Abstract
Population-based allele frequencies and genotype prevalence are important for measuring the contribution of genetic variation to human disease susceptibility, progression, and outcomes. Population-based prevalence estimates also provide the basis for epidemiologic studies of gene–disease associations, for estimating population attributable risk, and for informing health policy and clinical and public health practice. However, such prevalence estimates for genotypes important to public health remain undetermined for the major racial and ethnic groups in the US population. DNA was collected from 7,159 participants aged 12 years or older in Phase 2 (1991–1994) of the Third National Health and Nutrition Examination Survey (NHANES III). Certain age and minority groups were oversampled in this weighted, population-based US survey. Estimates of allele frequency and genotype prevalence for 90 variants in 50 genes chosen for their potential public health significance were calculated by age, sex, and race/ethnicity among non-Hispanic whites, non-Hispanic blacks, and Mexican Americans. These nationally representative data on allele frequency and genotype prevalence provide a valuable resource for future epidemiologic studies in public health in the United States.
Keywords: alleles; continental population groups; ethnic groups; genetics, population; genotype; nutrition surveys; polymorphism, genetic; prevalence
Completion of the human genome sequence (1–3) and recent advances in the analysis of genome-wide associations for several common diseases (4–20) are generating tremendous opportunities for epidemiologic studies to evaluate the role of genetic variants in the etiology of common human diseases. Identification of allelic variants has accelerated as a result of the cataloging and mapping of single nucleotide polymorphisms (SNPs) throughout the genome by the International HapMap Project (21–23) and characterization of the scope of structural variation, including copy number variants, in the genome (24–27). Application of these advances to improve public health requires assessing the frequency of these variants in distinct populations, identifying diseases influenced by these variants, determining the magnitude of the associated risks, and elucidating gene–gene and gene–environment interactions. Although the number of published investigations in these areas of human genome epidemiology has increased rapidly, with publication of more than 6,000 reports yearly (28), methodological issues have made it difficult to integrate the evidence and, thus, to easily translate the findings into public health improvements (29–31).
Early studies of genotype prevalence used samples that were convenient to obtain, and minimal information was provided on the selection of participants (31). In addition, most estimates were calculated from data on small study populations, which limited the accuracy of estimates of allele frequency and genotype prevalence. Furthermore, frequencies for most genetic polymorphisms have been measured only in select US racial and ethnic groups and have not been presented by age group or by sex. Although select polymorphism frequencies have been reported in large populations (32, 33), these studies were community based or controls from larger case-control studies. In contrast, data on genetic variants can be obtained from large, well-designed, epidemiologically well-characterized, and population-based US surveys such as the Third National Health and Nutrition Examination Survey (NHANES III) (34, 35). These data are a unique and unparalleled resource for epidemiologic research to assess genetic variation in the population, gene–disease associations, interactions between gene–gene and gene–environment factors, and population-attributable risk for genetic variants.
In particular, NHANES III offers the opportunity to assess genetic variation among major racial and ethnic groups in the United States, for whom multiple health disparities exist (36–40). Health disparities result from the complex interactions of social, environmental, behavioral, and genetic influences in a diverse population (36, 41, 42). Public health strategies to address health disparities are more likely to be effective when they are based on sound integration of such risk information at the population level. NHANES III is a paradigm for complex analysis of unbiased, population-based data on social, environmental, behavioral, and biologic characteristics—including genetic variation—in relation to health status.
MATERIALS AND METHODS
Survey design
NHANES III is a complex, multistage sample survey conducted by the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC) (35, 43), during 1988–1994. This cross-sectional study was designed to provide national statistics on the health and nutritional status of the civilian, noninstitutionalized population in the United States aged 2 months or older. Certain populations, including young children, older adults, non-Hispanic blacks, and Mexican Americans, were oversampled (35). As with standard NHANES analyses, race/ethnic groups were defined on the basis of the combination of the reported race (black, white, other) and reported ethnicity (not Hispanic, Mexican American, other Hispanic) of survey participants (35). Detailed household interviews were conducted to obtain information on sociodemographic variables, medical history, health-related behaviors, and use of medications. As part of the survey, physical examinations and laboratory and radiologic measurements were performed in special mobile examination centers (35).
NHANES III DNA bank
During Phase 2 of NHANES III (1991–1994), 10,052 participants aged 12 years or older were examined in the mobile examination centers. As part of the examination consent, participants agreed that their blood could be kept for long-term storage and future research, although genetic research was not mentioned specifically. In August 2001, the CDC/NCHS Ethics Review Board approved a revised plan for use of these specimens according to guidelines in the August 1999 National Bioethics Advisory Commission report on the use of stored biologic materials for research. This revised plan allows linkage of the genetic laboratory results to NHANES data through the NCHS Research Data Center to ensure that confidentiality of the study participants’ identities is maintained (44). Attempts were made to establish Epstein-Barr virus-transformed cell lines (35, 44) from white blood cells obtained from 8,200 of the Phase 2 participants. However, the final NHANES III DNA bank contains 7,159 participants because of the inability to transform and grow a successful immortalized cell line (n = 1,004), concerns regarding laboratory practice and quality assurance (n = 21), and exclusion of 16 individuals who were not genotyped. The bank is jointly maintained by both NCHS and the National Center for Environmental Health at CDC. Demographic characteristics of participants in the DNA bank are included in Table 1. Sixty-two percent of participants were from households with multiple family members (average, 1.59 members per household; range, 1–11). This prevalence study was approved by the NCHS Ethics Review Board.
Table 1.
Demographic Characteristics of Participants, Third National Health and Nutrition Examination Survey, Phase 2 (1991–1994), DNA Bank
| Demographic Characteristics | All Participants |
Non-Hispanic White |
Non-Hispanic Black |
Mexican American |
Other |
||||||||||
| No. of Subjects | Unweighted, % | Weighted, % | No. of Subjects | Unweighted, % | Weighted, % | No. of Subjects | Unweighted, % | Weighted, % | No. of Subjects | Unweighted, % | Weighted, % | No. of Subjects | Unweighted, % | Weighted, % | |
| Total | 7,159 | 100 | 100 | 2,630 | 36.7 | 73.5 | 2,108 | 29.5 | 11.7 | 2,073 | 29.0 | 5.7 | 348 | 4.9 | 9.2 |
| Sex | |||||||||||||||
| Male | 3,102 | 43.3 | 48.1 | 1,052 | 40.0 | 48.6 | 897 | 42.6 | 45.2 | 1,024 | 49.4 | 51.2 | 129 | 37.1 | 46.2 |
| Female | 4,057 | 56.7 | 51.9 | 1,578 | 60.0 | 51.4 | 1,211 | 57.4 | 54.8 | 1,049 | 50.6 | 48.8 | 219 | 62.9 | 53.8 |
| Age, years | |||||||||||||||
| 12–19 | 1,211 | 16.9 | 13.6 | 266 | 10.1 | 12.0 | 483 | 22.9 | 18.2 | 393 | 19.0 | 20.1 | 69 | 19.8 | 16.4 |
| 20–39 | 2,597 | 36.3 | 39.3 | 709 | 27.0 | 37.1 | 861 | 40.8 | 42.9 | 892 | 43.0 | 50.1 | 135 | 38.8 | 45.5 |
| 40–59 | 1,552 | 21.7 | 27.7 | 600 | 22.8 | 29.0 | 469 | 22.2 | 24.8 | 403 | 19.4 | 21.6 | 80 | 23.0 | 24.6 |
| ≥60 | 1,799 | 25.1 | 19.4 | 1,055 | 40.1 | 21.9 | 295 | 14.0 | 14.0 | 385 | 18.6 | 8.2 | 64 | 18.4 | 13.5 |
Selection of candidate genes and variants
Members from a multidisciplinary working group reviewed available phenotype data from NHANES III, performed systematic literature reviews, and identified candidate genes and physiologic pathways thought to be associated with diseases of public health significance at the time of project initiation. The selection of polymorphisms for this study was also based upon input from the SNP500Cancer resource (45), which had already developed genotyping assays for numerous SNPs in the selected genes based on their potential importance to physiologic processes, epidemiologic studies, and health outcomes.
