Abstract
Gene duplication is a primary source of molecular substrate for the emergence of evolutionary novelties. The chances for redundant gene sequences to evolve new functions are small compared with the probability that the copies become disabled by deleterious mutations. Functional divergence after gene duplication can result in two alternative evolutionary fates: one copy acquires a novel function (neofunctionalization), or each copy adopts part of the tasks of their parental gene (subfunctionalization). The relative prevalence of each outcome is unknown. Similarly unknown is the relative importance of positive selection versus random fixation of neutral mutations. Aldehyde oxidase (Ao) and xanthine dehydrogenase (Xdh) genes encode two complex members of the xanthine oxidase family of molybdo-flavoenzymes that carry different functions. Ao is known to have originated from a duplicate of an Xdh gene in eukaryotes, before the origin of multicellularity. We show that (i) Ao evolved independently twice from two different Xdh paralogs, the second time in the chordates, before the diversification of vertebrates; (ii) after each duplication, the Ao duplicate underwent a period of rapid evolution during which identical sites across the two molecules, involving the flavin adenine dinucleotide and substrate-binding pockets, were subjected to intense positive Darwinian selection; and (iii) the second Ao gene likely endured two periods of redundancy, initially as a duplicate of Xdh and later as a functional equivalent of the old Ao, which is currently absent from the vertebrate genome. Caution is appropriate in structural genomics when using sequence similarity for assigning protein function.
Gene duplication and subsequent functional divergence of the descendant genes has long been recognized as a source of material for the origin of evolutionary novelties (1-3). The chances for a paralog to evolve a new function are small when compared with the fraction of duplicates that become silenced by degenerative mutations (4). If duplications eventually become a significant molecular source for evolutionary novelty, it is because they occur at a very high rate: on average, one per gene per 100 million years, estimated from eukaryote genomic surveys (4), which is comparable to the rate of mutation per nucleotide site in nuclear genomes of vertebrates (5).
Functional divergence after gene duplication can hypothetically result in two alternative evolutionary fates: (i) neofunctionalization, in which one copy acquires an entirely new function whereas the alternative copy maintains the original function (3, 7); and (ii) subfunctionalization, in which each descendant copy adopts part of the tasks of the ancestral gene (6-8). Theoretical results suggest that subfunctionalization should be a more common outcome of duplication than neofunctionalization under plausible conditions, specifically when subfunctionalizing mutations greatly outnumber neofunctionalizing mutations and the selective advantage of the neofunctional alleles is small (8). However, little is known about the relative importance of each evolutionary outcome from real data (9).
Two models have been most frequently invoked to account for functional divergence after gene duplication. The first model, referred to as the Dykhuizen-Hartl effect model (10), does not require positive Darwinian selection. According to this model, functional divergence occurs by random fixation of neutral mutations under relaxed purifying selection owing to reduced functional constraints of redundant genes. These fixed mutations can be complementary loss-of-subfunction mutations or mutations that later induce a change in gene function when the environment or the genetic background is altered (10). The second model requires positive Darwinian selection that accelerates the fixation of advantageous mutations (3). Favored changes can be mutational refinements to alternative gene subfunctions already present in the ancestral gene, or mutations that enhance the activity of a novel function (8, 10). Both models are consistent with accelerated rate of amino acid replacement often observed after gene duplication (9, 11). The two models make contrastable predictions about the nonsynonymous (dN) to synonymous (dS) substitution rates ratio (ω). Under relaxed selection, the rate of amino acid replacement will never exceed that of synonymous substitution; only positive Darwinian selection can produce ω values significantly larger than 1. Recent methodological developments within the maximum-likelihood framework make use of this property for identifying lineage-specific changes in selective pressure at specific amino acid sites (12).
