often situated in close proximity to the clathrin coat (region between arrowheads). Small arrows point to BSA-gold particles that were used in this study as endocytic tracers; their presence demonstrates an early position of the organelle in the endocytic pathway. (Bar, 60 nm.) (C) Model view of MVB from A and B, seen from two different angles, showing the association of the clathrin coat (yellow patches on the surface of the endosome) and the inward budding outer membrane. The arrows point at sites of inward budding profiles. This model is generated from the 3D reconstructions of two serial sections with a total thickness of ≈420 nm. Movie 2, which is published as supporting information on the PNAS web site, shows a movie of the 3D model. (D–G) Different stages of internal vesicle formation are shown in tomographic reconstructions of endosomes in RN cells. The arrows point at the site where the outer membrane is forming an inward budding vesicle.
Fig. 5.
Clathrin and Hrs in endosomal coat. (A) Early endosome in RN cell immunolabeled for clathrin. Protein A-gold particles are present in the coat at the endosomal outer membrane. (Bar, 200 nm.) (B) Endosomal coat in RN cell immunolabeled for Hrs, indicated by the 10-nm gold particles. (Bar, 200 nm.)
If indeed the clathrin/Hrs coat was instrumental in sorting of proteins into the internal vesicles, we should be able to show a spatial relationship between inward budding profiles and the coat itself. Fig. 4 D–G shows several images that may represent stages of inward budding. In the 3D reconstructions these profiles appeared as buds connected to the outer membrane. Interestingly, the inward budding profiles were often located at the edges of the coated areas. This is well illustrated in a movie of the 3D model of this MVB, where the arrows point at the buds (Movie 2). The close apposition of the coated areas and inward budding sites is in agreement with the proposed linkage between protein recruitment in the coats and internal vesicle formation (26, 41). In theory, we cannot exclude that the membrane indentations represent back fusion profiles. However, as maturing early endosomes characteristically accumulate internal vesicles, it is more likely that they are indeed sites of vesicle formation.
The need for an accurate understanding of the MVB membrane organization was prompted by the observation that MHC class II molecules are recruited from the MVB interior to the outer membrane for their transport to the cell surface. The main evidence for the existence of this pathway is the fact the presynthesized pool of MHC class II on the internal membranes of MVBs is the major source for the increased cell surface expression of MHC class II on DC activation (5, 42, 43). This was based on the findings that (i) steady levels of MHC class II on the outer membrane of the MVB are too low to account for the increased level of MHC class II on the surface of activated cells (ii) DC activation leads to a concomitant loss of internal vesicles and increase in the amount of MHC class II on the outer membrane (5), (iii) MHC class II de novo synthesis does not increase on activation, and (iv) the kinetics of MHC class II transport to the surface exceed the speed of degradation of late endosomal content (44). Our present data substantiate that the process whereby MHC class II from the internal membranes of MVBs eventually reaches the cell surface is comprised of a back fusion step of the free internal vesicles with the outer membrane. The nature of this fusion process, which is followed by the morphological change of MVBs into tubules (5), remains to be elucidated. When proteins reach the outer membrane, transport to the cell surface can take place. This may be achieved through a vesicular transport step direct fusion of tubules derived from late endosomes and lysosomes with the plasma membrane (7).
Other proteins that have been proposed to recycle from the intralumenal membranes of late endosomes are CD63 and MPR. In case of the tetraspanin CD63 in endothelial cells, evidence was obtained that it is recruited from the internal membranes to the secretory Weibel–Palade bodies. MPRs are known to recycle to the trans-Golgi network after delivery of their associated lysosomal enzymes in the lumen of MVBs. In a recent study it was shown that the egress of MPR from late endosomes depends on cholesterol (11). For both proteins, however, the underlying mechanism has remained unsolved, in part because of the lack of essential insight in MVB structure. Having established the membrane organization of MVBs, we now postulate that the back fusion pathway, which mediates the trafficking of MHC class II, could also play a role in the transport of CD63, MPR, and possibly other proteins.
The proposed back fusion requires an exceptional fusion mechanism. Membrane fusions of vesicles and organelles in the cell generally involve molecular machineries at the cytoplasmic leaflets of the two fusion partners (45). However, in the case of the internal vesicles and the outer membrane, the exoplasmic leaflets must fuse. To our knowledge, there are no precedents of such a fusion in the cell except for the pathway used by internalized viruses, which enter the cytosol through fusion with the outer membrane of endosomes (46). pH is an important regulator of this fusion event, because viral entry through endosomes depends on endosomal acidification. Because a recent study showed that the pH in the endocytic pathway of DCs decreases on activation (44), it is possible that endosomal pH is also critical for MHC class II transport by regulating the back fusion of internal vesicles in endosomes. Potential candidates to participate in the intra-endosomal fusion machinery are the tetraspanins on the internal vesicles. This is based on the fact that CD9 as well as CD81 mediate sperm-egg fusion, a process that also involves the fusion of exoplasmic leaflets (47, 48).