The selected variants are in genes that encode proteins in 6 major cellular and physiologic pathways: 1) nutrient metabolism (e.g., homocysteine, lipids, glucose, and alcohol); 2) immune and inflammatory responses; 3) xenobiotic metabolism (e.g., of drugs, carcinogens, or environmental contaminants); 4) DNA repair; 5) hemostasis and the renin-angiotensin-aldosterone system; and 6) oxidative stress. The variants are in pathways affecting the development of multiple diseases, including cardiovascular disease, diabetes, cancer, and infectious diseases, as well as modulation of the effects of environmental and occupational exposures.
Genotyping methods
DNA analysis for the project was performed at two facilities because neither lab had methodology developed to analyze all of the genetic variants: 1) the Core Genotyping Facility, National Cancer Institute (NCI), National Institutes of Health, Bethesda, Maryland (http://cgf.nci.nih.gov), and 2) the Division of Laboratory Sciences, National Center for Environmental Health, CDC, Atlanta, Georgia. Each lab analyzed all DNA specimens for each subset of genotyping assays performed.
Most polymorphisms were assayed by either the TaqMan assay (5′ nuclease assay; Applied Biosystems, Foster City, California) or the MGB Eclipse assay (3′ hybridization-triggered fluorescence reaction; Nanogen (formerly Epoch Biosciences), Bothell, Washington). Two polymorphisms were genotyped by pyrosequencing, and one was by capillary fragment analysis. Water controls and DNA samples with known genotypes, purchased from Coriell Cell Repositories (Camden, New Jersey), were included on each 384-well plate. Detailed genotyping methods, including primer and probe sequences, are described in Web Appendix 1 and Web Table 1, respectively. (This information is described in Web-only material that includes 8 Web appendixes, 1 Web table, and 1 Web figure; each is preceded by “Web” in the text. All are posted on the Journal’s website (http://aje.oxfordjournals.org/).)
Quality control
The NHANES III genotyping data were monitored by a quality assurance and quality control committee composed of experts in laboratory science at CDC and NCI. The group monitored results of NHANES III quality control genotyping to ensure that the data met quality control guidelines established by NCHS.
Initial quality assurance assessments determined that at least 7,128 specimens, depending on the laboratory, were suitable for genotyping analysis on the basis of sample quality. All polymorphisms with genotyping call rates below 95% completion did not meet quality control criteria and were removed from further analyses. NHANES provides 480 quality control specimens for all studies that use the NHANES III DNA bank samples. These include blind replicates of approximately 6% of the 7,159 samples, to determine the accuracy and reproducibility of the assays. Assays that passed the blind-replicate analyses (>98% concordance according to NCHS guidelines) were tested for deviation from Hardy-Weinberg proportions calculated separately for each race/ethnic group in a standard unweighted analysis (46). The threshold for a genetic variant to pass Hardy-Weinberg analysis was P ≥ 0.01 (2 sided) for at least 2 of the 3 main race/ethnic groups (i.e., non-Hispanic white, non-Hispanic black, and Mexican American), with use of a chi-square goodness-of-fit test. The race/ethnicity category “other” was not used in determining the deviation from Hardy-Weinberg proportions because of the genetic heterogeneity of this group. Data from 192 samples were removed from certain assays because of a sample handling issue discovered in one of the laboratories. Genetic variants that met all quality control guidelines were used for further analyses. The range of successful genotype identifications for these variants was 97.5%–99.9% (median, 99.2%). Results from the tests of deviations from Hardy-Weinberg proportions for these variants are listed in Web Appendix 2.
Overall, 90 variants in 50 genes were available for estimation of allele frequency and genotype prevalence. Nearly all (n = 87) of the variants genotyped are SNPs, and 3 are insertion/deletions. Various diseases or conditions for which these genes have a confirmed or purported association are shown in Web Appendix 3. This list is not comprehensive, but it demonstrates that the genes studied are involved in major pathways that have a role in the etiology of several diseases or conditions with public health significance.
Statistical analysis
Sample weights.
Because NHANES III is a multistage, complex sample survey, all statistical analyses must account for sample weights and the survey design to produce unbiased national estimates and appropriate standard errors. The variance in clustered data caused by households with multiple related study participants was accounted for by use of the appropriate sample weights and the survey design in SUDAAN software (SUDAAN Statistical Software Center, Research Triangle Park, North Carolina). Point estimates and variances were calculated by using sample weights recalculated (47) for the Genetic Component of NHANES III. These weights were derived from the appropriate NHANES III, Phase 2, mobile examination center (MEC) sample weights to adjust for participant refusal to consent to future research and from the inability to generate cell lines and obtain DNA as mentioned above. NHANES genetic weights are specifically estimated for the genetic component of the 7,159 DNA bank participants, and none of the other weights provided by NHANES is appropriate. More detailed information about statistical weights in NHANES III is available online (http://www.cdc.gov/nchs/about/major/nhanes/nh3data_genetic.htm).
Prevalence estimation.
Analyses were conducted by using SAS-callable SUDAAN, version 9.01, and SAS, version 9.1 (SAS Institute, Inc., Cary, North Carolina). Deviations from Hardy-Weinberg proportions were tested with a chi-square goodness-of-fit approach by using SAS/Genetics (SAS Institute, Inc.). Allele frequency and genotype prevalence were calculated in SUDAAN and weighted by using the NHANES III Genetic Component sample weights for each gene variant for all major race/ethnic groups (i.e., non-Hispanic white, non-Hispanic black, Mexican American, and other) (data for “other” are not shown), age groups, and sexes. Point estimates and 95% confidence intervals were calculated and weighted for each race/ethnic group in SUDAAN to obtain the nationally representative estimates for the US population. The Taylor series linearization approach (48, 49), which derives a linear approximation of variance estimates to develop corrected standard errors and confidence intervals, was implemented to estimate variances. Tests of the difference in allele frequencies among race/ethnic groups (“other” was excluded), age groups, and sexes were performed by using polytomous logistic regression. Tests of the differences in genotype prevalence among these groups were evaluated using the Wald chi-square method. Statistical significance was considered as P < 0.05. The differences in allele frequency and genotype prevalence by age and by sex were examined after adjustment for race/ethnicity by using the Cochrane-Mantel-Haenszel test at a significance level of 0.05.
RESULTS
Demographic characteristics of the 7,159 participants in the NHANES III DNA bank are described in Table 1. After adjustment for the NHANES III sample design and for nonresponse in the genetic component, there were slightly more women than men in the US population between 1991 and 1994. The weighted frequency for each of the 3 main race/ethnic groups was 73.5% non-Hispanic white, 11.7% non-Hispanic black, and 5.7% Mexican American. The highest weighted frequency of persons aged 60 or more years and the lowest weighted frequency of persons aged 12–19 years were observed in non-Hispanic whites.
