Abstract
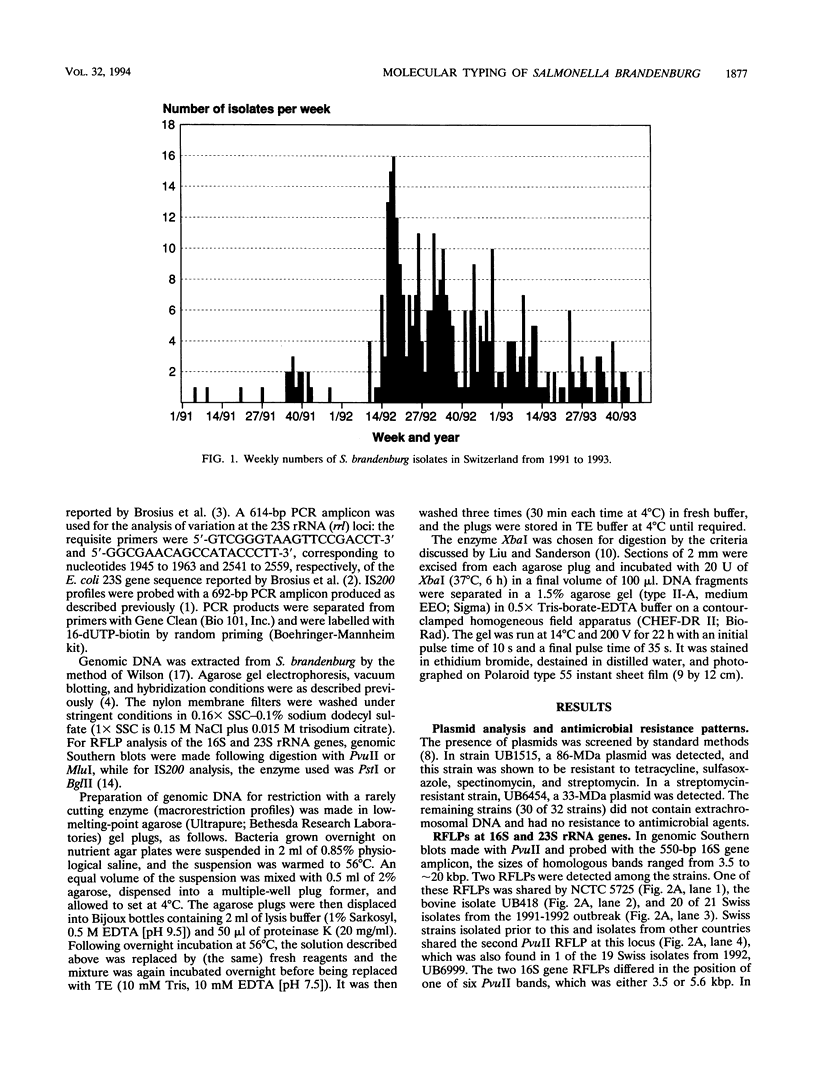
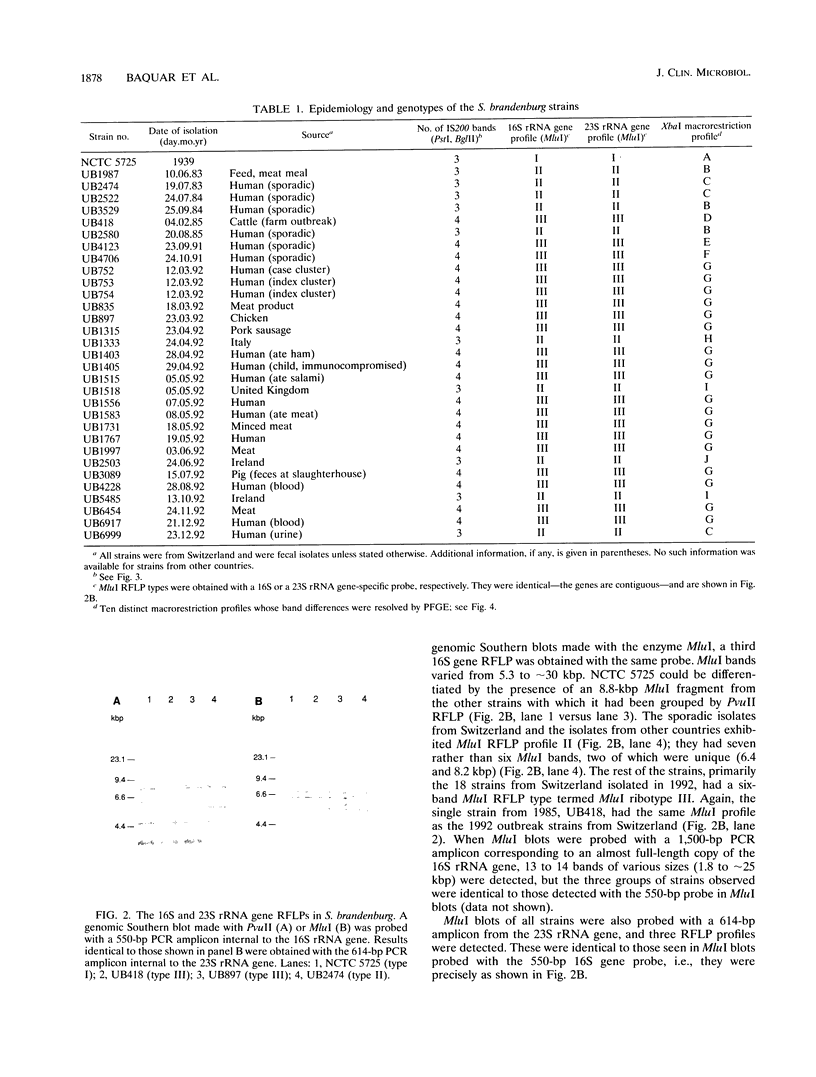
In Switzerland in 1992 there was a prolonged series of outbreaks of human salmonellosis caused by a previously rare serotype, Salmonella brandenburg. In order to examine the genotypic basis of the epidemic, molecular typing was applied to representative strains of this serovar isolated between 1983 and 1992. These included sporadic human isolates up to 1985, isolates from unrelated geographical areas, and Swiss isolates from humans, animals, and meat products isolated after 1991. Plasmid profiling was not found to be applicable to S. brandenburg, but chromosomal typing was accomplished by analyzing restriction fragment length polymorphisms with DNA probes for three marker loci; the 16S and 23S rRNA genes and sites of insertion of the mobile DNA element IS200. The macrorestriction profiles of the whole genome were examined by pulsed-field gel electrophoresis, which proved to be the most discriminatory of the typing methods. The study demonstrated the comparative value and complementary relationship between these typing methods for epidemiological purposes. All approaches concurred in identifying the 1992 isolates as a single genotypic clone, which was present in multiple (food) vehicles of infection. They were distinct from sporadic isolates of this serovar and from strains of S. brandenburg isolated in other countries.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baquar N., Threlfall E. J., Rowe B., Stanley J. Molecular subtyping within a single Salmonella typhimurium phage type, DT204c, with a PCR-generated probe for IS200. FEMS Microbiol Lett. 1993 Sep 1;112(2):217–221. doi: 10.1111/j.1574-6968.1993.tb06451.x. