Abstract
Motivation: We noted that the sumoylation site in C/EBP homologues is conserved beyond the canonical consensus sequence for sumoylation. Therefore, we investigated whether this pattern might define a more general protein motif.
Results: We undertook a survey of the human proteome using a regular expression based on the C/EBP motif. This revealed significant enrichment of the motif using different Gene Ontology terms (e.g. ‘transcription’) that pertain to the nucleus. When considering requirements for the motif to be functional (evolutionary conservation, structural accessibility of the motif and proper cell localization of the protein), more than 130 human proteins were retrieved from the UniProt/Swiss-Prot database. These candidates were particularly enriched in transcription factors, including FOS, JUN, Hif-1α, MLL2 and members of the KLF, MAF and NFATC families; chromatin modifiers like CHD-8, HDAC4 and DNA Top1; and the transcriptional regulatory kinases HIPK1 and HIPK2. The KEPEmotif appears to be restricted to the metazoan lineage and has three length variants—short, medium and long—which do not appear to interchange.
Contact: toby.gibson@embl.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Members of the ubiquitin multiprotein family function as covalent modifiers of other proteins. These post-translational modifications (PTMs) then cause the target protein to be relocated to another subcellular location (Dye and Schulman, 2007). In the case of SUMO (the small ubiquitin-like modifier), attachment can affect processes including gene transcription and cell-cycle progression, although the mechanisms by which relocation within the nucleus achieves this are far from clear (Geiss-Friedlander and Melchior, 2007). While sumoylation seems largely to be restricted to the nucleus, a few non-nuclear proteins have been proposed to be sumoylated (Watts, 2004). SUMO substrates are often difficult to validate due to the low stoichiometry of the SUMO modification: however, proteomic approaches have lead to the identification of many putative substrates (reviewed in Rosas-Acosta et al., 2005).
Like other PTMs, sumoylation occurs at accessible linear motifs (LMs), usually in regions of natively disordered polypeptide (reviewed in Diella et al., 2008). Sumoylation occurs on a lysine in a motif that can be described by the pattern φK.E where φ=hydrophobic (Girdwood et al., 2004; Rodriguez et al., 2001) or the regular expression [VILMAFP]K.E as used in the ELM linear motif resource (Puntervoll et al., 2003). Sumoylated proteins that do not have the classical consensus motif have also been reported (Zhou et al., 2006). The φK.E pattern matches nearly half of the proteins in Swiss-Prot (Yang et al., 2006), indicating that most of the matches are false positive. As a consequence, there have been several attempts to try to extend this motif to get more specificity, resulting in the identification of different extended SUMO consensus motifs. Thus, the phosphorylation-dependent sumoylation motif (PDSM) φK.E..SP has been described in a subset of substrates, mainly transcriptional regulators: the phosphorylation of the SP motif regulates the interaction between the substrates and the SUMO-conjugating machinery, promoting sumoylation of the substrates (Hietakangas et al., 2006). In a second analysis, a cluster of acidic residues downstream from the core of many SUMO sites has been shown to be important for substrate binding and subsequent sumoylation (Yang et al., 2006). The importance of negative charges was also identified in substrates like Elk-1 and LRH-1; this extended SUMO consensus motif was named NDSM, negatively charged amino acid-dependent sumoylation motif.
The C/EBP transcription factors regulate cellular proliferation and differentiation of a range of cell types. They have been described as both tumour promoters and tumour suppressors, indicating that their regulatory system is complex (Nerlov, 2008). In C/EBPα, a regulatory domain motif (RDM) has been shown to inhibit the activity of an activation domain in a position-independent, but dose-dependent manner. The RDM was characterized by the consensus [VIL]K.EP and it was shown that sumoylation of lysine at position 2 decreases its inhibitory function in vitro (Kim et al., 2002; Nerlov, 2008).
A major hindrance to bioinformatic investigation of LM occurrences is that simple database searches do not yield significant results, while the false instances of motifs vastly outnumber the true ones. However, with improved sequence database annotation, LMs can sometimes be significantly enriched with certain keywords. Thus, Copley used transcriptional keywords to detect and justify new examples of the EH1 transcriptional repressor motif (Copley, 2005). A similar approach in combination with disorder prediction and conservation scoring, has shown that KEN-box destruction motifs are significantly enriched in the set of UniProt/Swiss-Prot entries annotated with cell-cycle keywords and Gene Ontology (GO) terms (Michael et al., 2008).