Weighted allele frequency point estimates for the 90 genetic variants for each of the 3 major race/ethnic subgroups in the US population are shown in Table 2. Complete frequency estimates with confidence intervals are shown in Web Figure 1 and Web Appendix 4. Allele frequencies were significantly different across race/ethnic groups for 88 (97.8%) of the variants studied (P < 0.05, two tailed), except for rs4986893 (CYP2C19, no homozygotes for minor allele) and rs1801274 (FCGR2A). Summary allele frequencies among the three major race/ethnic groups are shown in Table 3. Of the 90 candidate gene variants, 80 (88.9%) among non-Hispanic whites, 79 (87.8%) among non-Hispanic blacks, and 80 (88.9%) among Mexican Americans had allele frequencies of 0.02 or greater.
Table 2.
Weighted Allele Frequencies of Genetic Variants in the US Population by Race/Ethnicity, Third National Health and Nutrition Examination Survey, Phase 2 (1991–1994), DNA Bank
| Gene Symbola | Gene Name [Chromosomal Position]a | Pathwayb | Variantc | Nucleotide Position [Amino Acid Change]d | Allelee | Total US, %f | Non-Hispanic White, % | Non-Hispanic Black, % | Mexican American, % | P Valueg |
| ABCB1 | ATP-binding cassette, subfamily B (MDR/TAP), member 1 [7q21.1] | 13 | rs1045642 | Ex27-55; 3435 [I1145I] | T (C) | 47.3 | 51.6 | 20.9 | 44.7 | <0.001 |
| ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 [17q23.3] | 2, 12 | rs4646994 | 289-bp Alu ins/del in intron 16 | ins (del) | 46.2 | 45.4 | 41.3 | 53.4 | <0.001 |
| ADH1B | Alcohol dehydrogenase IB (class I), beta polypeptide [4q21-q23] | 12, 13 | rs1229984 | Ex3 + 23 [R48H] | A (G) | 6.4 | 4.7 | 1.4 | 5.5 | <0.001 |
| rs17033 | Ex9 + 77 | G (A) | 10.0 | 8.5 | 6.9 | 30.0 | <0.001 | |||
| rs2066702 | Ex9 + 5 [R370C] | T (C) | 2.9 | 0.4 | 20.0 | 0.9 | <0.001 | |||
| ADH1C | Alcohol dehydrogenase 1C (class I), gamma polypeptide [4q21-q23] | 12, 13 | rs1693482 | Ex6-14 [R272Q] | A (G) | 33.8 | 38.5 | 14.6 | 34.1 | <0.001 |
| rs698 | Ex8-56 [I350V] | G (A) | 34.4 | 39.4 | 14.9 | 31.1h | <0.001 | |||
| ADRB1 | Adrenergic, beta-1-, receptor [10q24-q26] | 2, 3, 12 | rs1801252 | Ex1 + 231 [S49G] | G (A) | 15.7 | 13.2 | 22.5 | 28.0 | <0.001 |
| ADRB2 | Adrenergic, beta-2-, receptor, surface [5q31-q32] | 2, 3, 12 | rs1042713 | Ex1 + 265 [G16R] | A (G) | 40.5 | 38.9 | 49.7 | 41.8 | <0.001 |
| rs1042714 | Ex1 + 298 [E27Q] | G (C) | 36.0 | 41.9 | 18.0 | 21.3 | <0.001 | |||
| ADRB3 | Adrenergic, beta-3-, receptor [8p12-p11.2] | 12 | rs4994 | Ex1 + 387 [W64R] | C (T) | 8.6 | 6.9 | 11.7 | 16.6 | <0.001 |
| ALAD | Aminolevulinate, delta-, dehydratase [9q33.1] | 12, 13 | rs1800435 | Ex4 + 13; 177 [K68N; K88N] | C (G) | 6.5 | 8.1 | 1.3 | 4.5 | <0.001 |
| B9D2i | B9 protein domain 2 [19q13.2] | 14 | rs1800468 | Ex4-262; −800 of TGFB1 | A (G) | 6.7 | 7.6 | 2.8 | 4.6 | <0.001 |
| rs1800469 | 308 bp 3′ of STP; −509 of TGFB1 | T (C) | 32.1 | 31.4 | 23.8 | 44.8 | <0.001 | |||
| CAPN10 | Calpain 10 [2q37.3] | 1, 12 | rs3792267 | IVS3-176 | A (G) | 24.6 | 25.8 | 16.2 | 27.3 | <0.001 |
| CAT | Catalase [11p13] | 11, 12 | rs769214 | −843 | G (A) | 37.6 | 33.9 | 41.2 | 50.3 | <0.001 |
| CBS | Cystathionine-beta-synthase [21q22.3] | 12 | No rs number | 844ins68 (68-bp insertion in exon 8) | + (−) | 10.0 | 8.1 | 25.9 | 6.3 | <0.001 |
| CCL5 | Chemokine (C-C motif) ligand 5 [17q11.2-q12] | 5, 10 | rs2280788 | −95 | G (C) | 2.6 | 2.8 | 0.7 | 1.3 | <0.001 |
| CCR2 | Chemokine (C-C motif) receptor 2 [3p21.31] | 5, 10 | rs1799864 | Ex2 + 241 [V64I] | A (G) | 11.2 | 9.5 | 14.5 | 21.7 | <0.001 |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) [10q11.1] | 5, 10 | rs169097 | Ex5 + 709 | T (C) | 2.5 | 0.3h | 17.2 | 1.0 | <0.001 |
| CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 [15q22-q24] | 12, 13 | rs2472299 | −17961 | T (C) | 29.8 | 28.2 | 38.5 | 26.7 | <0.001 |
| rs2606345 | IVS1 + 606 | G (T) | 44.0 | 33.8 | 84.1 | 61.7 | <0.001 | |||
| CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 [15q24] | 12, 13 | rs11854147 | 5341 bp 3′ of STP | T (C) | 40.4 | 31.4h | 72.1 | 61.3 | <0.001 |
| rs2069514 | −3859 | A (G) | 8.4 | 1.6 | 26.3 | 33.7 | <0.001 | |||
| rs4886406 | 9773 bp 3′ of STP | G (T) | 29.4 | 27.8 | 38.3 | 26.5 | <0.001 | |||
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 [2p21] | 12, 13 | rs1056836 | Ex3 + 251 [V432L] | G (C) | 46.0 | 45.2 | 75.0 | 26.5 | <0.001 |
| rs1056837 | Ex3 + 304 [D449D] | T (C) | 45.5 | 45.0 | 72.7 | 26.3 | <0.001 | |||
| rs162557 | −2919 | T (C) | 21.4 | 23.2 | 21.6 | 13.1 | <0.001 | |||
| CYP2A6 | Cytochrome P450, family 2, subfamily A, polypeptide 6 [19q13.2] | 12, 13 | rs1801272 | Ex3-15 [L160H] | A (T) | 2.7 | 3.3 | 0.6 | 2.0 | <0.001 |
| CYP2C19 | Cytochrome P450, family 2, subfamily C, polypeptide 19 [10q24.1-q24.3] | 12, 13 | rs4986893 | Ex4-7 [W212*] | A (G) | 0.2 | 0.1 | 0.2 | 0.1 | 0.744 |
| rs4986894 | −97 | C (T) | 14.8 | 13.7 | 18.3 | 11.0 | <0.001 | |||
| CYP2C9 | Cytochrome P450, family 2, subfamily C, polypeptide 9 [10q24] | 12, 13 | rs1057910 | Ex7-75 [I359L] | C (A) | 5.8 | 6.8 | 1.1 | 3.9 | <0.001 |