In conclusion, our 3D reconstructions show that MVBs are compartmentalized organelles with distinct and segregated luminal and outer membrane domains. As a result, proteins can only leave the endosomal lumen on fusion of the free internal vesicles with the outer membrane.
Supplementary Material
Acknowledgments
We thank Dr. Catherine Rabouille for critically reading the manuscript and René Scriwanek and Marc van Peski for quality photographic work. We thank Drs. Paola Ricciardi-Castagnoli and Frank Verreck for the cell line D1, Dennis Langenberg for human DCs, and Dr. Misjaël L. Lebbink for helping with the 3D reconstructions. This work was supported by Netherlands Organization for Scientific Research Grants 805-48-014 and 901-28-142.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Hrs, hepatocyte growth factor-regulated tyrosine kinase substrate; MBV, multivesicular body; DC, dendritic cell; NRK, normal rat kindey.
References
- 1.Mellman, I. (1996) Annu. Rev. Cell Dev. Biol. 12, 575-625. [DOI] [PubMed] [Google Scholar]
- 2.Stoorvogel, W., Strous, G. J., Geuze, H. J., Oorschot, V. & Schwartz, A. L. (1991) Cell 65, 417-427. [DOI] [PubMed] [Google Scholar]
- 3.Mullock, B. M., Bright, N. A., Fearon, C. W., Gray, S. R. & Luzio, J. P. (1998) J. Cell Biol. 140, 591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raposo, G., Vidal, M. & Geuze, H. J. (1997) in Unusual Secretory Pathways: From Batceria to Man, eds. Kuchler, K., Rubartelli, A. & Holland, B. (R. G. Landes, Washington, DC), pp. 161-184.
- 5.Kleijmeer, M., Ramm, G., Schuurhuis, D., Griffith, J., Rescigno, M., Ricciardi-Castagnoli, P., Rudensky, A. Y., Ossendorp, F., Melief, C. J., Stoorvogel, W. & Geuze, H. J. (2001) J. Cell Biol. 155, 53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi, T., Vischer, U. M., Rosnoblet, C., Lebrand, C., Lindsay, M., Parton, R. G., Kruithof, E. K. & Gruenberg, J. (2000) Mol. Biol. Cell 11, 1829-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, A., Toomre, D., Garrett, W. & Mellman, I. (2002) Nature 418, 988-994. [DOI] [PubMed] [Google Scholar]
- 8.Boes, M., Cerny, J., Massol, R., Op den Brouw, M., Kirchhausen, T., Chen, J. & Ploegh, H. L. (2002) Nature 418, 983-988. [DOI] [PubMed] [Google Scholar]
- 9.Geuze, H. J. (1998) Immunol. Today 19, 282-287. [DOI] [PubMed] [Google Scholar]
- 10.Mellman, I., Turley, S. J. & Steinman, R. M. (1998) Trends Cell Biol. 8, 231-237. [DOI] [PubMed] [Google Scholar]
- 11.Miwako, I., Yamamoto, A., Kitamura, T., Nagayama, K. & Ohashi, M. (2001) J. Cell Sci. 114, 1765-1776. [DOI] [PubMed] [Google Scholar]
- 12.Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J. & Geuze, H. J. (1996) J. Exp. Med. 183, 1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuser, J. E., Reese, T. S., Dennis, M. J., Jan, Y., Jan, L. & Evans, L. (1979) J. Cell Biol. 81, 275-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoll, G., Braun, C. & Plattner, H. (1991) J. Cell Biol. 113, 1295-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winzler, C., Rovere, P., Rescigno, M., Granucci, F., Penna, G., Adorini, L., Zimmermann, V. S., Davoust, J. & Ricciardi-Castagnoli, P. (1997) J. Exp. Med. 185, 317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleijmeer, M. J., Morkowski, S., Griffith, J. M., Rudensky, A. Y. & Geuze, H. J. (1997) J. Cell Biol. 139, 639-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Möbius, W., Ohno-Iwashita, Y., van Donselaar, E. G., Oorschot, V. M., Shimada, Y., Fujimoto, T., Heijnen, H. F., Geuze, H. J. & Slot, J. W. (2002) J. Histochem. Cytochem. 50, 43-56. [DOI] [PubMed] [Google Scholar]
- 19.Möbius, W., Van Donselaar, E., Ohno-Iwashita, Y., Shimada, Y., Heijnen, H. F., Slot, J. W. & Geuze, H. J. (2003) Traffic 4, 222-231. [DOI] [PubMed] [Google Scholar]
- 20.Murk, J. L., Posthuma, G. P., Koster, A. J., Geuze, H. J., Verkleij, A. J., Kleijmeer, M. J. & Humbel, B. M. (2003) J. Microsc. 212, 81-90. [DOI] [PubMed] [Google Scholar]