In this article, we report a computational investigation based on the RDM of C/EBPs. We refined the motif in the aligned C/EBP RDM sequence segments and then deployed a protocol involving keyword enrichment, native disorder prediction and conservation scoring in a survey of protein sequence databases. Highly significant results for motif matches were obtained with sets of entries annotated with keywords, such as nucleus, transcription and chromatin. The conservation pattern of the C/EBP motif was found to be the archetype of a linear motif, which we term KEPE, which is present in many nuclear proteins of the metazoa.
2 METHODS
2.1 Motif search
The interactive motif search tool SIRW combines regular expression searches with keyword searches of text annotation (Ramu, 2003). SIRW (sirw.embl.de/) was used to explore the human UniProt/Swiss-Prot database (release 54.7; 18054 entries for Homo sapiens) with the C/EBP-derived KEPE regular expression, [MILVFT]K.EP.{1,4}[DE]. To limit the search space, relevant annotation terms, such as nucleus, transcription and chromatin were used to search the fields for the GO terms [the Gene Ontology annotation (Harris et al., 2004; The_Gene_Ontology_Consortium, 2008)] as well as the separate keyword (KW) fields for keywords. (Note that the GO and KW searches are not formally equivalent because the GO terms include longer phrases in the definitions, while the annotators are also likely to have used different guidelines.) In order to assess the significance of the relative enrichment, we calculated P-values using Fisher's exact test (available in Excel and the R package).
2.2 Motif permutation controls
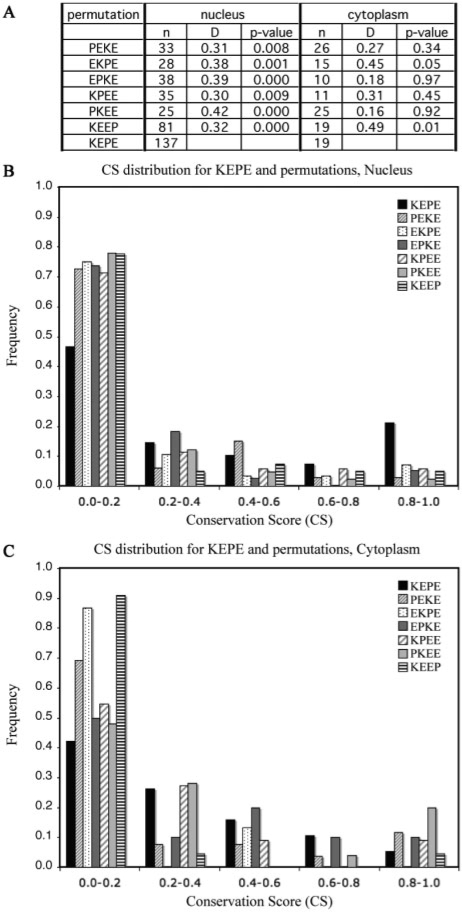
It is important to exclude artefactual or trivial reasons for motif enrichment, such as a bias in favour of the amino acids K, E and P. Therefore, in order to control for the background frequency, we permuted the three fully conserved residues in the KEPE motifs (KPEE; EKPE; EPKE; PEKE; PKEE) as well as a specific permutation (KEEP) for the two residues P and [DE] that extend the SUMO motif. Then we examined them for keyword association and conservation score (CS) as for the KEPE. Results for the controls are presented in detail in the Supplementary Material.
2.3 Modular protein architecture context
Motif matches were evaluated for presence in known globular domains using the Pfam and SMART domain databases (Letunic et al., 2006; Sammut et al., 2008). IUPred (http://iupred.enzim.hu/) (Dosztanyi et al., 2005) was used to test whether the motifs were found in predicted globular or natively disordered regions (also known as IUP, intrinsically unstructured polypeptide). Using the IUPred ‘long’ parameter setting to predict longer stretches of disorder, flanking regions of 15 amino acids upstream and downstream of the motif were scored, applying a value of 0.4 as the cut-off threshold.
2.4 Evolutionary conservation
Each match of the KEPE and the permuted motifs in Swiss-Prot proteins was also scored for conservation in homologous proteins using the CS method described in Chica et al. (2008). This approach has already been applied for the KEN box motif (Michael et al., 2008). The dataset used for calculating the CS included proteins (i) that are annotated to be in the nuclear or cytoplasmic compartment and (ii) whose motif match is found in a disordered/unstructured region according to the IUPred prediction. To compare the CS distribution between KEPE and the permutation controls, we used the Kolmogorov–Smirnov (KS) goodness of fit test.
