Abstract
Defining the transcriptome, the repertoire of transcribed regions encoded in the genome, is a challenging experimental task. Current approaches, relying on sequencing of ESTs or cDNA libraries, are expensive and labor-intensive. Here, we present a general approach for ab initio discovery of the complete transcriptome of the budding yeast, based only on the unannotated genome sequence and millions of short reads from a single massively parallel sequencing run. Using novel algorithms, we automatically construct a highly accurate transcript catalog. Our approach automatically and fully defines 86% of the genes expressed under the given conditions, and discovers 160 previously undescribed transcription units of 250 bp or longer. It correctly demarcates the 5′ and 3′ UTR boundaries of 86 and 77% of expressed genes, respectively. The method further identifies 83% of known splice junctions in expressed genes, and discovers 25 previously uncharacterized introns, including 2 cases of condition-dependent intron retention. Our framework is applicable to poorly understood organisms, and can lead to greater understanding of the transcribed elements in an explored genome.
Keywords: computational biology, RNAseq, next generation sequencing, transcriptome profiling, Saccharomyces cerevisiae
Experimentally defining the complete transcriptome of eukaryotic organisms has traditionally been a challenging task, involving large, costly, and slow experimental efforts for sequencing of ESTs and full-length cDNA libraries. Unlike the genome, RNA transcripts are not present at equimolar concentrations, and are typically expressed in a context-specific manner. Thus, despite the fact that the genomes of >1,000 species have been sequenced, only few transcriptomes have been extensively characterized.
Recent advances in massively parallel sequencing technology (1, 2) offer new and powerful approaches to the study of transcriptomes. Recent studies (3–7) have shown that, by sequencing the mRNA content of cells, one can quantify the expression levels of known genes (by counting how often sequences from a given gene are observed) and refine their boundaries. For example, Nagalakshmi et al. (3) studied the Saccharomyces cerevisiae transcriptome by mapping reads to the location of known genes to quantify expression, and to known splice sites to measure their occurrence. Similarly, Mortazavi et al. (5) studied the mouse transcriptome by mapping reads to known exons and known splice junctions, as well as to “putative” junctions between known exons. Thus, in both cases (and in additional studies, see refs. 4–7) the analysis critically depended on existing annotation.
A more challenging problem is to define a transcriptome ab initio, based only on the unannotated genome sequence and millions of short reads from cDNA samples. Rapid and efficient methods to do so would transform our ability to define transcripts and study transcription in any genome. This ability would be particularly important in a new genome project involving phylogenetically isolated species and in cancer genome projects, where the genome annotation may fail to reflect pathological aberrations. The full goal would include: (i) identification of all regions encoding transcripts (coding and noncoding RNAs) in a given condition or cell type; (ii) demarcation of the 5′- and 3′- ends of transcripts; (iii) determination of splice junctions and identification of different splice variants; and (iv) identification of posttranscriptional transcript editing.
Here, we present a general approach to accomplish all of these goals, based solely on an unannotated genome sequence and data from a single sequencing run on an Illumina sequencer (2). To test our approach, we apply it to the budding yeast S. cerevisiae, and compare our ab initio results to the known transcript annotation (8). Our approach automatically and fully defines 86% of the genes expressed under the given conditions, and discovers 160 previously undescribed transcription units of 250 bp or longer. The approach correctly demarcates the correct 5′ and 3′ UTR boundaries of 86 and 77% of expressed genes, respectively. The method identifies 83% of known splice junctions in expressed genes, and discovers 25 previously uncharacterized introns, including evidence for 2 rare cases of condition-dependent “alternative splicing.” Last, we use the data to quantify absolute and relative expression levels of each transcript, showing remarkable agreement with well-established microarray technologies.
Our results demonstrate that massive, cost-efficient, and fast sequencing can be used to accurately define and quantify a transcriptome ab initio. To evaluate the strength of our approach, we have refrained from using other sets of data and gene predictions methods. However, in many practical cases, these methods can be incorporated into a single bioinformatics pipeline for a more powerful outcome. This framework can be readily applied to study poorly understood organisms, for which only the genomic sequence is known.
Results
Sequencing the Budding Yeast Transcriptome.
To define the budding yeast transcriptome ab initio, we generated cDNA libraries from poly(A)+ mRNA from the budding yeast S. cerevisiae under 2 growth conditions: in rich medium (YPD) and after heat shock (HS). We used a cDNA preparation procedure that combines a random priming step with a shearing step (see Materials and Methods). This approach has 2 benefits, which are essential for ab initio predictions. First, unlike other methods that provide a signal only in the 5′ or the 3′ end of transcripts, our method results in signal that covers the whole transcript (Fig. 1A). Second, for sequencing with short reads, random priming alone results in extensive nonuniformity in the start sites (9), whereas we obtain better uniformity.