- 21.Müller, M., Meister, N. & Moor, H. (1980) Mikroskopie 36, 129-140. [PubMed] [Google Scholar]
- 22.Wild, P., Schraner, E. M., Adler, H. & Humbel, B. M. (2001) Microsc. Res. Tech. 53, 313-321. [DOI] [PubMed] [Google Scholar]
- 23.Ziese, U., Janssen, A. H., Murk, J. L., Geerts, W. J., Van der Krift, T., Verkleij, A. J. & Koster, A. J. (2002) J. Microsc. 205, 187-200. [DOI] [PubMed] [Google Scholar]
- 24.Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. (1996) J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- 25.Raposo, G., Kleijmeer, M. J., Posthuma, G. P., Slot, J. W. & Geuze, H. J. (1997) in Handbook of Experimental Immunology, eds. Herzenberg, L. A., Weir, D. & Blackwell, C. (Blackwell Science, Cambridge, U.K.), Vol. 4, pp. 1-11. [Google Scholar]
- 26.Sachse, M., Urbe, S., Oorschot, V., Strous, G. J. & Klumperman, J. (2002) Mol. Biol. Cell 13, 1313-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rescigno, M., Martino, M., Sutherland, C. L., Gold, M. R. & Ricciardi-Castagnoli, P. (1998) J. Exp. Med. 188, 2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths, G., Matteoni, R., Back, R. & Hoflack, B. (1990) J. Cell Sci. 95, 441-461. [DOI] [PubMed] [Google Scholar]
- 29.van Deurs, B., Holm, P. K., Kayser, L., Sandvig, K. & Hansen, S. H. (1993) Eur. J. Cell Biol. 61, 208-224. [PubMed] [Google Scholar]
- 30.Fernandez-Borja, M., Wubbolts, R., Calafat, J., Janssen, H., Divecha, N., Dusseljee, S. & Neefjes, J. (1999) Curr. Biol. 9, 55-58. [DOI] [PubMed] [Google Scholar]
- 31.Muller, O., Sattler, T., Flotenmeyer, M., Schwarz, H., Plattner, H. & Mayer, A. (2000) J. Cell Biol. 151, 519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murk, J., Stoorvogel, W., Kleijmeer, M. & Geuze, H. (2002) Semin. Cell Dev. Biol. 13, 303. [DOI] [PubMed] [Google Scholar]
- 33.Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L. & Turbide, C. (1987) J. Biol. Chem. 262, 9412-9420. [PubMed] [Google Scholar]
- 34.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- 35.Raiborg, C., Rusten, T. E. & Stenmark, H. (2003) Curr. Opin. Cell Biol. 15, 446-455. [DOI] [PubMed] [Google Scholar]
- 36.Raposo, G., Tenza, D., Murphy, D. M., Berson, J. F. & Marks, M. S. (2001) J. Cell Biol. 152, 809-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoorvogel, W., Kleijmeer, M. J., Geuze, H. J. & Raposo, G. (2002) Traffic 3, 321-330. [DOI] [PubMed] [Google Scholar]
- 38.Gillooly, D. J., Morrow, I. C., Lindsay, M., Gould, R., Bryant, N. J., Gaullier, J. M., Parton, R. G. & Stenmark, H. (2000) EMBO J. 19, 4577-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi, T., Beuchat, M. H., Lindsay, M., Frias, S., Palmiter, R. D., Sakuraba, H., Parton, R. G. & Gruenberg, J. (1999) Nat. Cell Biol. 1, 113-118. [DOI] [PubMed] [Google Scholar]
- 40.Futter, C. E., Collinson, L. M., Backer, J. M. & Hopkins, C. R. (2001) J. Cell Biol. 155, 1251-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clague, M. J. (2002) Curr. Biol. 12, R529-31. [DOI] [PubMed] [Google Scholar]
- 42.Sallusto, F., Cella, M., Danieli, C. & Lanzavecchia, A. (1995) J. Exp. Med. 182, 389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rescigno, M., Citterio, S., Thery, C., Rittig, M., Medaglini, D., Pozzi, G., Amigorena, S. & Ricciardi-Castagnoli, P. (1998) Proc. Natl. Acad. Sci. USA 95, 5229-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trombetta, E. S., Ebersold, M., Garrett, W., Pypaert, M. & Mellman, I. (2003) Science 299, 1400-1403. [DOI] [PubMed] [Google Scholar]
- 45.Chen, Y. A. & Scheller, R. H. (2001) Nat. Rev. Mol. Cell Biol. 2, 98-106. [DOI] [PubMed] [Google Scholar]
- 46.Stegmann, T. (2000) Traffic 1, 598-604. [DOI] [PubMed] [Google Scholar]
- 47.Miller, B. J., Georges-Labouesse, E., Primakoff, P. & Myles, D. G. (2000) J. Cell Biol. 149, 1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaji, K., Oda, S., Miyazaki, S. & Kudo, A. (2002) Dev. Biol. 247, 327-334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.