2.5 Proteome analysis
We wrote a script to analyse the frequency of the motif and its permutations in human and yeast proteomes. We downloaded all proteins having associated GO terms from the EnsEMBL resource for H.sapiens (Hubbard et al., 2007) and from the SGD resource (Hong et al., 2008) for Saccharomyces cerevisiae and we obtained two datasets of 16 504 and 5327 proteins, respectively. Subsequently, we ran IUPred using the ‘long’ parameter as before (Dosztanyi et al., 2005). The ELM conservation filter (Chica et al., 2008) was then applied to assess the conservation of the matches.
3 RESULTS
3.1 Survey of Swiss-Prot with the KEPE regular expression
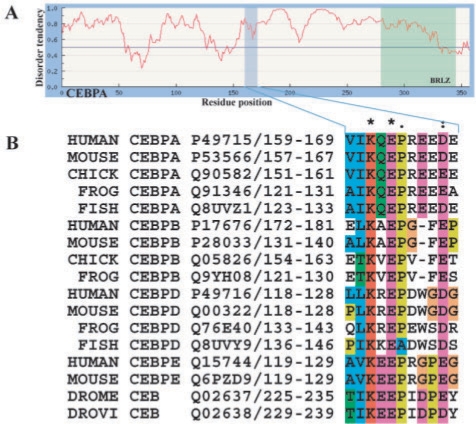
Using an alignment of Drosophila C/EBP and the four vertebrate paralogues C/EBPα, -β -δ and -ε (Fig. 1, Supplementary Fig. 1) we noted that downstream of the RDM motif [VIL]K.EP there are some additional conserved acidic residues, especially in positions 3 and 4 after the proline. Earlier studies have partially described the motif that matches the observed sequence conservation (Kim et al., 2002). We term the motif KEPE after the conserved residues. The motif is not in a known globular domain but rather in a region predicted to be natively disordered (Fig. 1A). To investigate if the observed motif could be present in other proteins, we undertook a survey for the KEPE-bearing proteins in the human entries of the UniProt/Swiss-Prot database (The_UniProt_Consortium, 2008) using the motif [MLIVFT]K.EP.{1,4}[DE]. We evaluated whether the matching proteins were found in the nuclear compartment or more widely. Since most LMs are known to be in natively disordered polypeptide segments (Fuxreiter et al., 2007), the KEPE matches were also evaluated for a clash with known globular domains using the SMART server (Letunic et al., 2006) or in predicted globular structure reported by IUPred (Dosztanyi et al., 2005).
Fig. 1.
The KEPE motif in C/EBP transcription factors. (A) The IUPred plot predicts human C/EBPα to be almost entirely natively disordered (the higher the peak, the more disordered). Like the KEPE motif, the leucine zipper (BRLZ) is also predicted as natively disordered (correctly so, since it must dimerise as coiled coil to acquire a stable folded structure). (B) C/EBP KEPE motif: an alignment of the RDM motif from Drosophila C/EBP and four vertebrate paralogues C/EBPα, -β, -δ and -ε (CEBPA, CEBPB, CEBPD and CEBPE) show the conservation of the motif (K is the sumoylated residue).
Of 331 human proteins matching the KEPE regular expression, 168 were annotated as localized in the ‘nuclear compartment’. Of those, more then 130 had KEPE matches localized in non-globular regions according to the IUPred prediction. These sequences were enriched in the functional classes ‘transcription factor’ or ‘chromatin modifier’ and in the GO class ‘protein function’ related to ‘transcription’. In only three cases was the KEPE motif found within a known globular domain according to SMART prediction. In these three paralogous bromodomain and PHD-finger containing proteins BRD1, BRF1 and BRF3 (Swiss-Prot:O95696, P55201, Q9ULD4), the KEPE motifs fell within PHD-finger domains. Since these KEPE motifs were found in the most variable loop of the PHD-finger (where an insertion of 15 residues or more is often found, SMART: SM00249), they could be potentially accessible for interaction.