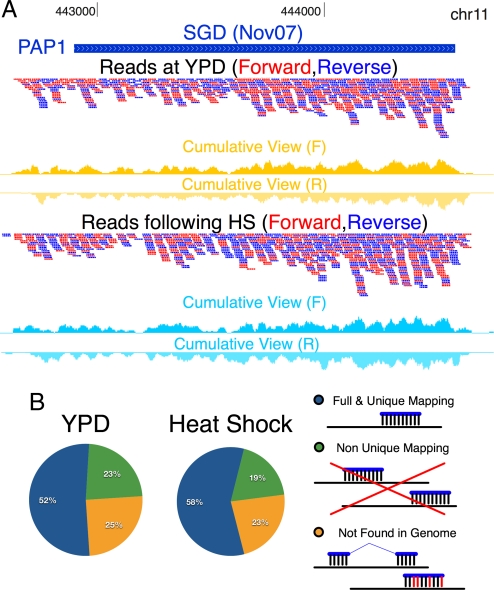
Fig. 1.
Unbiased sequencing of the yeast transcriptome. (A) Distribution of reads mapped to the PAP1 locus. Shown are SGD annotations (downloaded at November 2007) (8), and mapped reads (red, W strand; blue, C strand). Additional tracks plot the cumulative number of reads covering each base position (yellow, YPD; light blue, HS). Full data can be accessed at http://compbio.cs.huji.ac.il/RNASeq, and is visualized using the University of California, Santa Cruz, genome browser (22). (B) Distribution of reads matched to the genome. Of the 26,050,414 reads sequenced in YPD (Left), 13,424,957 (52%, blue) were uniquely mapped to a single genomic locus, 6,144,595 (23%, green) were mapped to several locations, and 6,480,862 (25%, yellow) could not have been aligned, and were later used to detect splice junctions. Similar numbers were found after a HS (Right).
We sequenced each library using an Illumina 1G Analyzer to generate 36-bp long reads. We obtained 25,043,976 reads from the YPD sample (2 biological replicates) and 11,776,251 reads after HS (see Materials and Methods). The entire experiment (RNA extraction, library preparation, and sequencing) required <14 workdays.
Then, we developed an accurate method to map reads to their genomic locations. The sequence matching approach used in previous studies (3, 4) may fail due to errors in the sequencing process or repetitive genomic regions (as a result of low-complexity or homology). Therefore, we developed a detailed probabilistic error model that scores the genomic matches of reads according to the position-specific probability of sequencing errors [see Materials and Methods; also, supporting information (SI) Fig. S1 and Dataset S1]. To minimize mapping errors, a read should match a specific genomic sequence at a strict threshold and should not match any other genomic location, even at a more relaxed threshold (see Materials and Methods). Applying this strategy to our data, we uniquely mapped 52% of the reads in YPD. We discarded an additional 23% of the reads that mapped to >1 genomic locus; this proportion is consistent with expectations due to genomic repeats (25.5% for 36-bp reads based on simulation). The remaining 25% reads did not map to any genomic locus at the required stringency (Fig. 1B). A minority is due to posttranscriptional modifications, such as splicing (see below). We obtained similar results with the reads in the HS experiment (Fig. 1B).
Ab Initio Construction of a Transcript Catalog for S. cerevisiae.
We next developed a procedure to ab initio define all of the transcriptional units expressed under the 2 conditions, using only the mapped cDNA reads and the (unannotated) genome sequence of S. cerevisiae (Fig. 2A). Based on the current annotation of the yeast genome and microarray-based expression studies (8, 10), we expect 4,630 known genes to be expressed in YPD (at >0.2 transcripts per cell; see Materials and Methods). We started by identifying contiguous regions with a density of cDNA reads above a given threshold. Because genes are densely packed in the S. cerevisiae genome, such regions can span several genes. Thus, we developed a procedure that breaks these regions into segments of consistent read density, reflecting the expectation that transcript levels should be much more consistent within genes, than between genes (see Materials and Methods and Fig. 2B; also, Fig. S2). Last, we predicted transcription orientation based on different read densities between ends of genes (even in our relatively uniform libraries, there is a higher read density toward the 3′ end, which may be due to the library preparation protocol; see Materials and Methods and Fig. 2B). In total, we identified 6,248 segments, demarcating putative transcribed regions.