| CYP2E1 | Cytochrome P450, family 2, subfamily E, polypeptide 1 [10q24.3-qter] | 12, 13 | rs2031920 | −1054; −1053 | T (C) | 3.1 | 2.2 | 0.8 | 10.7 | <0.001 |
| CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 [7q21.1] | 12, 13 | rs2740574 | −391 | G (A) | 11.7 | 4.1 | 64.0 | 7.7 | <0.001 |
| F2 | Coagulation factor II (thrombin) [11p11-q12] | 5, 9 | rs1799963 | Ex14-1 | A (G) | 1.0 | 1.1 | 0.3 | 1.1 | 0.001 |
| F5 | Coagulation factor V (proaccelerin, labile factor) [1q23] | 9 | rs6025 | Ex10-11 [R534Q] | A (G) | 2.2 | 2.6 | 0.6 | 1.0 | <0.001 |
| FAM82Aj | Family with sequence similarity 82, member A [2p22.2] | 14 | rs163086 | IVS10-1363 | T (C) | 21.3 | 22.8 | 19.1 | 14.5 | <0.001 |
| FCGR2A | Fc fragment of IgG, low affinity IIa, receptor (CD32) [1q23] | 10 | rs1801274 | Ex4−120 [H166R; H167R] | A (G) | 50.0 | 49.4 | 47.4 | 48.6 | 0.096 |
| FGB | Fibrinogen beta chain [4q28] | 6, 9 | rs1800790 | −462 | A (G) | 17.6 | 19.4 | 5.5 | 15.0 | <0.001 |
| IL10 | Interleukin 10 [1q31-q32] | 1, 4, 7, 10 | rs1800871 | −853; −819 | T (C) | 29.0 | 24.2 | 39.6 | 38.1 | <0.001 |
| rs1800872 | −626; −592 | A (C) | 29.0 | 24.3 | 39.3 | 37.9 | <0.001 | |||
| rs1800896 | −1116; −1082 | G (A) | 42.3 | 46.9 | 35.6 | 30.5 | <0.001 | |||
| IL1B | Interleukin 1, beta [2q14] | 1, 7, 10 | rs1143623 | −2022 | C (G) | 28.7 | 28.5 | 11.2 | 42.9 | <0.001 |
| IL4 | Interleukin 4 [5q31.1] | 1, 7, 10 | rs2243248 | −1098 | G (T) | 9.0 | 7.5 | 15.7 | 12.0 | <0.001 |
| rs2243250 | −588; −524; −590 | T (C) | 25.5 | 16.0 | 64.0 | 42.3 | <0.001 | |||
| rs2243270 | IVS2-1297 | G (A) | 25.7 | 16.4 | 63.7 | 41.8 | <0.001 | |||
| IL4R | Interleukin 4 receptor [16p11.2-12.1] | 1, 7, 10 | rs1801275 | Ex12 + 828 [Q576R] | G (A) | 26.8 | 20.8 | 67.1h | 28.6 | <0.001 |
| rs1805015 | Ex12 + 608 [S503P] | C (T) | 17.8 | 15.7 | 36.7 | 15.7 | <0.001 | |||
| ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) [5q23-q31] | 5, 6, 9 | rs1126643 | Ex7-21 [F253F] | T (C) | 39.7 | 41.2 | 29.6 | 44.6 | <0.001 |
| ITGB3 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) [17q21.32] | 1, 5, 6, 9 | rs5918 | Ex3 + 11 [L59P] | C (T) | 14.4 | 16.3 | 10.1 | 9.7 | <0.001 |
| MBL2 | Mannose-binding lectin (protein C) 2, soluble (opsonic defect) [10q11.2-q21] | 10 | rs11003125 | −618; −550 | G (C) | 34.9 | 36.4 | 12.8 | 51.0 | <0.001 |
| rs1800450 | Ex1-27 [G54D] | A(G) | 13.5 | 14.6 | 4.0 | 14.6 | <0.001 | |||
| rs1800451 | Ex1-18 [G57E] | A (G) | 4.6 | 2.0 | 23.3 | 2.5 | <0.001 | |||
| rs5030737 | Ex1-34 [R52C] | T (C) | 6.1 | 7.4 | 1.1 | 2.9h | <0.001 | |||
| rs7096206 | −289; −221 | C (G) | 20.4 | 22.4 | 15.1 | 11.4h | <0.001 | |||
| MTHFR | 5,10-Methylenetetrahydrofolate reductase (NADPH) [1p36.3] | 12 | rs1801131 | Ex8-62; 1298 [E429A] | C (A) | 28.4 | 31.1 | 17.9 | 18.8 | <0.001 |
| rs1801133 | Ex5 + 79; 677 [A222V] | T (C) | 30.8 | 32.6 | 11.6 | 44.6 | <0.001 | |||
| rs2066470 | Ex2-120 [P39P] | T (C) | 9.7 | 9.6 | 8.7 | 6.5 | 0.042 | |||
| MTRR | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase [5p15.3-p15.2] | 12 | rs1801394 | Ex2-64 [I22M; I49M] | G (A) | 46.9 | 52.8 | 28.7 | 26.1 | <0.001 |
| NAT2 | N-Acetyltransferase 2 (arylamine N-acetyltransferase) [8p22] | 13 | rs1041983 | Ex2 + 288 [Y94Y] | T (C) | 36.4 | 35.4 | 46.1 | 31.9 | <0.001 |
| rs1208 | Ex2-367 [K268R] | G (A) | 39.4 | 41.0 | 38.3 | 35.7 | <0.001 | |||
| rs1799930 | Ex2-580 [R197Q] | A (G) | 30.2 | 32.3 | 28.0 | 18.0 | <0.001 | |||
| rs1801279 | Ex2 + 197 [R64Q] | A (G) | 1.0 | 0.0 | 7.8 | 0.5h | <0.001 | |||
| rs1801280 | Ex2 + 347 [I114T] | C (T) | 38.9 | 42.1 | 29.3 | 32.4 | <0.001 | |||
| NOS2A | Nitric oxide synthase 2A (inducible, hepatocytes) [17q11.2-q12] | 2, 10, 11, 12 | rs1800482 | G>C in promoter | C (G) | 1.0 | 0.1 | 6.3 | 0.3h | <0.001 |
| rs9282799 | −2892; −1173 | T (C) | 0.7 | 0.0 | 4.5 | 0.4 | <0.001 | |||
| NOS3 | Nitric oxide synthase 3 (endothelial cell) [7q36] | 2, 3, 9, 11, 12 | rs1799983 | Ex8-63 [E298D] | T (G) | 28.5 | 32.5 | 13.1 | 19.6 | <0.001 |
| rs2070744 | IVS1-762; −786 | C (T) | 34.2 | 39.1 | 15.1 | 25.1 | <0.001 | |||
| NQO1 | NAD(P)H dehydrogenase, quinone 1 [16q22.1] | 12, 13 | rs10517 | Ex6-457 | T (C) | 13.6 | 13.2 | 13.9 | 6.5 | <0.001 |
| rs1800566 | Ex6 + 40; 609 [P187S; P149S; P153S] | T (C) | 21.5 | 19.4 | 18.8 | 36.8 | <0.001 | |||
| rs34755915 | IVS3 + 20 | A (G) | 1.4 | 1.8 | 0.4 | 0.5 | <0.001 | |||
| rs689452 | IVS1-27 | G (C) | 12.5 | 11.9 | 13.7 | 6.3 | <0.001 | |||
| rs689453 | Ex2 + 65 [E24E] | A (G) | 7.0 | 7.9 | 5.1 | 5.2 | <0.001 | |||
| OGG1 | 8-Oxoguanine DNA glycosylase [3p26.2] | 8 | rs1052133 | Ex6-315 [S326C; P332A] | G (C) | 23.0 | 21.5 | 16.4 | 33.0 | <0.001 |
| PON1 | Paraoxonase 1 [7q21.3] | 11, 13 | rs662 | Ex6 + 78 [Q192R] | G (A) | 38.5 | 31.5 | 67.1 | 46.5 | <0.001 |
| rs854560 | Ex3 + 18 [L55M] | A (T) | 31.5 | 35.6 | 17.9 | 23.0 | <0.001 | |||
| PPARG | Peroxisome proliferator-activated receptor gamma [3p25] | 7, 10, 12 | rs1801282 | Ex4-49 [P12A] | G (C) | 11.6 | 13.2 | 2.5 | 12.4 | <0.001 |
| SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 [7q21.3-q22] | 5, 6, 9, 12 | rs1799762, rs1799768, rs1799889 | 4G/5G ins/del in promoter | 4G (5G) | 47.9 | 52.5 | 26.7 | 34.0 | <0.001 |
| TGFB1 | Transforming growth factor, beta 1 [19q13.1] | 1, 4, 7, 10 | rs1982073 | Ex1-327 [P10L] | C (T) | 40.4 | 38.5 | 44.0 | 50.3 | <0.001 |
| TLR4 | Toll-like receptor 4 [9q32-q33] | 10 | rs4986790 | Ex4 + 636 [D299G] | G (A) | 6.4 | 6.8 | 7.5 | 2.5 | <0.001 |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) [6p21.3] | 1, 4, 7, 10, 12 | rs1800629 | −487; −308 | A (G) | 15.3 | 17.2 | 13.1 | 7.3 | <0.001 |
| rs1800750 | −555 | A (G) | 1.6 | 1.3h | 2.3 | 2.5 | 0.07 | |||
| rs361525 | −417; −238 | A (G) | 5.5 | 5.8 | 4.1 | 5.8 | 0.05 | |||
| VDR | Vitamin D (1,25- dihydroxyvitamin D3) receptor [12q13.11] | 7, 10, 12 | rs2239185 | IVS8-3968 | C (T) | 49.3 | 47.8h | 42.9 | 58.2 | <0.001 |
| rs731236 | Ex11 + 32 [I352I (TaqI variant)] | C (T) | 34.5 | 38.1 | 28.2 | 23.8 | <0.001 | |||
| XRCC1 | X-ray repair complementing defective repair in Chinese hamster cells 1 [19q13.2] | 8 | rs1001581 | IVS2-216 | A (G) | 38.9 | 39.9 | 36.7 | 30.2 | <0.001 |
| rs1799782 | Ex6-22 [R194W] | T (C) | 7.0 | 5.0 | 6.2 | 14.8 | <0.001 | |||
| rs25486 | IVS9-59 | G (A) | 33.6 | 36.0 | 24.1 | 25.8 | <0.001 | |||
| rs25487 | Ex10-4 [R399Q] | A (G) | 32.6 | 36.1 | 15.9 | 25.6 | <0.001 | |||
| rs25489 | Ex9 + 16 [R280H] | A (G) | 4.7 | 4.0 | 3.5 | 12.4 | <0.001 |
Abbreviations: ATP, adenosine triphosphate; bp, base pair(s); del, deletion; ex, exon; HUGO, Human Genome Organisation; IgG, immunoglobulin G; ins, insertion; IVS, intervening sequence; MDR, multidrug resistance; NADPH, nicotinamide adenine dinucleotide phosphate; rs, reference SNP; SNP, single nucleotide polymorphism; STP, STOP codon; TAP, transporter-associated antigen processing.
Official gene symbols and names are from the HUGO Gene Nomenclature Committee (http://www.genenames.org/). All chromosomal positions are from Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez?db = gene).
Inclusion of genes in pathways is based on information gathered from the GeneCards database (http://www.genecards.org), the KEGG GENES database (http://www.genome.jp/kegg/genes.html), and selected publications for ACE, CAPN10, and SERPINE1 (refer to references in Web Appendix 3): 1, apoptosis; 2, blood pressure regulation; 3, cardiac function; 4, cell cycle; 5, cell migration/motility; 6, cellular adhesion; 7, cellular growth and differentiation; 8, DNA repair; 9, hemostasis; 10, immunity and inflammation; 11, metabolism of free radicals/oxidative stress; 12, nutrient metabolism; 13, xenobiotic metabolism; 14, unknown.
Unique identifier in the Entrez SNP database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Nucleotide and amino acid change information are from the SNP500Cancer database (http://snp500cancer.nci.nih.gov) and dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). Alternate designations and additional locus information are given, if available.
The major allele (determined by the frequency in the total population) is in parentheses.
Includes the “other” race/ethnicity category.
The “other” race/ethnicity category was not included in chi-square tests.
The variant deviates from Hardy-Weinberg proportions in the race/ethnicity group at P < 0.01 in an unweighted chi-square goodness-of-fit test.
B9D2 is a protein that lies partially within the TGFB1 promoter. Although within B9D2, these two SNPs are believed to be promoter polymorphisms of TGFB1.
FAM82A is a hypothetical protein that lies immediately 5′ of CYP1B1. This variant may be an intronic polymorphism of FAM82A or a CYP1B1 variant that lies 3′ of the gene.
Table 3.
Range of Allele Frequencies for Study Polymorphisms by Race/Ethnicity, Third National Health and Nutrition Examination Survey, Phase 2 (1991–1994), DNA Bank
| Population | Allele Frequency |
|||||||
| 0–<0.01 |
0.01–<0.02 |
0.02–<0.05 |
≥0.05 |
|||||
| No. of Variants With Allele Frequency | % | No. of Variants With Allele Frequency | % | No. of Variants With Allele Frequency | % | No. of Variants With Allele Frequency | % | |
| Non-Hispanic white | 6 | 6.7 | 4 | 4.4 | 8 | 8.9 | 72 | 80.0 |
| Non-Hispanic black | 7 | 7.8 | 4 | 4.4 | 7 | 7.8 | 72 | 80.0 |
| Mexican American | 6 | 6.7 | 4 | 4.4 | 8 | 8.9 | 72 | 80.0 |
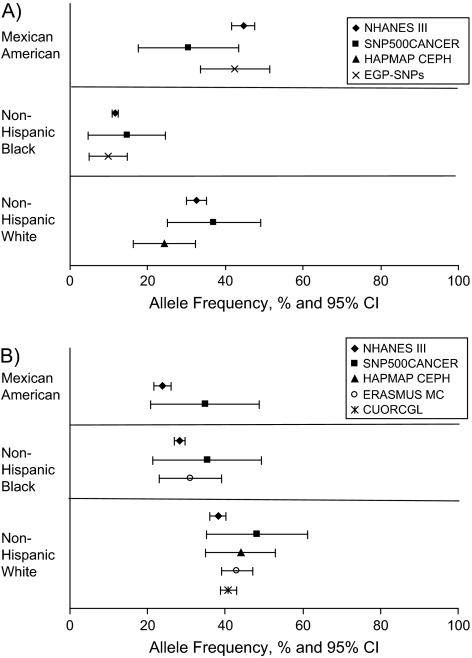
Differences in minor allele frequency of more than 20% (absolute value) compared with non-Hispanic whites are 22.4% (22 of 90 polymorphisms) for non-Hispanic blacks and 7.8% (2 of 90) for Mexican Americans (data not shown). Comparisons between NHANES III allele frequency estimates and other publicly available data sources are shown in Figure 1, with variants in MTHFR and VDR as examples. As observed, the NHANES III study includes much larger sample sizes, resulting in frequency estimates with small confidence intervals.