Aldehyde oxidase (Ao; E.C. 1.2.3.1) and xanthine dehydrogenase (Xdh; E.C. 1.1.1.204) provide a prototype case of neofunctionalization after gene duplication. Ao and Xdh encode two large (generally, >1,330 codons), structurally complex oxidoreductases (AOX and XDH, respectively) of the xanthine oxidase family of molybdo-flavoenzymes (13). AOX and XDH are homodimers with a molecular mass of ≈290 kDa, with each monomer acting independently in catalysis. Each monomer comprises three consecutive domains linked by short interdomains: one ≈20-kDa N-terminal domain that contains two distinct iron sulfur redox centers (2FeS), an ≈40-kDa flavin adenine dinucleotide (FAD)-binding domain, and an ≈85-kDa C-terminal molybdo-pterin (Mo-pt)-binding domain, also containing the substrate binding sites (13). XDH has long been recognized as the key enzyme in the catabolism of purines, oxidizing hypoxanthine into xanthine, and xanthine into uric acid. Comparatively, AOX has been much less studied, although it is known to catalyze the oxidation of aldehydes into acids and not to show reactivity with hypoxanthine. A physiological substrate for the enzyme has not yet been established (13). In AOX and XDH, oxidization of the substrate occurs at the Mo-pt active site [or molybdenum cofactor (MoCo)-binding site], which is located within the substrate binding-pocket in the Mo-pt domain. The electrons thus introduced into the enzyme are transferred via two 2FeS centers to FAD, and from this to the final electron acceptor, which in AOX is dioxygen, whereas in XDH it is NAD+. In mammals, XDH interconverts with an oxidase form [xanthine oxidase (XO)], which, like AOX, uses dioxygen as the final electron acceptor. Interconversion is caused by dislocation of the active-site loop, a stretch of several consecutive amino acid residues (Gln 423-Lys 433, in bovine XDH) that surrounds the FAD cofactor (14, 15). AOX and XDH can easily be aligned along their entire lengths. This, jointly with the fact that Xdh is ubiquitous in the tree of life, whereas Ao is circumscribed to, but pervasive through multicellular eukaryotes, indicates that Ao evolved from a eukaryotic copy of Xdh some time before the origin of multicellularity.
Several biochemical studies have noted some unexpected features of AOX (reviewed in ref. 13). In particular, at the level of primary sequence, mammalian AOXs are more similar to mammalian XDHs, with which they share a large fraction of their many introns, than to invertebrate AOXs, which have very few introns, raising the possibility that AOX had evolved not once, but two independent times from XDH. This hypothesis has not been previously explored from an evolutionary standpoint. If correct, this observation would raise additional questions concerning both the timing of the duplications and the fate of the ancestral mammalian AOX gene. Even more important, it would provide a naturally replicated experiment to investigate the process of molecular adaptation.
Materials and Methods
Species and Sequences. We use a data set consisting of 17 species comprising three bacteria and 14 eukaryote representatives. Bacteria have only Xdh, and their corresponding protein sequences are included for rooting the Ao-Xdh tree. To minimize biases induced by unbalances in phylogenetic sampling, we selected 14 eukaryotes such that the Ao and Xdh sequences are represented basically by the same sets of species (see Fig. 1). The GenBank accession numbers are: NP_105850, Rhizobium loti; AJ001013, Rhodobacter capsulatus; and NP_285502, Deinococcus radiodurans, for bacterial XDHs; and AF009441 and U06117, Homo sapiens (human); AF121945 and X62932, Mus musculus (mouse); NM_019363 and NM_017154, Rattus norvegicus (rat); NM_176668 and NM_173972, Bos taurus (cow); AB009345 for Oryctolagus cuniculus (rabbit); AF286379, Felis catus (cat); D13221, Gallus gallus (chicken); AY034103, Poecilia reticulata (guppy); NM_142218 and Y00308, Drosophila melanogaster (fruit fly); and BAA28624 and AL079347, Arabidopsis thaliana, for eukaryotes Ao and/or Xdh codon sequences, respectively. Putative Ao and Xdh sequences for Fugu rubripes (puffer fish); Ciona intestinalis (sea squirt), Anopheles gambiae (mosquito), and Oryza sativa (rice), were obtained by conducting BLAST searches using known AOX and XDH amino acid sequences against available genome databases, and unambiguously corroborated by phylogenetic criterion. Ao and Xdh codon sequences were aligned by using the protein clustalx (16) alignment.