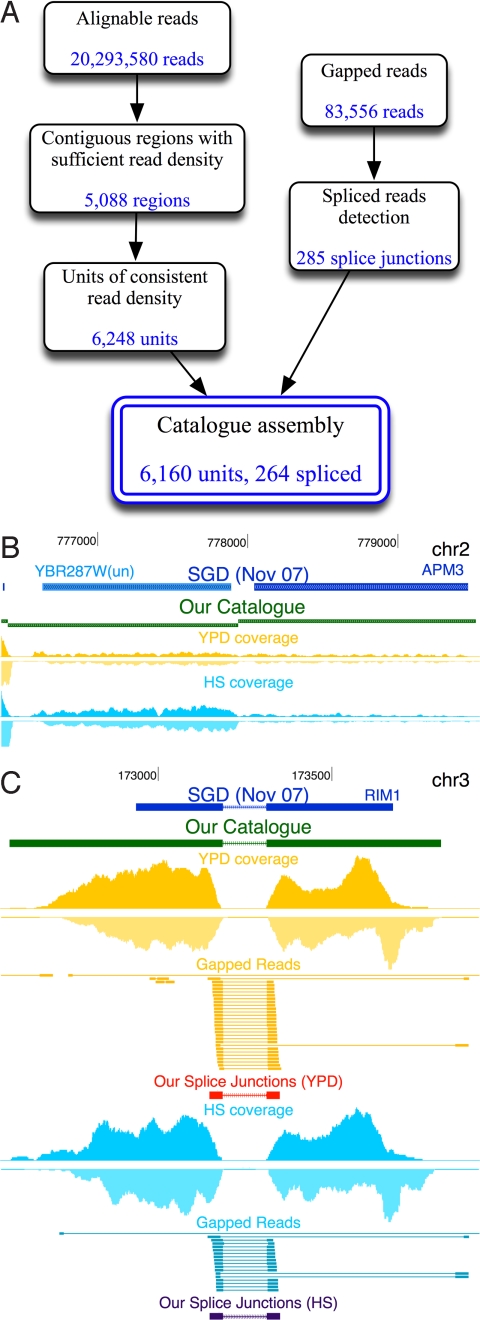
Fig. 2.
Ab initio assembly of a transcript catalog. (A) Outline of steps in the catalog construction pipeline. (B) Segmentation of a contiguously transcribed region into 2 regions of distinct expression levels corresponding to the genes YBR287W and APM3. When using YPD reads alone, both genes exhibit similar coverage and thus cannot be segmented. However, in HS, they are differentially expressed, and hence by combining observations from both conditions the automatic segmentation procedure (see Materials and Methods) correctly separates them to 2 units. Tracks from top to bottom: SGD annotations (blue), our catalog (green), read coverage at YPD (yellow), and read coverage at HS (blue). (C) Detection of splice junctions. Full and gapped reads mapped to the RIM1 genomic locus. Tracks are as in B, together with gapped reads (connected segments), our putative splice junctions (in red and blue), including the junction orientations as estimated by donor and acceptor sequence motifs (arrows). As shown, our procedure identifies the exact coordinates and orientation of the known splice site.
Before assembling a gene catalog, we next searched for splicing events. We analyzed the 25% of reads (9,212,859) that did not match the genome to identify those that may originate from splicing events. In such events, sequences from 2 exons that are separated in the genomic sequence are adjacent in the mature mRNA, yielding reads with a “gapped alignment” (Fig. 2C).
We developed an automatic method to systematically discover splice junctions. First, we identified reads with a gapped alignment, involving 2 sites of at least 10 bp each separated by at most 2 Kb (and together adding up to 36 bp). We required the same noise thresholds as before to filter out mismatches and nonunique matches (see Materials and Methods). Because we allow only a single gap, the probability of finding a spurious match is extremely low, although the precise gap location might be ambiguous by 1 or 2 base pairs, depending on the exact sequence at the gap boundaries. To eliminate spurious events, we required splice junctions to be supported by multiple observations. Specifically, we included all putative junctions that were either (i) supported by at least 5 independent reads (possibly starting at different locations; 243 junctions); (ii) supported by at least 3 independent reads and contain donor (5′) and acceptor (3′) splice site motifs (263 junctions; see http://compbio.cs.huji.ac.il/RNASeq); or (iii) supported by 2 independent reads and contain very strong splice motifs (13 junctions). This scoring allows us to resolve ambiguities, increase confidence in gapped reads, and assign an orientation to the junction (see Materials and Methods and Fig. 2C). The remaining putative junctions had little support and were discarded. In particular, shorter junctions are likely due to short deletions in the genomic DNA of the particular strain, consistent with Illumina sequencing of the DNA of this specific strain (data not shown). The resulting set had 285 junctions of 40 bp or longer. Notably, the majority of these junctions (243/285) were identified by the first criterion (5 strong junctions lack canonical splice site signals altogether; see http://compbio.cs.huji.ac.il/RNASeq), demonstrating the power of ab initio detection.
Joining the putative transcribed units based on the splice junctions, we built a final catalog of the yeast transcriptome in the 2 measured conditions (Fig. 2A; Dataset S2). This catalog includes 6,160 transcripts, 264 of them with at least 1 splicing junction.
Assessment of Transcription Units of the Catalog.
We compared our ab initio catalog of transcripts with the current annotated transcriptional catalog of the yeast genome. Approaches based on sequencing of mRNAs cannot discover genes that are not expressed. Also, because we rely on short reads, we are limited to identifying transcripts in alignable (nonrepetitive) genomic regions. By using conservative thresholds, there are 5,437 (94%) known genes (classified as “verified” or “uncharacterized” ORF genes; see ref. 8) in the yeast genome that are “alignable” (at 50% coverage or more) with 36-bp reads, of which 4,784 are expressed in YPD (see Materials and Methods).
