Abstract
Prokaryotic genomes are substantially diverse, even when from closely related species, with the resulting phenotypic diversity representing a repertoire of adaptations to specific constraints. Within the microbial population, genome content may not be fixed, as changing selective forces favor particular phenotypes; however, organisms well adapted to particular niches may have evolved mechanisms to facilitate such plasticity. The highly diverse Helicobacter pylori is a model for studying genome plasticity in the colonization of individual hosts. For H. pylori, neither point mutation, nor intergenic recombination requiring the presence of multiple colonizing strains, is sufficient to fully explain the observed diversity. Here we demonstrate that H. pylori has extensive, nonrandomly distributed repetitive chromosomal sequences, and that recombination between identical repeats contributes to the variation within individual hosts. That H. pylori is representative of prokaryotes, especially those with smaller (<2 megabases) genomes, that have similarly extensive direct repeats, suggests that recombination between such direct DNA repeats is a widely conserved mechanism to promote genome diversification.
Prokaryotes that successfully colonize niches for extended periods of time must be capable of adapting to changing environmental stresses (1). Genomic diversity, caused by spontaneous point mutation, intragenomic rearrangement, and horizontal gene acquisition (2), creates a phenotypically diverse population from which the most-fit variants are selected. Colonization of the primate stomach by Helicobacter pylori is prolonged, usually persisting for decades without host clearance (3). That essentially all H. pylori isolates obtained from unrelated individuals are genetically distinguishable (4), and that diverse subclones have been identified in individual hosts (5-7) suggest that persistence may be facilitated by the organism's ability to adapt to dynamic environments by continuous change (8). HIV and other pathogenic viruses that persistently infect hosts employ similar strategies of “quasispecies” development to increase fitness success (9).
For H. pylori, neither spontaneous point mutation (10) nor recombination with other bacterial cells (11) is sufficient to explain observed intrahost genetic variation (5, 7). Computational analysis of the fully sequenced H. pylori strain 26695 (12) indicates numerous direct DNA repeats (13, 14); however, these studies do not demonstrate a role for repeats through experimental analyses, provide evidence that these repeat structures are involved in any biologic function, or assess how the nonrandom distribution of repeats may affect the physiology or evolution of the host organism. Because recombination involving direct repeats allows for deletion or duplication of intervening sequences (15), we sought to determine the extent to which H. pylori uses repetitive DNA sequences to generate diversity. Our observations provide evidence that the presence of extensive, nonrandomly distributed repetitive DNA in H. pylori serves to enhance programmed diversification at particular loci, which appears to be a highly conserved mechanism in prokaryotes.
Methods
Computational Analyses. For 51 fully sequenced bacterial genomes representing 44 species (Table 2, which is published as supporting information on the PNAS web site) obtained from GenBank, the repeat visualization program reputer (16) was used to identify all identical DNA repeats of lengths with expected occurrence values <0.01 (13, 14).
Analysis of Potential Hotspots Identified by Computational Analyses. PCR was performed on chromosomal DNA from 33 strains by using primers amiAF and amiAR (Table 3, which is published as supporting information on the PNAS web site) that flank the amiA repeat regions (see Fig. 2) as described (17).
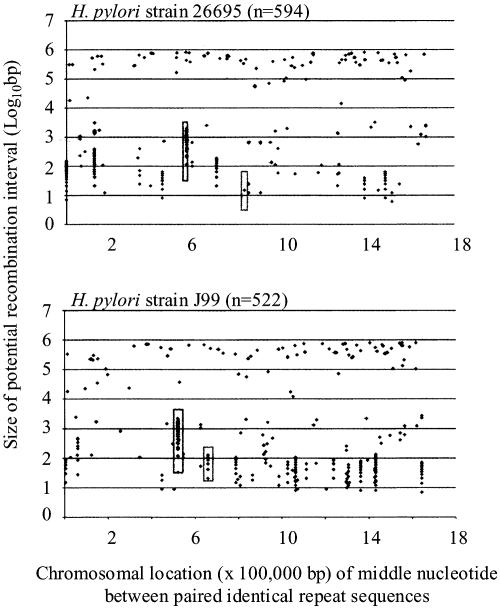
Fig. 2.
Genomic locations of paired repeats in H. pylori strains 26695 and J99. The abscissa represents the chromosomal location (×100,000 bp) of the middle nucleotide of the segment flanked by the paired repeats, and the ordinate represents the size of the potential recombination interval. The H. pylori cagY and amiA loci are in long and short boxes, respectively.