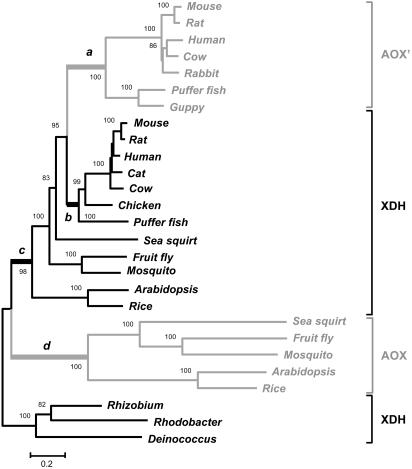
Fig. 1.
Neighbor-joining tree based on the protein gamma distance of mega (20) with α = 1.10 estimated by maximum likelihood. Numbers on the nodes are percent bootstrap values based on 1,000 pseudoreplications. AOX′ and AOX clades are drawn in gray. Thicker branches labeled a and d sprouting right after each Xdh duplication event are those specified a priori to have been subjected to positive selection in the branch-site codon-based maximum likelihood analysis. Species scientific names are given in Materials and Methods.
Phylogenetic Inferences. The evolutionary relationships between Ao and Xdh were determined by using the encoded amino acid sequences. First we obtained a maximum-likelihood estimate of among-site rate variation by using the discrete-gamma model (setting eight categories of rates) of Yang et al. (17-18), which is based on the matrix of Jones et al. (19), with amino acid frequencies set as free parameters, and the topology of Fig. 1 with the relationships among the mammalian orders set as a polytomy. The estimate of the gamma shape parameter α was used to build the neighbor-joining tree using the protein gamma distance implemented in MEGA (20), retaining only nodes present in >50% of 1,000 bootstrap replications.
Study of Molecular Adaptation. Tests for the occurrence of positive selection along the branches that sprout right after the duplication events, called foreground branches (i.e., branches a and d in the tree of Fig. 1; all other branches are called background branches), were conducted by using the branch-site codon-based maximum-likelihood approach of Yang and Nielsen (12), assuming the neighbor-joining relationships of Fig. 1. Briefly, this approach is based on comparing the fit of alternative nested categorizations of the nonsynonymous (dN) to synonymous (dS) substitution rates ratio (ω) by means of a likelihood ratio test, such that the operation of positive Darwinian selection at a particular codon along a specific branch can be inferred if the corresponding ω value is significantly larger than 1. Specifically, we set as the null hypothesis the site-specific Yang and Nielsen's two-site classes M3 model (12), which allows for highly constrained site (i.e., class 1, with ω = ω0 << 1) and quasi-neutralsite classes (i.e., class 2, with ω = ω1 ≈ 1), which are uniform over the entire phylogeny. As the alternative hypothesis, we set the Yang and Nielsen's four-site classes Model B (12), which allows some sites with ω0 and ω1 to change to positive selection (i.e., ω = ω2 > 1) in the foreground branch (i.e., classes 3 and 4, with ω0 → ω2 and ω1 → ω2). For a given tree topology (e.g., Fig. 1), Model B, with P = 5 free parameters (including p0 or the proportion of highly constrained sites, p1 or the proportion of quasi-neutral sites, plus ω0, ω1, and ω2) and log-likelihood L1 fits the data significantly better than the nested submodel M3 with q = 3 free parameters (including p0, ω0 and ω1) and likelihood L0 if the deviance D = -2 log Λ = -2(log L1 - log L0) falls in the rejection region of a χ2 distribution with n = p - q = 2 df. Specific classes 3 and 4 sites were identified by using the empirical maximum-likelihood-based Bayes method of Nielsen and Yang (21). We focused on branches a priori specified to have been under positive selection, i.e., the branches that offshoot right after each gene duplication. Bacteria were excluded from these analyses, and vertebrate and nonvertebrate sequences were assessed separately after removing all gaps from their joint alignment, so that exactly the same 1,109 codon positions were considered for each subset of species. These analyses were conducted with the paml program package (22).