Overall, the ab initio transcriptional units in our catalog cover 99% of these expressed genes over >80% of the length of genes (Fig. 3A). For 86% of the genes, the transcriptional units fully cover the known genes (4096/4784; see Fig. 3A). For the remaining 13% of genes, the genes are largely covered, but correspond to multiple transcriptional units that have not been confidently connected (due to gaps or unevenness in coverage, particularly for highly expressed genes); this problem should be largely eliminated by connecting transcribing units through the use of “paired-end” reads, which are now becoming routinely available on the Illumina platform (11). Last, we correctly assigned orientation to 3,432 genes (84%), based solely on the pattern of increasing read density from 5′ to 3′-end. Overall, these results demonstrate that we can reconstruct the compendium of transcripts with great sensitivity and specificity.
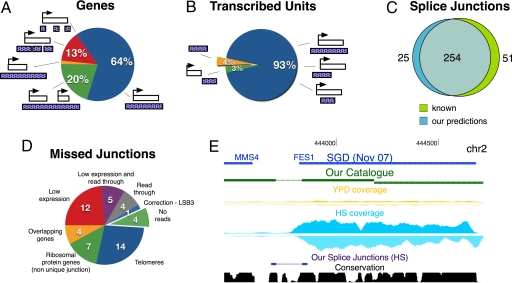
Fig. 3.
Validation of the transcript catalog. (A) Coverage of the top 86% expressed genes by our predicted transcribed units, based on different patterns of coverage. (B) Relationship between found transcribed units and annotated transcribed features from SGD. In both A and B, white boxes denote genes, and purple boxes denote transcribed units. (C) Comparison of our putative splice junctions (blue) to known ones (green). (D) The 51 known introns missed by our predictions are partitioned into 8 categories. (E) Validation of splicing read-through in the gene FES1. Tracks are as in Fig. 2C, including the evolutionary conservation of each position across 7 yeast species (15).
Notably, our analysis indicates transcription from some “dubious ORFs” loci (62 of 206 expressed alignable dubious ORFs that do not overlap any other gene). In comparison, only 1% of nontranscribed loci based on ultradense tiling arrays (12) are covered by transcription units in YPD. This observation suggests that these are less likely to be spurious transcription events, and that some of these loci encode for functional transcripts (possibly noncoding RNAs).
The transcripts in our catalog assign the correct gene structure in terms of boundaries (and splicing; see below). Notably, because RNA-sequencing only samples short reads from transcripts, it has limited ability to accurately determine transcript boundaries in a highly compact genome (as compared with 5′ sequencing methods). Nevertheless, our transcript boundaries reasonably match several previous annotations of transcript boundaries in S. cerevisiae. These include the known annotations (SGD) as well as start site definitions based on previous full-length cDNA sequencing (13) and ultradense tiling arrays (12). In particular, our 5′ UTR positions match 80% of previous definitions within 50 bp, but have limited agreement in higher resolution [47% with Miura et al. (13); 22% with David et al. (12) in 10-bp resolution]. This latter result may be because our protocol likely misses 8–21 nt at the 5′ end of the transcript (14). Notably, we correctly predict the 3′ boundaries of 307 of 501 (60%) pairs of converging genes, and miss the boundary by at most 50 bp for an additional 58 cases (11%). Differential expression is a major contributor to correct detection. For correctly predicted pairs, the mean differential expression ratio is 8.5, whereas for those pairs that we cannot correctly differentiate, the mean differential expression ratio is 2.9. By considering the predicted ORFs within our transcripts, we estimate the typical lengths of 5′ and 3′ UTRs as 153 bp (SD of 145 bp), and 169 bp (SD of 142 bp), respectively (see http://compbio.cs.huji.ac.il/RNASeq; also, Dataset S3).
To our surprise, although 93% of our catalog corresponds to known genes (Fig. 3B; Dataset S2), we also discovered 160 transcription units of length ≥250 bp that did not overlap any previously annotated transcripts (Dataset S2; see ref. 8). Many of these units are clearly transcribed, for example, a ≈3,694-bp region at Chromosome 1, coordinates 196277–199970, that we also validated experimentally (see below). Many of these transcripts have supporting evidence in the raw data from hybridization to tiling arrays (129 units overlap; see ref. 12) and cDNA sequencing (92 units overlap; see ref. 13); although these previous studies did not report them as transcriptional units per se. Some of the units are differentially expressed between YPD and HS (Dataset S2). Most notably, 12/160 novel units have are induced 10-fold or higher in HS vs. YPD, and 2 of those are not detected at all in YPD.
Overall, the previously undescribed units are mostly short (mean length of 713 bp, SD of 431 bp), and many are likely not coding for a protein. Several lines of evidence support this conclusion. First, the predicted ORFs are usually short (mean predicted ORF of 51 aa, SD of 19 aa, 20 units >80 aa; see Dataset S2), and do not match predicted or known proteins in other fungal species. Second, when sampling regions of the same length at random from intergenic regions, the median length of predicted ORFs is 146 aa, in contrast to the much shorter median length of predicted ORFs in these transcription units (48 aa). Last, relatively few of the units are evolutionary conserved (28/160 units >50% conservation; see ref. 15), which is not significant when compared with random (P = 0.059).