Construction of H. pylori Mutants Used to Assess Deletion Frequencies. Plasmids pAT40,5,15,50,100 each containing a kanamycin resistance (aphA) cassette interrupted by a chloramphenicol resistance (catgc) cassette, were constructed as shown (Fig. 5, which is published as supporting information on the PNAS web site) by using primers listed in Table 3. Correct cassette organization of each plasmid was confirmed after each ligation by digestions with appropriate restriction endonucleases. Each plasmid was transformed into H. pylori strains JP26, B128, J166, 9999, and 9886 as described (17); transformants were selected on media containing 8 μg/ml chloramphenicol, and correct insertion of each deletion construct within the chromosomal vacA locus was verified by PCR. To examine deletion frequencies in other H. pylori genomic loci, the recA (18), ureA (12), and uvrB (19) ORFs were PCR-amplified from H. pylori strain JP26 and cloned into pGEM-T-EZ to create pRECA, pUREA, and pUVRB, respectively. Deletion cassettes containing 0-, 50-, or 100-bp identical DNA sequences (IDS) were PCR-amplified from pAT40, pAT450, and pAT4100, respectively, digested with BamHI (New England Biolabs, Beverly, MA), ligated into pRECA, pUREA, or pUVRB BamHI-digested inverse PCR products, and transformed into separate DH5α cells. Each resulting plasmid (pRECA0,50,100, pUREA0,50,100, pUVRB0,50,100) was transformed into H. pylori strain JP26 as described (20); transformants were selected on media containing 8 μg/ml chloramphenicol, and correct insertion of each deletion construct was verified by PCR. RecA- and UvrB-negative phenotypes were verified by sensitivity to methyl methanesulphonate (18) and UV radiation exposure (19).
Deletion Frequency Assays in H. pylori. To assess recombination frequencies in H. pylori strains containing deletion constructs (transformed by pAT40,5,15,50,100), the cells were grown on trypticase soy agar (TSA) plates for 48 h at 37°C (5% CO2), allowing for deletion to occur, then harvested and washed twice in saline, and 25-, 100-, and 200-μl aliquots were plated onto Brucella agar (BA) plates supplemented with newborn calf serum (NBCS) and 25 μg/ml kanamycin. As controls, 200 μl from each suspension was plated onto BA plates containing NBCS, kanamycin, and chloramphenicol. Total colony-forming units (cfu) and number of deletion mutants were determined by plating serial dilutions onto TSA plates or TSA plates with kanamycin. Plates were incubated at 37°C in a 5% CO2 environment for 96 h, colonies were counted, and deletion frequencies were calculated.
To assess deletion frequencies in Escherichia coli, RR1 (RecA+) or HB101 (RecA-) strains (20) containing deletion-assay plasmids were grown in Mueller Hinton broth cultures supplemented with ampicillin (100 μg/ml) at 37°C for 12 h. Cells were harvested and resuspended in 1 ml of saline. The remainder of the assay was identical to that for H. pylori, except that cells were plated at frequencies of 10-6, 10-7, and 10-8, and incubated at 37°C for 24 h.
Results
Characteristics of Direct DNA Repeats in 51 Prokaryotic Genomes. When the characteristics of direct identical DNA repeats with expected occurrence probability <0.01 (13, 14) in H. pylori were compared with those in 49 other genome sequences (from 44 prokaryotic species), the sizes of the smallest significant repeats and median identical repeat size varied within a relatively narrow range (Table 2). In contrast, there was substantial variation between organisms in the maximum repeat size [from 29 bp (Buchnera aphidicola) to 14,317 bp (Xylella fastidiosa)] and repeat density/megabase (Mb) [from 12 (Chlamydia trachomatis) to 5,069 (Streptococcus pneumoniae)]. Linear and logarithmic regression analyses showed no significant correlations between genome size and either median repeat size or density (data not shown). H. pylori strains 26695 and J99 were little different from other organisms with respect to these criteria.
Chromosomal Distribution of Direct DNA Repeats Facilitates Intragenomic Recombination in Small (<2 Mb) Genomes. Because the distance between repeats may influence recombination frequency (15), for the 51 genomes studied, the median potential recombination sizes (MPRS), defined as the distance between paired identical repeat sequences plus the length of one repeat, were determined. The MPRS ranged from 185 bp (C. trachomatis) to 1,024,894 bp (Nostoc sp. PCC 7120). Although there are notable exceptions (e.g., Rickettsia conorii, Mycobacterium tuberculosis), organisms with genomes <2 Mb had significantly (P < 0.001, Mann-Whitney U test) smaller MPRS (n = 20; 59.6 ± 20.0 kb) than genomes ≥2 Mb (n = 31; 348.3 ± 62.3 kb). These data are consistent with our hypothesis that small genome organisms preferentially use intragenomic recombination between repeat sequences to modulate gene content.