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdry N., Threlfall E. J., Rowe B., Stanley J. Genotype analysis of faecal and blood isolates of Salmonella dublin from humans in England and Wales. Epidemiol Infect. 1993 Apr;110(2):217–225. doi: 10.1017/s0950268800068138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein B. I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990 Apr;161(4):595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S., Roth J. R. IS200: a Salmonella-specific insertion sequence. Cell. 1983 Oct;34(3):951–960. doi: 10.1016/0092-8674(83)90552-4. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Sanderson K. E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992 Mar;174(5):1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslow J. N., Mulligan M. E., Arbeit R. D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993 Aug;17(2):153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- Stanley J., Baquar N., Threlfall E. J. Genotypes and phylogenetic relationships of Salmonella typhimurium are defined by molecular fingerprinting of IS200 and 16S rrn loci. J Gen Microbiol. 1993 Jun;139(Pt 6):1133–1140. doi: 10.1099/00221287-139-6-1133. [DOI] [PubMed] [Google Scholar]
- Stanley J., Burnens A., Powell N., Chowdry N., Jones C. The insertion sequence IS200 fingerprints chromosomal genotypes and epidemiological relationships in Salmonella heidelberg. J Gen Microbiol. 1992 Nov;138(11):2329–2336. doi: 10.1099/00221287-138-11-2329. [DOI] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moissac Y. R., Ronald S. L., Peppler M. S. Use of pulsed-field gel electrophoresis for epidemiological study of Bordetella pertussis in a whooping cough outbreak. J Clin Microbiol. 1994 Feb;32(2):398–402. doi: 10.1128/jcm.32.2.398-402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]