We experimentally tested and verified 4 of these novel transcripts by RT-PCR followed by sequencing. These included: (i) the novel ≈3,694-bp transcript discussed above (Chromosome 1, 196277–199970; see Fig. S3A); (ii) a transcribed pseudogene at Chromosome 15, coordinates 36742–38650 (Fig. S3B); (iii) a novel transcription unit at the YMR194C locus that spans both a dubious ORF (YMR194C-B) and the gene YMR194C-A (Fig. S3C); and (iv) a predicted 900-bp 3′ UTR for the FEN2 gene. In the latter 2 cases, the novel transcriptional units overlap, expand, or modify dubious ORFs or pseudogenes. For example, the novel transcription unit at the YMR194C locus also includes a 200-bp 3′ UTR past the predicted stop codon of YMR194C-A, suggesting a recent pseudogene.
Validation of Splice Junctions.
Our splice site predictions are also highly accurate and sensitive, as compared with the known annotated junctions. The 285 ab initio detected splice junctions include most of the annotated junctions in the yeast genome (Fig. 3C; Dataset S2). We predict 254 (83%) out of 305 known junctions within 5-bp resolution. Of the 51 missed junctions, 21 are in non unique “unalignable” regions (telomeres and ribosomal protein genes), and 21 have very low read coverage (Fig. 3D). From the remaining 9 cases, we see read-through transcription in 4 undetected junctions, whose introns are matched by a significant number of reads (see http://compbio.cs.huji.ac.il/RNASeq), and determine a corrected location for 1 junction (LSB3 gene; see below). Thus, in only 4 of the 51 cases, we do not detect spliced reads for unknown reasons.
We also discovered 25 previously uncharacterized splice junctions that are not close to any annotated ones (one is an “artifact” caused by the HIS3 deletion in this strain). To study the implications of these splice junctions, we examined their effect on transcript structure. We found that 11 of the putative junctions are within annotated coding regions and affect the encoded protein, either by modifying existing introns, or by introducing additional ones (Dataset S3). For example, in the LSB3 gene, our putative intron is 24-bp shorter than the known one, adding 8 aa to the translated protein. When compared with other yeast species, the 8-aa stretch shows clear evolutionary conservation in the orthologous proteins (Fig. S4; see ref. 16); thus, it appears to be a conserved part of the protein.
In 6 of these junctions, we see evidence for alternative splicing (intron retention), because 3 junctions appear only in YPD and 3 only in HS (while taking into consideration the number of full reads aligned in both conditions; see http://compbio.cs.huji.ac.il/RNASeq). For example, in the MRM2 gene, the discovered intron is spliced out only in YPD; thus, creating a shorter protein, which perfectly aligns with orthologs of this gene in Kluyveromyces lactis, Candida lusitaniae, Debopriya hansenii, Candida guilleromondi, Candida tropicalis, and Candida albicans. In C. albicans, for example, the intronic sequence is completely missing from the genome, strongly supporting the functionality of this spliced form. Similarly, in the APE2 gene, the HS intron is slightly shorter, which creates a protein that is 6-aa shorter than the regular one. This modified protein has a domain that fits orthologs of this gene in Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus.
We experimentally tested 6 predicted splicing events and validated 4 of them (in the genes FES1, YMR148W, RPS22B, and AGA2) using RT-PCR and sequencing (Fig. 3E; Fig. S5). For example, in the FES1 gene, our catalog identified a previously uncharacterized intron with full reads through the splice junction and inside the intron, suggesting alternative splicing (Fig. 3E). In the spliced variant, the annotated stop codon is abolished and a later stop codon is introduced, resulting in a 10-aa extension. Validation by RT-PCR shows bands consistent with both the spliced and unspliced variants (sequencing of these bands confirmed the splice site). Another example of alternative splicing is the SUS1 gene, where, in addition to the 2 known introns, we also observe clear read-through at both junctions (Fig. S5A). Experimental validation confirms our predictions by revealing 3 bands, 2 bands consistent with just 1 intron spliced, and a stronger band consistent with both introns spliced out. A third example is an intron from the end of the snoRNA, SNR44, to the acceptor site of its hosting intron, inside RPS22B (Fig. S5B). All experimental validations were performed by RT-PCR followed by sequencing of the bands to verify the exact splice site. The predicted splice junctions that we could not validate may be in low-abundance or represent partial splicing.
Inferring Expression from Massively Parallel Sequencing.
Having defined a gene catalog, we then examined the ability to infer quantitative expression levels from sequence abundance. We estimated the mRNA abundance of known annotated ORFs by calculating the average density of reads along each ORF and compared the results with expression data from microarrays. We converted the read densities per gene to rough assessments of absolute mRNA copy numbers per cell, using a conservative estimation of 15,000 transcripts per yeast cell (17). This analysis reveals at least 4 orders of magnitude differences in mRNA copy number among genes. For example, we find an average of 26 mRNA copies per cell for the top 5% of expressed genes, in contrast to an average of 0.0026 copies per cell for the bottom 5% (Fig. S6A). The top 5% of expressed genes in YPD account for 58% of the transcriptome, mostly comprised of transcripts encoding protein biosynthesis proteins and central carbon metabolism enzymes. Our mRNA copy number estimates are consistent with previous estimates using DNA microarrays (Pearson correlations of 0.67, P < 10−300; 0.72, P < 10−300; and 0.83, P < 10−300, respectively; see Dataset S3 and Fig. S7) (3, 10, 18).