Examination of Potential Recombination Intervals in H. pylori and Representative Genomes. Genomes of the two H. pylori strains (Fig. 1 and Tables 4 and 5, which are published as supporting information on the PNAS web site) and four representative organisms (Fig. 6, which is published as supporting information on the PNAS web site) were further examined to determine relationships between repeat length, genomic location, and potential recombination events. H. pylori had the highest proportion (58.5% and 54.8% for strains 26695 and J99, respectively) of repeats <5 kb apart, whereas for E. coli K12 and O157:H7, 85.1% and 85.6% (not shown), respectively, of repeats were >100 kb apart. In Bacillus subtilis, repeats >5 kb apart and in Campylobacter jejuni, repeats <5 kb apart are overrepresented. That these computational analyses identified nonrandom and characteristic spacing patterns between paired direct DNA repeats suggests that these repetitive sequences are serving species-specific, but possibly parallel, biological functions.
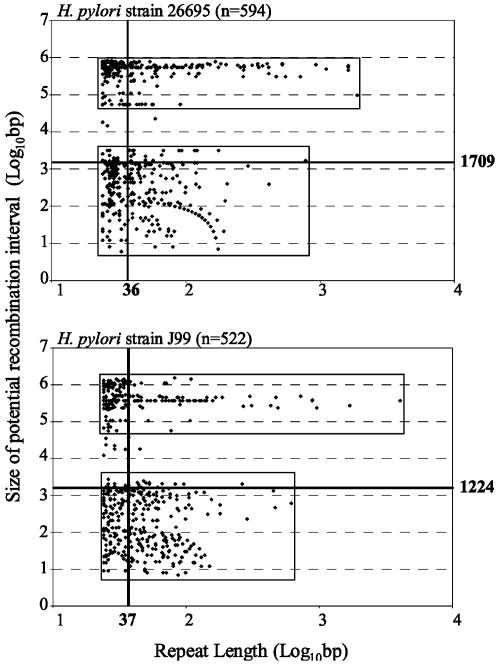
Fig. 1.
Relation between size of paired repeats and length of chromosomal regions subject to recombination for H. pylori strains 26695 and J99. The abscissa represents the length of the repeated elements, and the ordinate represents the size of the recombination interval (segment between paired repeats plus one repeat); the bold lines represent the median values, with lengths (in bp) indicated.
Analyses of locations of chromosomal repeats provide further evidence for their nonrandom distribution (Fig. 2 and Fig. 7, which is published as supporting information on the PNAS web site). In both H. pylori genomes, there was clustering of the locations of the repeats <5 kb apart (boxed areas are illustrative) but not >10 kb apart (Fig. 2), suggesting that these clustered loci may be “recombination hotspots,” potentially resulting in chromosomal deletions or duplications. These computational analyses are consistent with experimental data indicating that for H. pylori, intragenomic recombination between direct DNA repeats ≤3.6 kb apart promote deletions of intervening sequences (17). We hypothesize that deletions between direct DNA repeats separated by >5 kb occur, but that isolates containing these deletions may be selected against, because the probability of essential genes between the repeat sequences increases as their distance apart increases. Clustering patterns differed among the other organisms; potential hotspots we identified (Fig. 7, green boxes are illustrative) reflect regions known for their variation (21-24). Analysis of clustered repeats in M. tuberculosis (Fig. 3) identified known hypervariable loci (22, 23), uncovering a pattern (“beehives flanking diamond”) that we believe is characteristic of hypervariable regions. This pattern supports the hypothesis that the identified recombination hotspots are regions of genome plasticity, exemplifying selection for highly nonrandom repeat distributions within prokaryotic genomes, and indicating that this phenomenon extends beyond H. pylori.
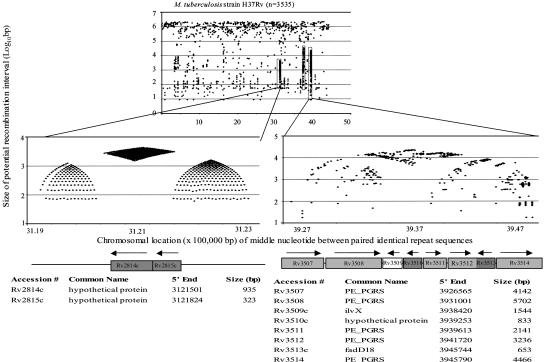
Fig. 3.