Results and Discussion
Aldehyde Oxidase Evolved Twice from Independent Xdh Paralogs. The evolutionary tree of Fig. 1 shows that aldehyde oxidase has a disjoint phylogenetic distribution. All vertebrate enzymes (here-after denoted as AOX′) cluster closer to vertebrate XDH than to their nonvertebrate AOX counterparts. This branching pattern is present in all bootstrap replicates and is unambiguously supported when contrasted by means of a likelihood ratio test (P < 10-6, df = 1) against a topology in which the AOX′ clade is connected to the protochordate Ciona intestinalis (sea squirt). The disjoint distribution of aldehyde oxidase is not an artifact created by convergence in the amino acid compositions of AOX′ and vertebrate XDH because the pattern of amino acid compositions of the sequences does not match the relationships shown in Fig. 1 (see refs. 23 and 24). In addition, vertebrate Ao possesses 17 of the 28 intron positions of Ciona Xdh, whereas it only exhibits 2 of the 27 intron positions of Ciona Ao. AOX′ is absent in the protochordate Ciona but is present in all vertebrate chordates (see Fig. 1), which indicates that the Xdh duplication event that gave place to AOX′ occurred at some time point between the origin of protochordates and the split of ray-finned fishes.
The Ancestral Vertebrate Ao Gene Was Lost. To ascertain the fate of the ancestral vertebrate Ao gene, i.e., the one that was functioning at the time AOX′ arose, we conducted blast searches against the complete mouse, human, and puffer fish genomes, using the closest extant relative, Ciona AOX as the query. If an ancestral Ao sequence would presently exist, it should be easily detected given the large size and complexity of the molybdo-flavoenzyme. However, our searches were unsuccessful, implying that AOX was probably lost before the diversification of vertebrates. It is not possible to tell whether AOX was still functional at the time AOX′ evolved. If AOX were still functional, this would imply that, during the second transition to aldehyde oxidase, the Xdh paralog would have endured two periods of functional redundancy, the first as XDH and the second as AOX′. Alternatively, the “newborn” AOX′ could have outcompeted and rapidly supplanted the old AOX.
Both AOX and AOX′ Evolved by Reorganization of the XDH FAD and Substrate-Binding Pockets. Common models of gene duplication predict an acceleration of the rate of amino acid replacement associated with neofunctionalization (see ref. 9). We investigated this issue separately for AOX′ and AOX by means of likelihood ratio tests. Specifically, for the case of AOX′ we tested the null hypothesis (H0) that after the duplication event the copy that acquired the Ao function (i.e., branch a in Fig. 1) evolved at the same rate as the copy that retained the ancestral Xdh function (i.e., branch b in Fig. 1), i.e., H0: a = b. Analogously, for the case of AOX we set as the null hypothesis H0: d = c (see Fig. 1). Likelihood ratio tests were conducted assuming the topology in Fig. 1 and the discrete-gamma amino acid substitution model of Yang et al. (10) (see Materials and Methods) as implemented in the hyphy package (25). The null hypothesis was rejected in the two cases (P < 10-6, df = 1). According to the branch length-ratios a:b and d:c, the sequences that acquired the Ao function evolved at rates 5.3 and 4.3 times faster after the duplications than those of their respective sister paralogs that retained the original Xdh function, for the cases of AOX′ and AOX, respectively. These values represent averages across long branches (see Fig. 1), so they are minimum estimates. The rate of amino acid replacement diminished after each Ao paralog acquired its new function, presumably because of increased effects of purifying selection. Nonetheless, the two Ao paralogs ostensibly continued to evolve faster than the Xdh paralogs, as reflected by the values that are obtained (1.4 and 1.9, respectively) when the branch length-ratios are taken of the sums across the corresponding a b and c d descendant internodal distances (considering only common descendant nodes for the case of vertebrates).