Results of combined motif–keyword searches with SIRW are summarized in Table 1: Several terms show enrichments that are highly significant according to the Fisher's exact test. The enrichment was particularly significant with the ‘transcription’, ‘bzip’ and ‘znf’ keywords as well as for ‘nuclear’ compartment. There is a possibility that this enrichment could be driven by the high background frequency of the embedded SUMO motif. In order to test this possibility, we calculated the enrichment using human sequences matching the SUMO motif as the background distribution. The P-values ‘S’ in the right column show that the enrichment is still significant. Significant enrichment is not, per se, proof of function and could be for a trivial reason, such as strong amino acid bias or 4-fold increased mean protein length in transcriptional proteins. This can be controlled for by using test motifs that contain the same amino acids and information complexity, which can be obtained by permuting residues in the motif. When we performed the same analysis using permuted motifs, we found moderate enrichment for some keywords (see Supplementary Tables 1 and 2) but KEPE enrichment is always greater.
Table 1.
Enrichment of KEPE motif matches with various term combinations from the KW and/or GO term fields in Swiss-Prot entries
| Keywordsa,b | No. | Total | Total | P-value | P-value |
|---|---|---|---|---|---|
| KEPEc | Hd | Sd | He | Se | |
| Homo sapiens | 331 | 18 054 | 7707 | – | – |
| GO=cytoplasm*!nucleus | 35 | 2303 | 1146 | 2.45E−01 | 2.67E–02 |
| or KW=cytoplasm*!nucleus* | |||||
| GO=nuclear*|nucleus| | 168 | 3608 | 1789 | 2.10E−36 | 9.41E–29 |
| nucleolus!cytoplasm* or | |||||
| KW=nuclear*!cytoplasm* | |||||
| |nucleus!cytoplasm* | |||||
| Link=znf* | 79 | 1577 | 799 | 6.82E−17 | 4.01E–13 |
| Link=bzip* | 17 | 9 | 35 | 7.58E−18 | 8.62E–15 |
| GO=transcription* | 90 | 1239 | 627 | 7.43E−31 | 2.36E–26 |
| GO=chromatin* or | 21 | 213 | 139 | 4.12E−10 | 4.08E–07 |
| KW=chromatin* | |||||
| GO:0003700 | 36 | 506 | 264 | 3.55E−12 | 4.65E–10 |
| (transcription factor | |||||
| activity) | |||||
| GO:0008270 | 9 | 164 | 74 | 3.32E−03 | 4.20E–03 |
| (zinc ion binding) | |||||
| GO:0006355 | 17 | 253 | 131 | 4.60E−06 | 4.11E–05 |
| (Regulation of transcription, | |||||
| DNA-dependent) |
aSearch terms which have been used to retrieve the KEPE sequences.
bIn Swiss-Prot the annotation ‘cytoplasm’ is used (incorrectly) as a synonym for ‘cytosol’.
cNumber of sequences matching the KEPE pattern [MLIVTF]K.EP.{1,4}[DE] used in combination with the various search terms.
dTotal number of sequences (as obtained with the SIRW search tool, in Swiss-Prot release 54.7) matching the search terms shown in the left column. (H): all human sequences; (S): human sequences with the SUMO motif [VILMTF]K.E.
eThe P-value for the relative enrichment was calculated by the Fisher's exact test from the R package (for total ‘H’ and total ‘S’).
Genuine LMs that function in cell regulation are found to be conserved in homologous proteins (Neduva and Russell, 2005). Therefore, we applied the ELM CS pipeline (Chica et al., 2008) to assess KEPE motif conservation. Figure 2 compares the distributions of CS values for matches to KEPE and its permuted motifs; the comparison was repeated for nuclear and cytoplasmic proteins. In the nuclear set, the KEPE motif shows much stronger conservation than the permuted motifs. Furthermore the CS distributions of all permuted instances are significantly different to the KEPE distribution with P-values ranging from 0.00 to 0.01 (Fig. 2A). This result strongly supports a predicted function for KEPE in a nuclear role.
Fig. 2.
Conservation score distributions for the KEPE motif and the six permutations comparing the nuclear and cytoplasmic compartments. KEPE bearing proteins were retrieved from UniProt/Swiss-Prot with the compartment keyword expressions in Table 1, processed for IUPred disorder prediction and evaluated for the ELM CS score. To enable comparison, the sets have been normalized into percentages and sorted into five CS score bins. The table in (A) shows that the KEPE matches have a significantly different conservation distribution in the nucleus compared with the controls. n=number of instances, D=maximum difference between the cumulative distributions, P-value=significance of the difference D, according to the KS test. KEPE matches also show a peak of strong conservation (unmatched by the controls) in the nucleus (B) but not in the cytoplasm (C). These results are consistent with a lack of functionality of the KEPE motif in the cytosol (since this is the implied meaning of ‘cytoplasm’ in Swiss-Prot).