Repeat organization in M. tuberculosis H37Rv potential recombination hotspots. The potential M. tuberculosis recombination hotspots show highly nonrandom distributions of repetitive DNA; the “beehives flanking diamond” pattern observed on the left also is present on the right, but across a wider distance and with less definition. M. tuberculosis strain CDC1551 shows highly similar phenomena (not shown). Below each panel are the annotated gene designations by tigr (www.tigr.org) and a schematic of the locus. Paralogous ORFs are shaded the same color.
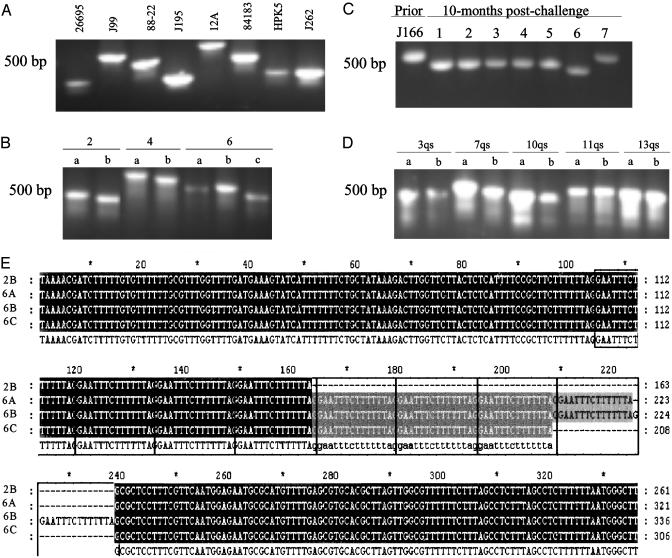
Recombination Hotspots Identified Through in Silico Analyses Map to Hypervariable Chromosome Sequences Subject to High-Frequency Deletion and Duplications. To test the hypothesis that the nonrandom distribution of paired repeat sequences within prokaryotic genomes promotes genome plasticity, for the two H. pylori genome sequences, we examined potential recombination hotspots, defined as chromosome regions ≤5 kb that contained ≥4 paired repeat sequences ≥24 bp. Strains 26695 and J99 contained 12 and 20 such sites, respectively. Examination of repeat distribution within potential recombination hotspots indicated that 50% (6 of 12) in strain 26695 and 45% (9 of 20) in strain J99 contained tandem repeats. The differing locations of potential recombination hotspots between strains results from differences in the number of repeats at a particular locus and from differences in genome location of a particular locus (25). Average G+C content of these sites in strains 26695 and J99 (32.2 ± 8.0% and 31 ± 10%, respectively) were lower than that of the whole chromosome (39%); 75% and 80%, respectively, of the sites were within ORFs. Nine of the sites in strain 26695 had conserved sequences in strain J99, and in most, sequence variation between homologous regions resulted from intragenomic deletions or duplication between paired repeats. Similarly, of the 17 sites in strain J99 with homology to 26695 sequences, most of the sequence variation between the two strains was caused by differences in the number of repeat sequences. To further assess recombination hotspot variation, we examined the nucleotide sequences of one site (Fig. 2, short boxes) differing between H. pylori strains 26695 and J99 and for which size variation could be reliably assessed by PCR. This locus mapped to orthologous genes, HP0772 (1,320 nt) in strain 26695 and JHP0709 (1,407 nt) in strain J99, that encode putative N-acetylmuramoyl-l-alanine amidases (amiA) (12, 25), and size differences between HP0772 and JHP0709 were caused by variation in 15-bp tandem repeats number. To assess variation of the repeat-containing region, PCR analysis was performed for H. pylori isolates obtained (i) from differing geographic locations (8 isolates, Fig. 4A); (ii) from a single clonal grouping (7) (7 isolates, Fig. 4B); (iii) 10 months after experimental rhesus inoculation (26) (8 isolates, Fig. 4C); and (iv) 7-10 years apart from individual hosts (6) (10 isolates, Fig. 4D). Results indicate extensive size variation between both unrelated and related isolates producing differing amiA alleles, that size variation is ongoing, and that variation is host-specific. Sequence analysis confirmed that the observed amiA differences within hosts were caused by variation in 15-bp repeat number (Fig. 4E). The lack of variation identified in PCR products amplified from two separate loci (glmM and flaA) without paired repeats confirms substantially lower background rates of recombination in the absence of repeats. That the deletions and duplications in all cases yield in-frame amiA suggests selection for expression. PCR and sequence analysis of other H. pylori hotspots, cagY (Fig. 2, long boxes) (27) and cagA (28) yielded parallel results, affirming that the sites identified in silico map to chromosomal regions with high levels of sequence variation resulting from intragenomic deletions or duplications.
Fig. 4.