Figure 1.
Comparison of minor allele frequencies between the Third National Health and Nutrition Examination Survey (NHANES III), Phase 2 (1991–1994), and other sources. A, T allele of MTHFR rs1801133; B, C allele of VDR rs731236. CI, confidence interval; CUORCGL, Creighton University Osteoporosis Research Center Genetics Lab; EGP-SNPs, National Institute of Environmental Health Sciences Environmental Genome Project; ERASMUS MC, Erasmus University Medical Center; HAPMAP CEPH, International HapMap Project Centre d'Etude du Polymorphisme Humain; rs, reference SNP; SNP, single nucleotide polymorphism; SNP500CANCER, SNP500Cancer database. All data except those for NHANES III have been deposited in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Significant differences in genotype prevalence across race/ethnic groups were seen for all variants except three: rs4986893 (CYP2C19), rs1801274 (FCGR2A), and rs2066470 (MTHFR) (Web Appendix 4). Deviations from Hardy-Weinberg proportions were seen for 4 of the 90 polymorphisms (4.4%) among non-Hispanic whites, 1 (1.1%) among non-Hispanic blacks, and 5 (5.6%) among Mexican Americans (P < 0.01) (Web Appendix 2).
Weighted allele and genotype frequencies did not differ significantly by age group in the US population for the majority of the polymorphisms. However, 16 variants (17.8%) differed significantly in allele frequency, and 21 (23.3%) differed significantly in genotype prevalence by age (data not shown). After adjustment for race/ethnicity, these numbers decreased dramatically to 5 (5.6%) polymorphisms for allele frequency (Web Appendix 5) and to 14 (15.6%) variants for genotype prevalence (Web Appendix 6). However, we found that some of the race/ethnicity-adjusted tests may not be reliable because of zero cell counts.
There were no significant differences in allele frequencies or genotype prevalence by sex, except for rs1800451 (MBL2) and rs1800482 (NOS2A) (data not shown). After adjustment for race/ethnicity, the allele frequencies of 3 variants were statistically significant by sex— rs2243248 (IL4), rs1800482 (NOS2A), and rs361525 (TNF) (Web Appendix 7). After adjustment for race/ethnicity, the genotype prevalence of two variants was significantly different between men and women—rs2031920 (CYP2E1) and rs2243248 (IL4) (Web Appendix 8). (All results presented in this study are available online from the website of the National Office of Public Health Genomics at CDC (http://www.cdc.gov/genomics/).)
DISCUSSION
Our study evaluated the allele frequency and genotype prevalence of polymorphisms that have known or proposed associations with common diseases in a large, minority-enriched, and nationally representative sample of the US population. This is the first relatively large-scale, population-based effort in the United States to obtain such data by race/ethnic group. These data and future planned analyses will serve as an important reference for investigations into US population structure, for examinations of gene–disease associations in other investigations of the NHANES data set, for calculation of attributable risk, and for use as a reference by researchers in the design of further studies to discover associations of alleles and genotypes with common diseases.
Estimates of allele frequency and genotype prevalence are available from a number of existing gene variant databases, including the International HapMap Project (21–23) (http://www.hapmap.org) and the SNP500Cancer Database (45) (http://snp500cancer.nci.nih.gov). However, comparisons between NHANES III and such databases are limited because of significant differences in inclusion criteria, study populations, and classification of racial and ethnic groups between NHANES III and the other studies. Especially important is that these public databases function as genomic discovery tools. Consequently, their study populations were drawn largely from a small number of non-population-based samples. These small numbers of participants preclude accurate estimation of allele frequency and genotype prevalence, especially for rare variants or those that vary significantly by race and ethnicity. We compared two variants in MTHFR and VDR with other data resources and found substantial differences in allele frequencies (Figure 1). SNP500Cancer reports the C allele frequency of rs731236 (VDR) as 48.3% (95% confidence interval (CI): 35.3, 61.3) in non-Hispanic whites and as 35.4% (95% CI: 21.4, 49.4) in the African-American population. However, NHANES III estimates are 38.1% (95% CI: 36.0, 40.3) and 28.2% (95% CI: 26.8, 29.6), respectively. In conclusion, the NHANES III estimates of allele frequency and genotype prevalence in the US population are more representative and stable than are those calculated from previously available data.
In this study, allele frequency (in 88 of 90 genetic variants) and genotype prevalence (in 87 of 90 variants) differed significantly by race/ethnic group. Non-Hispanic blacks had considerable differences in minor allele frequency compared with non-Hispanic whites, with almost one-quarter of variants differing by at least 20% (absolute difference). In contrast, less than 10% of variants differed by at least 20% in allele frequency between Mexican Americans and non-Hispanic whites. Differences in allele and genotype frequency could partially contribute to differences in disease occurrence between population subgroups. As an example, the Pro12Ala variant of PPARG (rs1801282) has been studied extensively in relation to type 2 diabetes, with the Pro allele (C) being associated with increased disease prevalence (50). This finding has been duplicated in some genome-wide association studies (13–15), although not in all populations (51, 52). The higher CC genotype prevalence in non-Hispanic blacks (95.0%) compared with non-Hispanic whites (75.8%) may be a strong contributing factor to the increased risk of type 2 diabetes among non-Hispanic blacks, as this PPARG variant has been estimated to have a large population attributable risk of ∼25% (50). Because differences in the occurrence of common human diseases among populations reflect variation in genetic factors, environmental factors, and their interaction, population-based genotype data, when coupled with other disease risk factors, will give us better insight into the causes of population differences in the occurrence of various diseases.
On the other hand, allele frequency and genotype prevalence did not differ significantly between men and women for most of the genetic variants studied (≥97.8%). Similar findings on allele frequency or genotype prevalence by sex have also been reported in some large studies (32, 33). Although we report statistically significant differences by age for approximately one-fifth of the genetic variants studied, most of these differences were no longer present after adjustment for race/ethnicity. This finding is likely attributable to the differences in age distribution between the race/ethnic groups (Table 1). Some of the significant differences in allele frequencies by age may indicate survival advantage, and other studies have found variants in or near MTHFR (53, 54), PON1 (55, 56), TLR4 (55, 57), and TNF (58) associated with aging or longevity. However, few genes have been reproducibly shown to do so (59, 60), and our results could be due to insufficient sample sizes or due to statistical chance in analyses.
There has been a concern that multiple individuals from a household were included (average household, 1.59 individuals; range, 1–11) in NHANES III for the estimation of allele and genotype frequencies. However, the estimates were calculated by using methods specifically designed to analyze data from surveys with complex designs. These methods adopt NHANES III sample weights and adjust the variance of the estimate among the correlated observations. NHANES III is a population-based survey that reflects the actual and overall genetic structure of the general US population, which contains many related individuals within or between subpopulations. Thus, inclusion of related individuals in the NHANES III survey should enhance the generalizability of estimates derived from these data.