Acceleration of the rate of amino acid replacement of the paralogs that gained the Ao function could occur because of positive Darwinian selection. Alternatively, it could be a reflection of the random fixation of neutral mutations under relaxed functional constraints. But it seems unlikely that random accumulation of neutral substitutions could result in two independent paralogs acquiring the same function. We conducted tests of positive selection separately for branches a and d, i.e., the branches reflecting increased evolutionary rate immediately after the duplication events (see Fig. 1), using the branch-site codon-based maximum likelihood approach of ref. 12. The results of these tests indicate that branches a and d were under intense positive selection, apparently stronger for branch d (≈21% positively selected sites out of 1,109 codon sites, with estimated intensity of positive selection, ω2 = 30.17; P < 10-6, df = 2), than for branch a (≈10% positively selected sites with ω2 = 3.27; P < 0.000, df = 2). A greater AOX than AOX′ ω2 value might have occurred because the acquisition of the aldehyde oxidase function may have followed alternative adaptive paths in the two paralogs, each involving different intensities of positive selection. After removing the sites inferred to have been favored by selection (see below) from each corresponding data set (i.e., nonvertebrate and vertebrate data sets for AOX and AOX′, respectively; see Materials and Methods), the length difference between the branches that offshoot immediately after the duplications disappears for the case of AOX (i.e., H0: a = b; P = 0.24, df = 1), but remains for AOX′ (i.e., H0: d = c; P < 0.001, df = 1), suggesting that purifying selection was less important for the paralog that gave place to AOX′. Reduced purifying selection for AOX′ might have occurred if this paralog underwent a longer period of functional redundancy. Note that unlike AOX, which represented the emergence of a novel function, AOX′ may have acquired its function when AOX was still functional, so that AOX′ would have endured two periods of redundancy, first as a paralog of XDH and then as a paralog of AOX. Alternatively, AOX′ may have experienced one single, but more prolonged period of redundancy as a paralog of XDH (e.g., if AOX had already lost its function by the time AOX′ arose). Be that as it may, it cannot be ruled out that the increase in the rate of amino acid replacement of AOX′ not ascribable to positive selection is a reflection of the accumulation of neutral substitutions previously to the acquisition of the Ao activity. Some of those replacements could have later been recruited for the novel function.
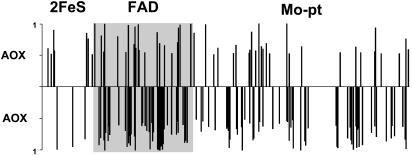
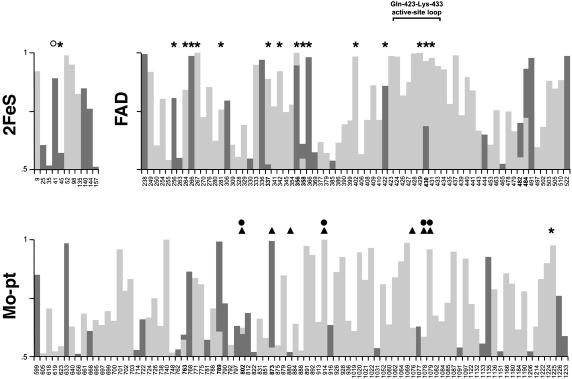
AOX and XDH differ in the substrate (aldehydes and hypoxanthine, respectively) they act upon, and in the molecule they use as the final acceptor of electrons (dioxygen and NAD+). It would, therefore, be expected that positive selection for the evolution of the aldehyde oxidase function from XDH would have affected XDH residues concerning the substrate and the FAD-binding pockets. Fig. 2 represents the distribution of AOX′ and AOX sites identified by the empirical, maximum-likelihood-based Bayesian method of Nielsen and Yang (21) to have been under positive selection along branches a and d, mapped against the bovine XDH domains (as defined in the Pfam protein families database; ref. 26), so far the only eukaryotic molybdo-flavoprotein for which a crystal structure has been obtained (14) (PDB entry 1F04). The two enzymes exhibit an excess of positively selected sites in the FAD domain (22 and 58, respectively) over the expected numbers (15.2 and 34.7, respectively) if adaptive sites were randomly distributed across the three protein domains (2FeS, FAD, and Mo-pt). The excess is highly significant for the case of AOX (χ2 = 26.9, P ≈10≈5, df = 2). The Protein Data Bank provides the specific bovine XDH amino acid residues that are predicted with the ligand-protein contact (LPC) software (27) to be in direct contact with the 2FeS, FAD, and Mo ligands, and salicylate, a competitive inhibitor that fills the channel that leads into the buried Mo-pterin active site, blocking the access of the substrate to the enzyme (13, 28). A significantly greater than expected number of positively selected sites hits FAD and salicylate-protein contact residues in both AOX′ and AOX (see Table 1 and Fig. 3). The inferred accelerated rate of amino acid replacement at those specific sites (and others nearby; see Fig. 3) is doubtless the footprint of adaptive processes involving the change from NAD+ to dioxygen as the final electron acceptor at the FAD reactive site, and modification of substrate affinities that led to the acquisition of the aldehyde oxidase function from Xdh. For example, bovine XDH Trp-336 (and likely Phe-337; ref. 13) is implicated in the dislocation of the active-site loop Gln-423-Lys-433 around the FAD cofactor, which causes the conversion of XDH into the xanthine oxidase form in mammals (14, 15). This structural rearrangement blocks the approach of NAD+ to FAD, changes the electrostatic potential around FAD, and opens the gate for the solvent channel, making it easier for dioxygen to reach the reduced cofactor (15). Nine of the 11 sites of this loop, residues 423-424, 426-431, and 433 in Fig. 3, are positively selected in AOX. Analogously, bovine XDH Glu-802 and Arg-880 are known to be critical for the positioning of the purine substrate in the Mo active site (13).