PCR and sequence analysis of H. pylori amiA region with multiple direct repeats. PCR primers (AmiF/AmiR) flank repeat region in A-D. (A) Eight unrelated wild-type strains from different locales. (B) Seven closely related isolates from three members of an extended family. (C) Eight isolates of strain J166 obtained before and 10 months after experimental challenge of a rhesus monkey. (D) Paired isolates obtained 7-10 years apart from five patients. (E) Sequence analysis of isolates 2B, 6A, 6B, and 6C (B) indicates their identity, except that the PCR product size variation is caused by differences in number (4, 8, 9, and 7, respectively) of direct 15-bp repeats (boxed).
Intragenomic Deletion Frequencies for H. pylori. The direct repeats identified in H. pylori and found to be significant (13, 14) ranged from 24 to 4,051 bp (Table 3). To determine whether repeat length affects recombination frequency, H. pylori mutants were constructed with a uniform cassette flanked by direct repeats of 5-100 bp of the aphA (kanamycin resistance) ORF (Fig. 5). We hypothesized that recombination between the repeat sequences would lead to deletion of the cassette and one copy of the direct repeat, restoring an uninterrupted aphA, that could be selected on the basis of kanamycin resistance. This deletion cassette was inserted into the vacA locus of five H. pylori strains because vacA encodes a secreted toxin important in virulence (29), with no evidence of involvement in H. pylori metabolism, recombination, or repair. For strain JP26, in the absence of flanking repeat sequences (Table 1) or with 5-bp repeats (data not shown), no deletions were observed; however, deletion was observed in the mutant with 15-bp flanking direct repeats (deletion frequency = 1.1 × 10-8 ± 0.3 × 10-8). H. pylori strains with longer (50 or 100 bp) repeat sequences showed a progressive increase in deletion frequency (Table 1): >10-fold frequency differences among the strains' indicated strain specificity. To determine whether frequencies observed were due to insertion in vacA, in parallel experiments, the identical deletion cassette was inserted into the uvrB locus (19) or ureA (12) locus, which are 237 kb downstream and 860 kb upstream of vacA, respectively, in strain 26695 (12). As expected in the absence of flanking repeat sequences, no deletions were observed (Table 1). For mutants with 50- or 100-bp repeat sequences, the same patterns were observed for the vacA, uvrB, and vacA mutants, suggesting that, within a particular strain, intragenomic deletions are not affected by genome location. Furthermore, the uvrB-mutant results indicate that the H. pylori nucleotide excision repair (NER) pathway (22) is not involved in intragenomic recombination.
Table 1. Detection of a deletion between paired identical direct repeats from the H. pylori chromosome, according to host strain, gene locus of cassette, and repeat size.
|
H. pylori strain
|
Locus of deletion cassette*
|
Deletion frequency (× 10−6)†
|
||
|---|---|---|---|---|
| 0-bp | 50-bp | 100-bp | ||
| J166 | vacA | <0.01 | 2.4 ± 1.7 | 9.0 ± 0.9 |
| B128 | vacA | <0.01 | 1.8 ± 0.9 | 12.0 ± 6.5 |
| 9999 | vacA | <0.01 | 2.6 ± 1.1 | 10.9 ± 3.4 |
| 9886 | vacA | <0.01 | 0.6 ± 0.1 | 3.6 ± 1.7 |
| JP26 | vacA | <0.01 | 0.4 ± 0.3 | 1.1 ± 0.7 |
| JP26 | uvrB | <0.01 | 0.4 ± 0.3 | 1.0 ± 0.3 |
| JP26 | ureA | <0.01 | 1.6 ± 1.1 | 4.6 ± 1.3 |
| JP26 | recA | <0.01 | 0.7 ± 0.3 | 0.7 ± 0.6 |
See Fig. 5 for schematic of the deletion cassette.
Cassettes constructed in the identical manner except for size of repeat sequences present.
To examine whether the correlation between repeat length and deletion frequency was specific for H. pylori, parallel experiments were performed in E. coli strain RR1 transformed with the pAT40-100 plasmid series. Whereas deletions were not observed in the absence of repeat sequences or with 5 bp repeats, frequencies increased progressively from ≈10-9 to 10-6 deletions per total cfu as repeat length increased from 15 to 100 bp (data not shown). Although the E. coli studies used multicopy plasmids, and, H. pylori a single chromosomal locus, the parallel findings indicate conserved recombination mechanisms that are repeat sequence length-specific.