To calculate the relative expression level of each gene in HS vs. YPD, we compared the read densities in the 2 conditions. We compared the result with relative expression levels for the same mRNA samples inferred by commercial 2-dye microarrays (see Materials and Methods). Indeed, these ratios show strong agreement (Pearson correlation coefficient of 0.87, P < 10−300; see Fig. S6B). These results were reproducible across sequencing and microarray replicates (Dataset S3; http://compbio.cs.huji.ac.il/RNASeq), consistent with recent studies (5).
Discussion
We set out to test whether it is possible to define a complete yeast transcriptome ab initio using only the (unannotated) genome sequence and massively parallel sequencing of cDNA from 1 or more experimental conditions. Our approach independently identifies the vast majority of known genes transcribed under the tested conditions, correctly infers splicing events, and detects the correct gene structure. Also, it corrects a number of current annotations and identifies previously undescribed transcriptional units and splice junctions, several of which we validated experimentally. Last, the method can also accurately quantify the expression levels of transcripts.
There are several crucial steps in the strategy. First, the creation of the cDNA fragments determines the transcript coverage. The laboratory protocol that we used here is only mildly biased toward the 3′ end of the transcript and thus provides efficient coverage throughout the transcript, allowing us to effectively assemble transcripts from short reads. Second, to accurately map reads to the reference genome, we created a sequencing noise model to limit the errors in mapping. Because the yeast genome has large unique regions, we can estimate the error model from the data without requiring calibration runs. Using this model, we correct for varying quality among batches. Unlike previous read mapping approaches (19), our method estimates the noise model separately for each batch; thus, it is more specific and, depending on the model, may allow for more mismatches if their probability is higher. Third, using the error model and sequence similarity tests, we reliably identify reads that are split between 2 genomic positions. This step is crucial for identifying splice junctions ab initio and defining correct gene structures, and is distinct from previous read mapping approaches (19).
Our approach has several limitations. First, we are unable to predict transcriptional units for low-copy transcripts and nonunique regions (e.g., at the telomeres). Although we can estimate relative expression of some low-copy transcripts, we cannot reliably determine splicing events or boundaries in such genes. We partially address this issue by creating libraries from YPD and HS. Deeper sequencing and libraries from additional conditions can further improve the completeness of the catalog. Second, we miss splicing events due to local nonuniqueness at the splice junction. We can alleviate this problem by sequencing either longer reads or paired-end fragments, both of which are becoming available (11). Last, our approach is limited in detecting and distinguishing antisense transcripts and differentiating between close divergent transcription units due to the lack of strand specificity. Although in most cases we can recover transcript orientation, we can further improve the predictions by constructing strand-specific cDNA libraries.
Unlike recent studies (3, 5), we demonstrate the use of massively parallel sequencing for complete, ab initio construction of a eukaryotic transcriptome, independent of any existing genome annotation. For example, Mortazavi et al. (5), and several similar approaches (3–7), use a step-wise mapping approach that relies on mapping reads to known gene models, exons and splice junctions. De novo discovery in these schemes is also limited, and is based on mapping reads to all possible combinations of known exons. Such approaches cannot detect splice junctions between unannotated exons. Also, they are not applicable to a genome for which there are poor (or no) gene predictions. In contrast, our approach searches for all the locations where a spliced version of an unaligned read can be mapped in the genome. Thus, our approach will be useful for both smaller more compact genomes, such as those of fungi or protists that often involve phylogenetically isolated groups for which there are poor gene predictions (20), as well as for aberrant cancer genomes.
Our work powerfully demonstrates the feasibility of constructing a transcriptome of an organism in a comprehensive, fast, and cheap way. To estimate the power of this approach, we conducted our analysis in isolation from any other source of data or gene prediction methods. Nevertheless, we anticipate that in many practical setups it can be powerfully combined with other gene prediction approaches. Applying our approach to explore the transcriptomes of less characterized organisms in an ab initio fashion, can have a significant impact on genomics studies.
Materials and Methods
Yeast Strains and Growth Conditions.
HS experiment.
The strain used was a derivative of the S. cerevisiae strain S288c (BY4741; see ref. 21). We grew 1-L cultures overnight in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) to an OD600 of ≈1.0. The cultures were split and 1 flask was submerged in a 37 °C water bath and the other in a 22 °C water bath; 50-mL samples were harvested after 0 and 15 min.
RNA extraction and library preparation.