Some potential limitations of this study are notable. First, NHANES III categorizes race and ethnicity according to self-reported affiliation, as do most epidemiologic studies. There is considerable literature on the accuracy of this social measure as a proxy for genetic ancestry (61–65). Despite the possible misclassification or oversimplification of genetic ancestry, these data may help to elucidate the uncertain contribution of genetic variation (65, 66) to the complex interactions among social, environmental, and behavioral influences in a diverse population that contribute to racial and ethnic health disparities. Another concern is that homozygotes were not detected for some rare polymorphisms in this study, and thus the statistical tests for these genetic variants may not be reliable. In addition, future studies of gene–disease associations and gene–environment interactions with rare variants may be limited by insufficient sample sizes when analyses are performed separately for each race/ethnic group and control for large numbers of variables.
In the near future, we plan to use race and ethnicity, as well as geographic information, to conduct a focused examination of the genetic substructure of the US population and subpopulations. This issue is generating increased interest, because latent population substructure has been discovered in populations previously thought to be relatively homogeneous (67, 68). Such analyses are, therefore, especially important for the heterogeneous US population and considering the high levels of admixture within African-American and Mexican-American populations (69–72). Multiple studies demonstrate that population substructure must be taken into account in the design and interpretation of genetic association studies (67, 68, 70, 73–75). Further research on population characteristics and genetic diversity will be invaluable in conducting genetic epidemiologic studies in the United States.
Determination of the prevalence of genetic polymorphisms associated with common diseases of public health importance in the US population and in subgroups of the population is a critical first step in evaluating the genetic epidemiology of complex diseases. These prevalence estimates can be used in predicting sample size requirements for future epidemiologic studies to evaluate genetic determinants of susceptibility to chronic and infectious diseases, the severity of disease, and interactions with other risk factors. Because data on genotype frequency are particularly sparse for non-Hispanic blacks and Mexican Americans, our estimates are useful in sample size calculations for studying the genomic contribution to the health of these populations. Investigations currently underway examine the associations of the reported genetic variants with select nutritional, biochemical, and clinical characteristics in the NHANES III data set that serve as markers or risk factors for numerous health outcomes. These outcomes include asthma and chronic obstructive pulmonary disease, diabetes, cardiovascular disease, viral infections, and osteoporosis.
With the recent successes of genome-wide association studies, the resource of the NHANES III DNA bank offers significant opportunities to move beyond investigations of candidate genes, as was done here. Many recent genome-wide association studies have uncovered replicable genetic associations with diseases such as breast (4–6), prostate (7, 9), and colorectal (8, 10) cancers; heart disease (11, 12); diabetes (13–17); and obesity (18–20). However, many of these large-scale, case-control studies did not use representative samples of the underlying populations from which the cases were derived. NHANES is the only nationally representative, population-based sample survey that systematically collects physical, physiologic, imaging, laboratory, and interview data on a large number of individuals in the United States. Use of a whole-genome approach to assess the prevalence of genetic polymorphisms, including copy number variants, in the NHANES III DNA bank will be an important next step toward identifying genetic variants that can help to predict disease susceptibility and progression. This approach will also provide the basis for estimating the numbers of people in the United States who may benefit from genome-based tools, such as risk factor reduction; disease screening efforts; or diagnostic tests, drugs, or other preventive or therapeutic interventions. Current and future NHANES III prevalence estimates will be deposited into a publicly accessible database for research.
Thus, this first effort in NHANES begins to lay a strong scientific foundation for studying the impact of genetic variation on common diseases in the United States and in the future evaluation of biomarkers and diagnostic tests. Information derived from NHANES will provide an important reference and will enhance the translation of genomic information into clinical and public health practice.
Supplementary Material
Acknowledgments
Author affiliations: National Office of Public Health Genomics, Centers for Disease Control and Prevention, Atlanta, Georgia (Man-huei Chang, Nicole F. Dowling, Ramal Moonesinghe, Renée M. Ned, Ajay Yesupriya, Muin J. Khoury); National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia (Mary Lou Lindegren, Mary R. Reichler); National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention, Cincinnati, Ohio (Mary A. Butler); National Cancer Institute, National Institutes of Health, Rockville, Maryland (Stephen J. Chanock); National Center for Environmental Health, Centers for Disease Control and Prevention, Atlanta, Georgia (Margaret Gallagher); National Center on Birth Defects and Developmental Disabilities, Centers for Disease Control and Prevention, Atlanta, Georgia (Cynthia A. Moore); National Center for Health Statistics, Centers for Disease Control and Prevention, Hyattsville, Maryland (Christopher L. Sanders); and Core Genotyping Facility, Division of Cancer Epidemiology and Genetics, Advanced Technology Program, SAIC Frederick, Inc., NCI-Frederick, Maryland (Robert Welch).
The authors would like to dedicate this article in memory of Bob Welch, a treasured collaborator and friend.
This work was supported by the Centers for Disease Control and Prevention, Atlanta, Georgia.
The authors would like to thank the following individuals from the CDC/NCI NHANES III Genomics Working Group: National Center on Birth Defects and Developmental Disabilities, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Craig Hooper, Dr. Quanhe Yang); National Center for Chronic Disease Prevention and Health Promotion, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Karon Abe, Dr. Heidi M. Blanck, Dr. Ingrid J. Hall, Dr. Guiseppina Imperatore, Dr. Ann Malarcher, Dr. Glen Satten); National Center for Environmental Health, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Amanda Brown, Dr. Omar Henderson, Dr. Deborah Koontz); National Center for Environmental Health, Agency for Toxic Substances and Disease Registry, Atlanta, Georgia (Olivia Harris); National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Michael Aidoo, Dr. Robert Chen, Dr. Janet McNicholl); National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Venkatachalam Udhayakumar); National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention, Morgantown, West Virginia (Dr. Ainsley Weston); National Office of Public Health Genomics, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Marta Gwinn, Tiebin Liu, Dr. Wei Yu); Office of the Director, Coordinating Center for Health Promotion, Centers for Disease Control and Prevention, Atlanta, Georgia (Dr. Karen Steinberg); National Cancer Institute, National Institutes of Health, Rockville, Maryland (Dr. Neil Caporaso, Amy Hutchinson); Office of the Medical Director, March of Dimes Foundation, New York, New York (Bruce Lin); Department of Medicine, University of Washington School of Medicine, Seattle, Washington (Dr. Jai Lingappa); Department of Epidemiology and Community Medicine, University of Ottawa, Ontario, Canada (Dr. Julian Little); Division of Medical Genetics, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah (Dr. Lorenzo Botto); Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, Georgia (Dr. Tom Hodge); Program in Health Decision Science, Harvard University, Cambridge, Massachusetts (Davene Wright).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Conflict of interest: none declared.