Because H. pylori cells are naturally competent (30), we asked whether the observed recombination was affected by horizontal transfer of deleted genotypes from subpopulations in which deletion already has occurred. In experiments conducted in the presence or absence of DNase, respectively, for JP26 vacA::IDS50 (2.3 × 10-6 ± 2.4 × 10-6 and 4.3 × 10-6 ± 5.0 × 10-6) and JP26 vacA::IDS100 (10.1 × 10-6 ± 7.9 × 10-6 and 17.7 × 10-6 ± 14.4 × 10-6), there were no significant differences in deletion frequencies. This finding confirms that the observed deletions are not mediated by intergenomic recombination.
RecA-Independence of H. pylori Intragenomic Recombination. For E. coli, as repeat sequence and intervening sequence lengths increase, intragenomic recombination becomes increasingly RecA-dependent (31). To examine RecA-dependence of the deletions in our experimental system, deletion frequencies were compared in RecA- E. coli strain, HB101 and its RecA+ counterpart, RR1 (20). For repeat sequences <100 bp, frequencies in the two strains were similar, but for 100-bp repeat sequences, significantly (P < 0.05) fewer deletions were observed in strain HB101 (3.7 × 10-7 ± 2.6 × 10-7) than in strain RR1 (2.7 × 10-6 ± 1.2 × 10-6), confirming previous reports (31). To determine whether the observed H. pylori deletions are dependent on RecA function, recA was inactivated in strain JP26 by insertion of the cassette (containing either 0-, 50-, or 100-bp IDS) into recA. Deletion frequencies in the JP26 recA::IDS50 and recA::IDS100 mutants did not significantly differ from deletion frequencies in parallel JP26 strains with RecA activity (Table 1). These results indicate that intragenomic recombination in H. pylori is RecA-independent for repeats ≤100 bp.
Restoration of Deleted DNA Segment Through Natural Transformation. For naturally competent organisms such as H. pylori, chromosomal DNA segments lost through deletion can be reacquired through natural transformation (17, 32). To examine whether the segment spontaneously deleted between the IDS segments could be restored via transformation, DNA from the parental CmRKms strains, JP26 vacA::IDS50 and JP26 vacA::IDS100, containing the full cassette (Fig. 5) was used to transform mutants JP26-50DM and JP26-100DM that had been selected for deletion of the intervening cat cassette. As expected, no CmR transformants were observed when no DNA was added or with donor DNA from wild-type strain JP26. However, incubation with donor DNA from JP26 vacA::IDS50 or JP26 vacA::IDS100 yielded transformants (Table 6, which is published as supporting information on the PNAS web site). That all CmR transformants also were Kms indicates that the cat cassette was restored to its engineered locus, and PCR studies confirmed these findings (data not shown). In total, these results indicate that within an H. pylori population, cells that lose chromosomal DNA segments through intragenomic deletions can have restoration of the lost segment through transformation, and suggest that the relative prevalence of the two genotypes will depend on the extant selective pressures.
Discussion
Colonic bacteria, such as E. coli, sharing niches with numerous other species, are subject to interspecies horizontal DNA transfer, facilitating adaptation to environmental stresses. How can H. pylori, essentially the only organisms that persistently colonize the highly acidic human stomach (3), diversify in the absence of cohabiting species with which to exchange DNA? H. pylori strains often have hypermutator phenotypes (10), allowing for high endogenous mutation rates. Because multiple H. pylori strains may cocolonize the stomach (11), their natural competence permits intergenomic recombination, although this is limited (32, 33) by the heterogeneity of restriction endonucleases present among H. pylori strains (34). Through in silico identification of potential recombination hotspots, experimental confirmation that these sites facilitate genome plasticity, and quantitation of deletion frequencies between repeat sequences of varying sizes in varying locations, we provide evidence that the nonrandom chromosomal distribution of repetitive DNA may have as important an influence on genomic variation as point mutation or transformation.
Comparison of five wild-type H. pylori strains indicate up to 10-fold differences in intragenomic deletion frequencies (Table 1). Because spontaneous mutation (10) and natural transformation (32, 33) rates also vary among H. pylori strains, our findings indicate that H. pylori strains differ in ability to diversify by all three important mechanisms. Current investigation is focused on defining the loci in H. pylori strains that influence plasticity, whether as separate systems or via a global “plasticity clock.” The presence of repetitive DNA within H. pylori virulence-associated loci (35-37), and identification in individual hosts of variants resulting from recombination between repeat sequences (17, 28) indicate the importance of this mechanism. Although the present studies examined identical H. pylori repeats, imperfect repeats also may be subject to recombination (17), further facilitating genomic plasticity. That intragenomic deletion events involving the 100-bp IDS in H. pylori are RecA-independent suggests that H. pylori may have different mechanisms than E. coli for intragenomic recombination between repeat sequences; mismatch repair systems present in E. coli (38), but not H. pylori (12), may affect recombination frequency. Alternatively, because the E. coli studies were performed on plasmids (31), whereas the present work examined chromosomal recombination, the specific mechanisms involved may differ.