We were worried that matches in multiprotein families might have skewed the results in favour of the KEPE motif. Therefore, the set of sequences matching the KEPE and the permuted motifs were checked for the number of paralogous assignments retrieved using EnsEMBL mappings (Hubbard et al., 2007). Most of the matches are in proteins with one or no paralogues (Supplementary Fig. 2). This result shows that the higher frequency observed for the KEPE matches in the maximum CS range is not artificially caused by a higher number of paralogues in the corresponding protein families. This implies that the number of KEPE matches appearing in paralogues of the same protein reflects their functional value and not a tendency of those protein families to have more paralogues.
The non-significant differences obtained for KEPE versus the motif permutations for proteins annotated as cytoplasmic serve as a negative control (Fig. 2C). Indeed, here the KEPE instances are as non-conserved as the permuted ones, consistent with a lack of functionality in the cytosol.
3.2 Surveys of human and yeast proteomes with the KEPE regular expression
Although Swiss-Prot GO terms are also mapped to the EnsEMBL human proteome via the GOA database (Camon et al., 2004), EnsEMBL provides additional electronically generated GO annotation. The EnsEMBL human proteome is also more complete than in Swiss-Prot. Since the Swiss-Prot searches were interactive, we wanted to evaluate whether a fully automated proteome pipeline could produce qualitatively similar results. As shown in Supplementary Fig. 3, an equivalent GO term—IUPred—CS assessment protocol yields an even stronger nuclear conservation plot than for the interactive Swiss-Prot survey (Fig. 2A). An automated pipeline allows the component stages to be evaluated separately. The keyword and the IUPred assignment steps each individually contributed clear enrichment of conserved motifs, affirming their individual and combinatorial relevance to motif prediction (Supplementary Fig. 3).
The annotation of the yeast S.cerevisiae proteome in the SGD project is also extensive. The CS distributions were obtained for the yeast proteome using the same pipeline. In this case neither the keywords, nor the IUPred assignments provide any support for the KEPE motif, relative to the permutation controls. Moreover Supplementary Figure 4 shows that most of the matches of KEPE and the permuted motifs are non-conserved in yeast. In addition, the number of matches to the KEPE motif in the yeast proteome is lower than expected (55) compared with the human proteome (331). This result is independent from the distributions of the K, P and E amino acids since their distributions are very similar in the human and yeast proteomes (Echols et al., 2002). Therefore, the difference in the number should depend only in the total sequence length of both proteomes. While the human to yeast proteome length ratio is 3.66, the ratio of the number of retrieved matches is nearly the double, 6.01. Thus, our protocol was unable to provide any evidence in favour of the existence of KEPE motifs in yeast. Manual screening of KEPE matches for plant, fungal and other non-metazoan protein entries in UniProt/Swiss-Prot likewise failed to provide evidence for plausible KEPE motifs. We surmise that KEPE motifs arose and proliferated in the metazoan lineage.
3.3 Three KEPE length variations
Inspection of the KEPE motif conservation in individual protein families showed that there are three length variants in the flexible gap after the P and preceding the last conserved negatively charged position. Typical KEPEs as in C/EBPs allow a 2–3 residue gap. Juns have longer variants and some Mafs have shorter variants (Supplementary Fig. 5). In all the alignments examined, the three variant motifs were never observed to interconvert during evolutionary change (although they are often found superimposed, e.g. in C/EBPα, NFATC1-3, TOP1, HDAC4, FOS, see Supplementary Table 3). This curious behaviour suggests that the length variants are functionally distinct, perhaps in a subtle way: for example, they could be recognized by different paralogous proteins; or they might all be recognized by the same protein but be modulated by interactions with distinct additional factors.