Fig. 2.
Sites that are subjected to selection in the branches a (AOX′) and d (AOX) of the tree of Fig. 1 along the three functional enzyme domains. The bar heights are proportional to the corresponding Bayes probabilities. Sites not subject to selection (Bayes probability <0.50) do not show vertical bars. The three enzyme domains are depicted proportionally to their length (after excluding alignment gaps) in the bovine XDH sequence as defined in the Pfam (26) protein families' database: 127, 262, and 594 codons for 2FeS, FAD, and Mo-pt domains, respectively. The figure shows a concentration of positively selected sites in the FAD domain.
Table 1. Number of AOX′ and AOX positively selected sites that match bovine XDH protein-ligant contacts.
| 2FeS, 19 | FAD, 38 | Mo, 9 | Substrate, 10 | |
|---|---|---|---|---|
| AOX′ | 1 (1.05) | 8** (3.19) | 1 (0.42) | 3** (0.47) |
| AOX | 0 (0.60) | 12* (8.41) | 4* (1.03) | 6*** (1.15) |
| Identical matches | 0 (0.00) | 7** (2.50) | 1 (0.30) | 2 (0.30) |
Numbers on the column headings are total predicted LPCs within the corresponding bovine XDH protein domains (after excluding 2FeS Asn-71, Mo Thr-1010, and Mo Val-1011 contacts because of alignment gaps; Mo and substrate ligands are in the Mo-pt domain); e.g., LPC software predicts 38 FAD-protein contacts within the FAD domain. In parentheses are the expected numbers of positively selected LPCs obtained assuming that the probability that a positively selected site match a LPC follows a binomial distribution. For AOX′ and AOX, the probability is obtained by dividing the number of positively selected sites in each domain (7, 22, and 28, and 4, 58, and 68 in the 2FeS, FAD, and Mo-pt domains, for AOX′ and AOX, respectively) by the length of the aligned domain (i.e., 127, 262, and 594 sites for 2FeS, FAD, and Mo-pt domains, respectively). For identical matches across AOX′ and AOX, the probability is obtained by dividing the number of positively selected sites matching LPCs (i.e., the values in the AOX′ and AOX rows of the table) by the total number of LPCs of each ligand. *, P < 0.05. **, P < 0.01. ***, P < 0.001.
Fig. 3.
Sites under positive selection along branches a (AOX′; dark gray) and d (AOX; light gray) in the 2FeS, FAD, and Mo-pt domains. Site numbers shaded in gray denote identical matches across AOX′ and AOX. Symbols above the bars represent 2FeS (open circle), FAD (asterisks), Mo (filled circles), and substrate (triangles) LPC sites. Bar heights indicate Bayes probabilities.
The selective changes of these protein regions occurred recurrently in two Ao paralogs that originated from Xdh by duplications separated by ≈1 billion years (i.e., the time span from the origin of multicellular eukaryotes to the last common ancestor of vertebrates). In the case of the FAD-binding pocket (the one involving the largest number of sites, so yielding the greatest statistical power), a greater than expected number of positively selected residues matches identical LPCs across AOX′ and AOX (see Table 1 and Fig. 3); in four of the seven sites, selection favored identical or chemically similar amino acids in AOX and AOX′, i.e., His at site 356, Ile and Leu at site 337, His and Asp at site 358, and Thr and Asp at site 484, respectively; however, in the remaining three sites, selection favored chemically different amino acids, i.e., Val and Ser at site 354, Ser and Ala at site 430, and Asn and Met at site 482, respectively, suggesting that acquisition of the aldehyde oxidase function from XDH was attained with different amino acid compositions in AOX and AOX′ (see ref. 29).