In-depth in silico analyses provided evidence that, for organisms that are obligately host-associated, intragenomic recombination between direct genomic repeats can modulate host responses. For example, within an individual host, intragenomic recombination of DNA repeats alters the number of phosphorylation sites in the cagA product, a type IV secretion system substrate injected into host cells where it undergoes tyrosine phosphorylation (39, 40). Through deletion or duplication of the cagA 3′ region-encoded tyrosine phosphorylation sites, H. pylori can differentially transduce signals in host cells (28), indicating that gene amplification modulates microbial signals and host responses.
We demonstrate that elements lost through recombination also can be regained, providing flexibility for microbes, such as H. pylori, that have no cocolonizing organisms but are naturally competent. Our data suggest that H. pylori is representative of small genome organisms in which recombination using direct DNA repeats is a paradigm for programmed regulation of gene content. Although repetitive DNA could lead to virtually unlimited expansion of intervening segments, environmental constraints select for the range of units at particular loci that have greatest fitness in the lifestyle contexts of a particular organism. In M. tuberculosis, which also occupies an ecological niche without cohabitants, potential recombination hotspots (Fig. 3) are loci of substantial interstrain variation (23, 24), supporting the hypothesis that nonrandomly distributed repetitive DNA promotes prokaryotic plasticity, regardless of genomic size. The extensive number of variants made possible by the repeat structure represented by the “beehives flanking diamonds” schema (Fig. 3) indicates the powerful selective forces at work; the critical role of this particular locus in host adaptation (41) further supports our thesis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01GM63270 and by the Medical Research Service of the Department of Veterans Affairs.
Abbreviations: Mb, mebabase(s); IDS, identical DNA sequences.
References
- 1.Dobrindt, U. & Hacker, J. (2001) Curr. Opin. Microbiol. 4, 550-557. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, Y., Nesbo, C. L. & Doolittle, W. F. (2001) Curr. Opin. Microbiol. 4, 285-289. [DOI] [PubMed] [Google Scholar]
- 3.Peek, R. M., Jr., & Blaser, M. J. (2002) Nat. Rev. Cancer 2, 28-37. [DOI] [PubMed] [Google Scholar]
- 4.Li, C., Ha, T., Chi, D. S., Ferguson, D. A., Jr., Jiang, C., Laffan, J. J. & Thomas, E. (1997) J. Clin. Microbiol. 35, 3021-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Israel, D. A. Salama, N., Krishna, U., Rieger, U. M., Atherton, J. C., Falkow, S. & Peek, R. M., Jr. (2001) Proc. Natl. Acad. Sci. USA 98, 14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuipers, E., Israel, D. A., Kusters, J. G., Gerrits, M. M., Weel, J., van Der Ende, A., van Der Hulst, R. W., Wirth, H. P., Hook-Nikanne, J., Thompson, S. A. & Blaser, M. J. (2000) J. Infect. Dis. 181, 273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Ende, A., Rauws, E. A., Feller, M., Mulder, C. J., Tytgat, G. N. & Dankert, J. (1996) Gastroenterology 111, 638-647. [DOI] [PubMed] [Google Scholar]
- 8.Webb, G. F. & Blaser, M. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3135-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodenaw, M., Huet, T., Saurin, W., Kwok, S., Sninsky, J. & Wain-Hobson, S. (1989) J. Acquired Immune Defic. Syndr. 2, 344-352. [PubMed] [Google Scholar]
- 10.Bjorkholm, B. Sjolund, M., Falk, P. G., Berg, O. G., Engstrand, L. & Andersson, D. I. (2001) Proc. Natl. Acad. Sci. USA 98, 14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kersulyte, D., Chalkauskas, H. & Berg, D. E. (1999) Mol. Microbiol. 31, 31-43. [DOI] [PubMed] [Google Scholar]
- 12.Tomb, J. F. White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539-547. [DOI] [PubMed] [Google Scholar]
- 13.Rocha, E. P. C., Danchin, A. & Viari, A. (1999) Mol. Biol. Evol. 16, 1219-1230. [DOI] [PubMed] [Google Scholar]