Total RNA and polyA+ RNA were isolated by using the RNeasy Midi Kit (Qiagen) and Poly(A) Purist kit (Ambion), respectively. Samples were quality controlled with the RNA 6000 Nano ll kit of the Bioanalyzer 2100 (Agilent). Sheared cDNA libraries were created for 6 samples (22 °C, 0 min; 22 °C, 15 min; 37 °C, 15 min; 2 replicates per condition; 150 ng of polyA+ RNA per sample). The cDNA was synthesized by using the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen) with SuperScript III (Invitrogen), 15-ng random hexamers (Invitrogen), and 20 units SUPERase·In (Ambion). Primer annealing was done at room temperature for 10 min followed by 1 h at 55 °C for first strand synthesis and 2 h at 16 °C for second strand synthesis; cDNA was sheared by sonication with 12 alternating cycles between “high intensity” (30 s; duty cycle, 20%; intensity, 10%; cycles per burst, 200) and “low intensity” (4 s; duty cycle, 5%; intensity, 10%; cycles per burst, 200) in the Frequency Sweeping mode (Covaris S2 machine). Adapters for Illumina sequencing were added following the instructions provided, except that 5 times less adapter mix was ligated to the cDNAs and PCR primers were removed by digestion with RecJ (New England Biolabs). Each library had an insert size of 60 to 110 bp. One lane of sequence (5.4 to 7.0 M reads) was generated for each sample on an Illumina 1G sequencer.
Genomic Mapping of Reads.
Error model.
We developed a detailed probabilistic model for scoring the quality of matching reads to the genome. Our score depends on the specific type of sequencing error made (e.g., genomic A sequenced as C) and its position within the sequenced read. Formally, the score of obtaining a read R originating from a genomic sequence G equals Σi=136log2(Pr(Ri|Gi,i)), where i is the position within the read, Ri is the sequenced nucleotide at position i, and Gi is the nucleotide at the corresponding genomic position. To estimate the error parameters, we identified reads with up to 4 mismatches to highly unique regions of the genome. Then, we estimated the fraction of errors at each position for each genomic nucleotide (Fig. S1).
Mapping method.
To map the sequenced reads to the genome with minimal errors, we devised the following strategy. Each read was compared with every possible 36-bp window in the genome and scored according to the error model above. We developed a procedure that uses suffix trees to efficiently finds all of the matches above a predefined threshold. To filter the matches, we require that the read matches the assigned genomic sequence with a threshold (−8.3) that assures correct mapping of 95% of reads (based on simulations). Also, to ensure uniqueness, we require the match to remain unique even when allowing a more relaxed threshold (−11.5).
Detection of Transcriptional Units.
Segmentation of transcriptional units.
To identify transcriptional units, we first artificially extended mapped reads to partially reconstruct the dsDNA segments they originated from. Because the segment size in our library varies between 60 and 110 bp, we chose a conservative approach and extended each read by an additional 40 bp (each read is now 76 bp). We then identified contiguously covered genomic regions. In many cases, these regions contained >1 gene, due to overlapping neighboring transcripts in the dense yeast genome. To refine these regions into single transcribed units, we developed an automated segmentation algorithm to fit the genomic patterns of mapped reads using piecewise linear regression (Fig. S2). Neighboring genes often exhibit different expression levels allowing an accurate partition. To achieve a coherent segmentation, we applied our algorithm to YPD and HS data simultaneously. This strategy also allows us to use the transcriptional differences of genes between the 2 conditions. For example, the 2 neighboring genes YBR287W and APM3 (Fig. 2B; Fig. S2) have similar expression levels at YPD; hence, preventing a proper segmentation to 2 transcription units. However, at HS, YBR287W is expressed in much higher levels than APM3, allowing us to position the boundary between the 2 genes.
Definition of nontranscribed loci.
For a negative control, we applied a sliding window of 75 bp over the data of David et al. (12), and identified 892 loci that presented the lowest mRNA to genome signal in YPD.
Automated determination of orientation.
As demonstrated in Fig. 2B, the typical density of reads is not completely uniform along the transcript with higher density toward the 3′ end. We use this pattern to estimate the orientation of each transcription unit. We use the slope of our piecewise linear fit to determine the orientation of each transcription unit. Specifically, we estimate a 95% confidence interval of the regressed slope parameters, and assign a forward or reverse orientation to the transcription unit if the entire interval is orientation-consistent (above or below zero, respectively).
Detection of splice junctions.
First, we map gapped reads by searching for coordinated partial matches to 2 genomic loci within 2 Kb, each one of at least 10 bp (and together adding up to 36 bp). We require the same noise thresholds to filter out mismatches and nonunique matches. Specifically, we score each putative match with the score described above, allowing a single gap in the genomic sequence. Second, we calculate the position-specific scoring matrix (PSSM) score for each gapped read, according to the splice motifs we learned from the known introns (see http://compbio.cs.huji.ac.il/RNASeq). Third, we cluster gapped reads by the genomic location of their gaps. Each cluster defines a putative junction in the transcriptome, and is characterized by the number of supporting reads and the PSSM score of the junction. We assign orientation to each putative junction using these asymmetric PSSM motifs. We define a threshold over the PSSM log-odd scores (2.78), such that 95% of the known splice junctions (based on SGD annotations, October 2007) are identified in the correct orientation.