If we were to assign AOX′ function on the basis of sequence similarity, we would arrive to the wrong conclusion that AOX′ is functionally more closely related to XDH than to AOX. Interconversion between members of the same protein family like those reported here suggests that caution must be exercised when using structural genomic approaches for assigning protein function on the basis of sequence similarity.
Acknowledgments
F.R.-T. and R.T. have received support from contracts Ramón y Cajal and Doctor I3P, respectively, from the Ministerio de Ciencia y Tecnología (Spain). This research was supported by National Institutes of Health Grant GM42397 (to F.J.A.).
Abbreviations: AOX, aldehyde oxidase; XDH, xanthine dehydrogenase; FAD, flavin adenine dinucleotide; Mo-pt, molybdo-pterin; LPC, ligand-protein contact.
References
- 1.Stephens, S. G. (1951) Adv. Genet. 4, 247-265. [DOI] [PubMed] [Google Scholar]
- 2.Nei, M. (1969) Nature 221, 40-42. [DOI] [PubMed] [Google Scholar]
- 3.Ohno, S. (1970) Evolution by Gene Duplication (Springer, Heidelberg).
- 4.Lynch, M. & Conery, J. S. (2000) Science 290, 1151-1155. [DOI] [PubMed] [Google Scholar]
- 5.Li, W.-H. (1997) Molecular Evolution (Sinauer, Sunderland, MA), pp. 179-181.
- 6.Hughes, A. L. (1994) Proc. R. Soc. London Ser. B 256, 119-124. [Google Scholar]
- 7.Force, A., Lynch, M., Pickett, B., Amores, A., Yan, Y.-L. & Postlethwait, J. (1999) Genetics 151, 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch, M., O'Hely, M., Walsh, B. & Force, A. (2001) Genetics 159, 1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, J. (2003) Trends Ecol. Evol. 18, 292-298. [Google Scholar]
- 10.Zhang, J., Rosenberg, H. F. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, Z. (2002) Curr. Opin. Genet. Dev. 12, 688-694. [DOI] [PubMed] [Google Scholar]
- 12.Yang, Z. & Nielsen, R. (2002) Mol. Biol. Evol. 19, 908-917. [DOI] [PubMed] [Google Scholar]
- 13.Garattini, E., Mendel, R., Romão, M. J., Wright, R. & Terao, M. (2003) Biochem. J. 372, 15-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enroth, C., Eger, B. T., Okamoto, K., Nishino, T., Nishino, T. & Pai, E. F. (2000) Proc. Natl. Acad. Sci. USA 97, 10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuwabara, Y., Nishino, T., Okamoto, K., Matsumura, T., Eger, B. T., Pai, E. F. & Nishino, T. (2003) Proc. Natl. Acad. Sci. USA 100, 8170-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmoungin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Z., Nielsen, R. & Hasegawa, M. (1998) Mol. Biol. Evol. 15, 1600-1611. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8, 275-282. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, R. & Yang, Z. (1998) Genetics 148, 929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (1999) Genetics 153, 339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarrío, R., Rodríguez-Trelles, F. & Ayala, F. J. (2001) Mol. Biol. Evol. 18, 1464-1473. [DOI] [PubMed] [Google Scholar]
- 25.Kosakovsky, S. L. & Muse, S. V. (2000) HYPHY: Hypothesis Testing Using Phylogenies (Program in Statistical Genetics, Dept. of Statistics, North Carolina State Univ., Raleigh), Version Beta 1.7.
- 26.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobolev, V., Sorokine, A., Prilusky, J., Abola, E. E. & Edelman, M. (1999) Bioinformatics 15, 327-332. [DOI] [PubMed] [Google Scholar]
- 28.Hille, R. (1996) Chem. Rev. 96, 2757-2816. [DOI] [PubMed] [Google Scholar]
- 29.Golding, G. B. & Dean, A. M. (1998) Mol. Biol. Evol. 15, 355-369. [DOI] [PubMed] [Google Scholar]