- 14.Achaz, G., Rocha, E. P., Netter, P. & Coissac, E. (2002) Nucleic Acids Res. 30, 2987-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chedin, F., Dervyn, E., Dervyn, R., Ehrlich, S. D. & Noirot, P. (1994) Mol. Microbiol. 12, 561-569. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz, S. & Schleiermacher, C. (1999) Bioinformatics 15, 426-427. [DOI] [PubMed] [Google Scholar]
- 17.Aras, R. A., Takata, T., Ando, T., van der Ende, A. & Blaser, M. J. (2001) Mol. Microbiol. 42, 369-382. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, S. A. & Blaser, M. J. (1995) Infect. Immun. 63, 2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, S. A., Latch, R. L. & Blaser, M. J. (1998) Gene 209, 113-122. [DOI] [PubMed] [Google Scholar]
- 20.Miyamoto, C., Byers, D., Graham, A. F. & Meighen, E. A. (1987) J. Bacteriol. 169, 247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorell, N., Mangan, J. A., Laing, K. G., Hinds, J., Linton, D., Al-Ghusein, H., Barrell, B. G., Parkhill, J., Stoker, N. G., Karlyshev, A. V., et al. (2001) Genome Res. 11, 1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, K. K., Zhang, X., Patibandla, A. S., Chien, P., Jr., & Laal, S. (2001) Infect. Immun. 69, 4185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen, D. de Haas, P. E., Hermans, P. W., Groenen, P. M. & van Embden, J. D. (1993) J. Clin. Microbiol. 31, 1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versalovic, J., Koeuth, T. & Lupski, J. R. (1991) Nucleic Acids Res. 19, 6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alm, R. A., Ling, L. S., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan, B., Guild, B. C., deJonge, B. L., et al. (1999) Nature 397, 176-180. [DOI] [PubMed] [Google Scholar]
- 26.Dubois, A., Berg, D. E., Incecik, E. T., Fiala, N., Heman-Ackah, L. M., Del Valle, J., Yang, M., Wirth, H. P., Perez-Perez, G. I. & Blaser, M. J. (1999) Gastroenterology 116, 90-96. [DOI] [PubMed] [Google Scholar]
- 27.Aras, R. A., Fischer, W., Perez-Perez, G. I., Crosatti, M., Ando, T., Haas, R. & Blaser, M. J. (2003) J. Exp. Med., in press. [DOI] [PMC free article] [PubMed]
- 28.Aras, R. A., Lee, Y., Kim, S. K., Israel, D., Peek, R. M., Jr., & Blaser, M. J. (2003) J. Infect. Dis. 188, 486-496. [DOI] [PubMed] [Google Scholar]
- 29.Ricci, V., Ciacci, C., Zarrilli, R., Sommi, P., Tummuru, M. K., Del Vecchio Blanco, C., Bruni, C. B., Cover, T. L., Blaser, M. J. & Romano, M. (1996) Infect. Immun. 64, 2829-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nedenskov-Sorensen, P., Bukholm, G. & Bovre, K. (1990) J. Infect. Dis. 161, 365-366. [DOI] [PubMed] [Google Scholar]
- 31.Bi, X. & Liu, L. F. (1994) J. Mol. Biol. 235, 414-423. [DOI] [PubMed] [Google Scholar]
- 32.Aras, R. A., Small, A. J., Ando, T. & Blaser, M. J. (2002) Nucleic Acids Res. 30, 5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ando, T., Xu, Q., Torres, M., Kusugami, K., Israel, D. A. & Blaser, M. J. (2000) Mol. Microbiol. 37, 1052-1065. [DOI] [PubMed] [Google Scholar]
- 34.Xu, Q., Morgan, R. D., Roberts, R. J. & Blaser, M. J. (2000) Proc. Natl. Acad. Sci. USA 97, 9671-9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. S. (1996) Proc. Natl. Acad. Sci. USA 93, 4648-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito, Y., Azuma, T., Ito, S., Suto, H., Miyaji, H., Yamazaki, Y., Kato, T., Kohli, Y., Keida, Y. & Kuriyama, M. (2000) J. Clin. Microbiol. 38, 483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaoka, Y., Kodama, T., Kashima, K., Graham, D. Y. & Sepulveda, A. R. (1998) J. Clin. Microbiol. 36, 2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radman, M. & Wagner, R. (1984) Curr. Top. Microbiol. Immunol. 108, 23-28. [DOI] [PubMed] [Google Scholar]
- 39.Asahi, M., Azuma, T., Ito, S., Ito, Y., Suto, H., Nagai, Y., Tsubokawa, M., Tohyama, Y., Maeda, S., Omata, M., Suzuki, T. & Sasakawa, C. (2000) J. Exp. Med. 191, 592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Backert, S., Moese, S., Selbach, M., Brinkmann, V. & Meyer, T. F. (2001) Mol. Microbiol. 42, 631-644. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishnan, L., Federspiel, N. A. & Falkow, S. (2000) Science 288, 1436-1439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.