Definition of Alignable Expressed Genes.
A genomic location is “nonalignable” if reads originating from that location will be mapped by our method to at least one other location in the genome; otherwise, we say that the location is alignable. We define a gene to be alignable if at least 50% of locations within its coding region are alignable. We define genes, known from previous studies to have at least 0.2 mRNA copies per cell (on average) (10), as “expressed,” reflecting 85% of the transcriptome at YPD condition.
Estimation of Gene Expression Levels.
Using annotations from SGD (October 2007), we calculate the number of reads mapped to each coding region. We approximate the expression level of each gene by the average density of reads along the unique (alignable) part of the coding region. This measure is expressed in arbitrary units of number of reads per lane per 1-K base pairs, and is assumed to be proportional to the actual number of mRNA molecules per cell. Assuming a conservative estimation of 15,000 transcripts per cell (17), we can assess the expected number of copies for each gene. Relative expression levels (HS vs. YPD) are calculated by comparing the average density of each gene at the 2 conditions.
Relative Gene Expression Using Commercial Arrays.
PolyA+ RNA samples from one replicate each of the 37 °C, 15 min (HS) and 22 °C, 15 min (YPD reference) were labeled with either Cy3 or Cy5 by using a modification of the protocol developed by De Risi (University of California, San Francisco) and Rosetta Inpharmatics that can be obtained at http://www.microarrays.org. For the detailed modified protocol see http://compbio.cs.huji.ac.il/RNASeq. Four technical replicates of the HS samples were hybridized against the reference on commercial S. cerevisiae (S288C strain) 2-color 60-mer oligo Agilent arrays in the 4 × 44 K format (Agilent). After hybridization and washing per Agilent instructions, arrays were scanned by using a scanner (Agilent) and analyzed with a feature extraction software (Agilent).
Validation of Novel Transcription Units and Splice Sites.
RNA from the HS and YPD reference samples was treated with TURBO DNA-free Kit (Ambion) to remove trace amounts of genomic DNA; cDNA was synthesized from this RNA by using a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). Assays were designed to detect predicted RNA species by the PCR. Reactions were performed under the conditions specified in the Amplitag gold polymerase product manual (Applied Biosystems) by using 10 ng of cDNA as template in a volume of 50 μL. For primer sequences, see http://compbio.cs.huji.ac.il/RNASeq. Products were amplified by using the following Thermocycler program: i, 95 °C for 5 min; ii, 95 °C for 30 s; iii, 56 °C for 30 s; iv, 70 °C for 45 s; go to step 2 for 40 cycles; v, 70 °C for 7 min; vi, 4 °C forever. PCR products were separated by using 3% Metaphor agarose (Cambrex) gels. The DNA fragments were isolated from the gel by using a QIAEX ll Gel extraction kit (Qiagen). These fragments were cloned by using a TOPO TA Cloning Kit for Sequencing (with pCR4-TOPO) with One Shot TOP10 Chemically Competent Escherichia coli and PureLink Quick Plasmid Miniprep Kit (Invitrogen). Insert containing constructs were sequenced at the Massachusetts Institute of Technology core facility. Sequences were verified by using the BLAST function at the Saccharomyces genome database (www.yeastgenome.org/).
Supplementary Web Site.
Raw data and additional notes and figures can be found at our supplementary web site (http://compbio.cs.huji.ac.il/RNASeq).
Supplementary Material
Acknowledgments.
We thank Ariel Jaimovich, Ilan Wapinski, Daphne Koller, Hanah Margalit, Jill Mesirov, Erin O'Shea, Oliver Rando, and especially Eric Lander, for comments and discussions; and Jason Funt, Toshiro Osumi, and the Broad Institute's Sequencing Platform for help with processing of Illumina sequence data. This work was supported by the Leibniz Research Center at the Hebrew University (T.K.), National Institutes of Health and Israel Science Foundation grants (to M.Y. and N.F.), a Human Frontiers Science Program grant (to D.-A.T.), funding from the Broad Institute (J.P.), and a Scientific Interface Career Award from the Burroughs Wellcome Fund (to A.R.).
Footnotes
Conflict of interest statement: G.S., S.L., and I.K. are from Illumina Corporation, which developed the sequencing technology we used, and thus have competing financial interests.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812841106/DCSupplemental.
References
- 1.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 5.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 6.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 7.Salehi-Ashtiani K, et al. Isoform discovery by targeted cloning, ‘deep-well’ pooling and parallel sequencing. Nat Methods. 2008;5:597–600. doi: 10.1038/nmeth.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherry JM, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 11.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 12.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura F, et al. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Alessio JM, Gerard GF. Second-strand cDNA synthesis with E. coli DNA polymerase I and RNase H: The fate of information at the mRNA 5′ terminus and the effect of E. coli DNA ligase. Nucleic Acids Res. 1988;16:1999–2014. doi: 10.1093/nar/16.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 17.Hereford LM, Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977;10:453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu CL, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.