Glossary
Abbreviations
- CDC
Centers for Disease Control and Prevention
- CI
confidence interval
- NCHS
National Center for Health Statistics
- NCI
National Cancer Institute
- NHANES III
Third National Health and Nutrition Examination Survey
- SNP
single nucleotide polymorphism
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 8.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 10.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 11.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 12.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 15.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakonarson H, Grant SF, Bradfield JP, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448(7153):591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 17.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312(5771):279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 19.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 25.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaja R, Zhang J, MacDonald JR, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38(12):1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am J Epidemiol. 2006;164(1):1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Ioannidis JP, Ntzani EE, Trikalinos TA, et al. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 31.Little J, Bradley L, Bray MS, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156(4):300–310. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 32.Garte S, Gaspari L, Alexandrie AK, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1239–1248. [PubMed] [Google Scholar]
- 33.Huang HY, Thuita L, Strickland P, et al. Frequencies of single nucleotide polymorphisms in genes regulating inflammatory responses in a community-based population. BMC Genet. 2007;8:7. doi: 10.1186/1471-2156-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyattsville, MD: National Center for Health Statistics; 2007. National Center for Health Statistics, Centers for Disease Control and Prevention. National Health and Nutrition Examination Survey. [Google Scholar]
- 35.National Center for Health Statistics. Plan and Operation of the Third National Health and Nutrition Examination Survey, 1988–94. Hyattsville, MD: National Center for Health Statistics; 1994. [PubMed] [Google Scholar]
- 36.National Healthcare Disparities Report, 2006. Rockville, MD: Agency for Healthcare Research and Quality; 2007. Agency for Healthcare Research and Quality, US Department of Health and Human Services. [Google Scholar]
- 37.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 38.Health disparities experienced by Hispanics—United States. MMWR Morb Mortal Wkly Rep. 2004;53(40):935–937. [PubMed] [Google Scholar]
- 39.Health disparities experienced by black or African Americans—United States. MMWR Morb Mortal Wkly Rep. 2005;54(1):1–3. [PubMed] [Google Scholar]
- 40.Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health and Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29(6):1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- 41.Olden K, White SL. Health-related disparities: influence of environmental factors. Med Clin North Am. 2005;89(4):721–738. doi: 10.1016/j.mcna.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Barnes KC. Genetic epidemiology of health disparities in allergy and clinical immunology. J Allergy Clin Immunol. 2006;117(2):243–254. doi: 10.1016/j.jaci.2005.11.030. quiz 255–256. [DOI] [PubMed] [Google Scholar]
- 43.Hyattsville, MD: National Center for Health Statistics; 2007. National Center for Health Statistics, Centers for Disease Control and Prevention. Third National Health and Nutrition Examination Survey (NHANES III) data files. [Google Scholar]
- 44.Hyattsville, MD: National Center for Health Statistics; 2007. National Center for Health Statistics, Centers for Disease Control and Prevention. NHANES III genetic data. [Google Scholar]
- 45.Packer BR, Yeager M, Burdett L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34(Database issue):D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trikalinos TA, Salanti G, Khoury MJ, et al. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163(4):300–309. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 47.Lohr SL. [Google Scholar]
- 48.Binder D. On the variance of asymptotically normal estimators from complex surveys. Int Stat Rev. 1983;51:279–292. [Google Scholar]
- 49.Woodruff R. A simple method for approximating the variance of a complicated estimate. J Am Stat Assoc. 1971;66:411–414. [Google Scholar]
- 50.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 51.Hayes MG, Pluzhnikov A, Miyake K, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56(12):3033–3044. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- 52.Meigs JB, Manning AK, Fox CS, et al. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet. 2007;8(suppl 1):S16. doi: 10.1186/1471-2350-8-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kluijtmans LA, Whitehead AS. Reduced frequency of the thermolabile methylenetetrahydrofolate reductase genotype in the elderly. Atherosclerosis. 1999;146(2):395–397. doi: 10.1016/s0021-9150(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 54.Stessman J, Maaravi Y, Hammerman-Rozenberg R, et al. Candidate genes associated with ageing and life expectancy in the Jerusalem longitudinal study. Mech Ageing Dev. 2005;126(2):333–339. doi: 10.1016/j.mad.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 55.Lunetta KL, D'Agostino RB, Sr, Karasik D, et al. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rea IM, McKeown PP, McMaster D, et al. Paraoxonase polymorphisms PON1 192 and 55 and longevity in Italian centenarians and Irish nonagenarians. A pooled analysis. Exp Gerontol. 2004;39(4):629–635. doi: 10.1016/j.exger.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 57.Balistreri CR, Candore G, Colonna-Romano G, et al. Role of Toll-like receptor 4 in acute myocardial infarction and longevity. JAMA. 2004;292(19):2339–2340. doi: 10.1001/jama.292.19.2339. [DOI] [PubMed] [Google Scholar]
- 58.Soto-Vega E, Richaud-Patin Y, Llorente L. Human leukocyte antigen class I, class II, and tumor necrosis factor-alpha polymorphisms in a healthy elder Mexican Mestizo population. Immun Ageing. 2005;2:13. doi: 10.1186/1742-4933-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7(6):436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capri M, Salvioli S, Sevini F, et al. The genetics of human longevity. Ann N Y Acad Sci. 2006;1067:252–263. doi: 10.1196/annals.1354.033. [DOI] [PubMed] [Google Scholar]
- 61.Foster MW, Sharp RR. Beyond race: towards a whole-genome perspective on human populations and genetic variation. Nat Rev Genet. 2004;5(10):790–796. doi: 10.1038/nrg1452. [DOI] [PubMed] [Google Scholar]
- 62.Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nat Genet. 2004;36(11 suppl):S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 63.Risch N, Burchard E, Ziv E, et al. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-comment2007. comment2007.1–comment2007.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bamshad M, Wooding S, Salisbury BA, et al. Deconstructing the relationship between genetics and race. Nat Rev Genet. 2004;5(8):598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- 65.Bamshad M. Genetic influences on health: does race matter? JAMA. 2005;294(8):937–946. doi: 10.1001/jama.294.8.937. [DOI] [PubMed] [Google Scholar]
- 66.Collins FS. What we do and don't know about ‘race’, ‘ethnicity’, genetics and health at the dawn of the genome era. Nat Genet. 2004;36(11 suppl):S13–S15. doi: 10.1038/ng1436. [DOI] [PubMed] [Google Scholar]
- 67.Helgason A, Yngvadottir B, Hrafnkelsson B, et al. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37(1):90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- 68.Feder J, Ovadia O, Glaser B, et al. Ashkenazi Jewish mtDNA haplogroup distribution varies among distinct subpopulations: lessons of population substructure in a closed group. Eur J Hum Genet. 2007;15(4):498–500. doi: 10.1038/sj.ejhg.5201764. [DOI] [PubMed] [Google Scholar]
- 69.Barnholtz-Sloan JS, Chakraborty R, Sellers TA, et al. Examining population stratification via individual ancestry estimates versus self-reported race. Cancer Epidemiol Biomarkers Prev. 2005;14(6):1545–1551. doi: 10.1158/1055-9965.EPI-04-0832. [DOI] [PubMed] [Google Scholar]
- 70.Choudhry S, Coyle NE, Tang H, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118(5):652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 71.Collins-Schramm HE, Chima B, Morii T. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet. 2004;114(3):263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- 72.Shriver MD, Parra EJ, Dios S, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112(4):387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 73.Ferreiros-Vidal I, D'Alfonso S, Papasteriades C, et al. Bias in association studies of systemic lupus erythematosus susceptibility due to geographical variation in the frequency of a programmed cell death 1 polymorphism across Europe. Genes Immun. 2007;8(2):138–146. doi: 10.1038/sj.gene.6364370. [DOI] [PubMed] [Google Scholar]
- 74.Freedman ML, Reich D, Penney KL, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36(4):388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 75.Marchini J, Cardon LR, Phillips MS, et al. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36(5):512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.