3.4 KEPE-bearing proteins
KEPE motifs are mostly found in transcription factors and proteins that are broadly involved in modifying chromatin conformation. Therefore, a role in modulating gene expression seems to be inevitable. The highest KEPE enrichment is in the leucine zipper class of transcription factor, where 30% possess the motif. As many KEPE sites are known to be sumoylated (Supplementary Table 3), a clear inference is that all KEPE sites are modified by sumoylation. The motif is sometimes found to be conserved in orthologous proteins for more then 500 million years, as in C/EBPs from Drosophila and vertebrates (Fig. 1B). However, in many paralogous gene families that originated with the genome expansion associated with the origin of the vertebrates (Gibson and Spring, 1998; Kasahara, 2007; Meyer and Van de Peer, 2005), KEPE motif evolution is much more dynamic. In the Fos transcription factor family, KEPE is conserved in cFos and Fra2 but absent from FosB and Fra1. It is found in HDAC4 and 9 but not in other histone deacetylases. There can be from 0 to 3 KEPE motifs in various NFATC transcription factor paralogues. Several Klf zinc-finger proteins have KEPEs in separate non-superposable locations: these motifs are likely to have independent origins by point mutation within large natively disordered polypeptide segments.
The KEPE motif is larger than the sequence conservation associated with sumoylation sites. It is possible that the additional conserved residues might be important to (i) be recognized by other binding proteins and/or (ii) in regulating the modification of the motif in other ways, e.g. by lysine acetylation, methylation or ubiquitinylation. [Thus, the tumour suppressor HIC1 can be sumoylated on a lysine which is also a target for acetylation, suggesting that this motif might represent a sumoylation/acetylation switch (Stankovic-Valentin et al., 2007).] The simplest model for KEPE function would be for a KEPE-binding protein to block access to the sumoylation site. Since sumoylation is reported to relieve transcriptional inhibition by the RDM element of C/EBP (Kim et al., 2002), unsumoylated KEPE should be bound by a protein that acts as a repressor (at least in this context). Since many of the KEPE proteins are assigned as chromatin modifiers, rather than as transcription factors per se, such a shared system of repression would be expected to be interlinked to chromatin conformational state. Experimental identification of the ligand proteins binding to the short, medium and long KEPEs may provide a new perspective on gene regulation.
4 CONCLUSIONS
LMs constitute nodes in cell regulatory networks that are acted upon by regulatory and signalling proteins and their domains. Here, we describe a new linear motif—KEPE—that is widespread in metazoan nuclear proteins classified as transcription factors or chromatin modulators. KEPE function is expected to regulate sumoylation, a proposal, which may be tested experimentally by biochemical and genetic means. Since KEPE is a common motif, elucidation of its function will have broad significance for understanding gene regulation in animals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the contributors to the ELM resource for making in silico linear motif discovery feasible, Pål Puntervoll, Rein Aasland and Manfred Koegl for checking interaction networks for any hints to the ligand, Evangelos Pafilis for help with the Ontology Lookup Service and Niall Haslam for critically reading the article.
Funding: EU EMBRACE (LHSG-CT-2004-512092).
Conflict of Interest: none declared.
REFERENCES
- Camon E, et al. The gene ontology annotation (GOA) database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chica C, et al. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9:229. doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley RR. The EH1 motif in metazoan transcription factors. BMC Genomics. 2005;6:169. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, et al. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural mechanisms underlying post-translational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Echols N, et al. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 2002;30:2515–2523. doi: 10.1093/nar/30.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, et al. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Spring J. Genetic redundancy in vertebrates: polyploidy and persistence of genes encoding multidomain proteins. Trends Genet. 1998;14:46–49. doi: 10.1016/s0168-9525(97)01367-x. [DOI] [PubMed] [Google Scholar]
- Girdwood DW, et al. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. The 2R hypothesis: an update. Curr. Opin. Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- Letunic I, et al. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Michael S, et al. Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Nerlov C. C/EBPs: recipients of extracellular signals through proteome modulation. Curr. Opin. Cell Biol. 2008;20:180–185. doi: 10.1016/j.ceb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Puntervoll P, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu C. SIRW: a web server for the simple indexing and retrieval system that combines sequence motif searches with keyword searches. Nucleic Acids Res. 2003;31:3771–3774. doi: 10.1093/nar/gkg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, et al. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta G, et al. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut SJ, et al. Pfam 10 years on: 10 000 families and still growing. Brief. Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Stankovic-Valentin N, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Gene_Ontology_Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_UniProt_Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts FZ. SUMO modification of proteins other than transcription factors. Semin. Cell Dev. Biol. 2004;15:211–220. doi: 10.1016/j.semcdb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Yang SH, et al. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, et al. A general user interface for prediction servers of proteins' post-translational modification sites. Nat. Protoc. 2006;3:1318–1321. doi: 10.1038/nprot.2006.209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.