Abstract
A new glutelin gene, designated GluD-1, has been discovered by comparing the seed storage proteins from 48 japonica and indica rice cultivars on SDS-PAGE gels. Evidence that GluD-1 is a member of the glutelin family was provided by Western blots using anti-glutelin antiserum and by mapping the gene to the chromosomal glutelin gene cluster. The limited GluD-1 size polymorphism among the rice varieties is due to amino acid substitutions rather than to post-transcriptional modification. GluD-1 is maximally expressed in the starchy endosperm starting at 5 d after flowering (DAF) and increasing through 30 DAF, a major difference from the other glutelins which are primarily expressed in the subaleurone from 10–16 DAF. Only about 0.2 kb of the GluD-1 promoter was sufficient to confer inner starchy endosperm-specific expression. The 0.2 kb truncated GluD-1 promoter contains a bifactorial endosperm box consisting of a truncated GCN4 motif (TGA(G/C)TCA) and AAAG Prolamin box (P box), and ACGT and AACA motifs as cis-regulatory elements. Gel retardation assays and trans-activation experiments indicated that the truncated GCN4 and P box are specifically recognized by RISBZ1 b-ZIP and RPBF Dof activators in vitro, respectively, and are synergistically transactivated, indicating that combinatorial interactions of these motifs are involved in essential endosperm-specific regulation. Furthermore, deviation from the cognate GCN4 motif alters tissue-specific expression in the inner starchy endosperm to include other endosperm tissues.
Keywords: bZIP, DOF, endosperm specific expression, glutelin, promoter, rice, seed storage protein, transgenic crop
Introduction
Rice is consumed as a staple food by about 60% of the world's population, and by more than 80% in Asian countries. Rice grain provides a major source of carbohydrates and proteins, especially in developing countries. Starch and storage proteins are predominantly deposited in the rice grain endosperm during seed maturation, and are used as a source of nitrogen and carbon for the germinating seedling.
Seed storage proteins (SSPs) can be grouped according to their physical properties based on solubility (Osborn, 1924). Albumins, globulins, prolamins, and glutelins make up the bulk of SSPs. In rice, glutelin is the major SSP, accounting for 60–80% by weight of total seed protein. Prolamin makes up about 20–30%, which is distinct from other cereal seeds, which have far more prolamin as a percentage of total protein. Glutelin and globulin are deposited in protein body II (PB-II) storage vacuoles, whereas prolamins accumulate in endoplasmic reticulum- (ER) derived protein body I (PB-I) structures that form within the lumen of the rough ER (Takaiwa et al., 1999). Rice glutelin originates from the same ancestral gene as 11S globulin. Rice glutelin is synthesized as a Mr 57 000 (57 kDa) precursor protein and is transported into the PB-II via ER and Golgi or directly from ER, where it is processed into mature Mr 37 000 (37 kDa) acidic and Mr 20 000 (20 kDa) basic subunits (Yamagata et al., 1982; Furuta et al., 1986; Krishnan and Okita, 1986).
Rice endosperm is an excellent platform for producing foreign recombinant proteins because rice crops are grown across very diverse agricultural regions, proteins can be produced in relatively abundant amounts, and targeted with considerable accuracy to specific tissues within the endosperm, and the proteins are stable at room temperature for long periods. A number of biopharmaceuticals, nutriceuticals containing health-promoting peptides, and mineral-binding peptides have been produced using rice endosperm as a bioreactor (Ye et al., 2000; Paine et al., 2005; Takagi et al., 2005a, b; Takaiwa, 2007; Takaiwa et al., 2007). The nutritional improvement of SSPs either by traditional breeding or by genetic engineering would be a substantial benefit to consumers because they are a major source of amino acids in the human diet, but are deficient in the essential amino acids lysine, threonine, and tryptophan. A parallel goal has been to reduce the production of native SSPs by mutation or gene suppression to allow for the production and accumulation of recombinant proteins, because low SSP mutants provide higher potential capacity for foreign gene products (Tada et al., 2003). Moreover, transgenes can be highly expressed in a specific tissue during a specific developmental stage by employing native SSP promoters (Qu and Takaiwa, 2004).
Many of the molecular mechanisms underlying endosperm-specific expression of cereal SSP genes have been determined. Several cis-elements, such as the prolamin box (P box: AAAG) and GCN4 (TGA(G/C)TCA), ACGT, and AACA motifs are conserved in many cereal SSP genes (Zheng et al., 1993; Takaiwa et al., 1996; Washida et al., 1999; Wu et al., 2000). Transcriptional factors that recognize these cis elements have also been isolated, such as Opaque2-like bZIP, Dof, and Myb proteins (Schmidt et al., 1992; Albani et al., 1997; Vicente-Carbajosa et al., 1997; Mena et al., 1998; Onate et al., 1999; Onodera et al., 2001; Diaz et al., 2002; Yamamoto et al., 2006). Understanding these gene regulation mechanisms in greater detail would provide useful information for fine-tuning the expression of recombinant products in transgenic crops.
In this study, a new glutelin was discovered by comparing the seed protein composition of various rice cultivars. The expression of this glutelin gene (GluD-1) has unique spatial and temporal features during seed development, although, like the other glutelins, its expression is endosperm-specific and GluD-1 is deposited in PB-II storage structures. GluD-1 was predominantly expressed in the inner starchy endosperm beginning about 5 DAF and steadily increased until maturity at 30 DAF. Such endosperm-specificity is controlled by combinatorial interactions of a GCN4-like motif and closely linked P box around position –200 from the ATG start site through binding to RISBZ1 and RPBF. Its spatial specificity may be caused by deviations within the GCN4 motif of the GluD-1 promoter.
Materials and methods
Plant materials
Forty-eight rice (Oryza sativa L.) accessions were obtained from the Genebank of the National Institute of Agrobiological Sciences (NIAS) (Table 1). Rice cultivars Nipponbare, Koshihikari, Nihonmasari, Kita-ake, Yamadanishiki, a-123, and Lgc-1, Koshihikari/Nona Bokra chromosomal segment substituted lines (CSSLs), and Nipponbare/Kasalath back-cross inbred lines (BILs) obtained from the Rice Genome Resource Center (RGRP) of NIAS (Lin et al., 1998; Takai et al., 2007), were also used in this study. Kita-ake and Nanjing 11 were crossed and progeny were cultivated in a greenhouse.
Table 1.
Rice accessions from Genebank of NIAS
| Lane no. | Stock no. | Accession name | Lane no. | Stock no. | Accession name |
| 1 | 6155 | Yoneshiro | 25 | 13530 | IR24 |
| 2 | 6612 | Shomokita | 26 | 12343 | Asominori |
| 3 | 6868 | Hatsukogane | 27 | 10484 | Wataribune 2 |
| 4 | 8479 | Kochihibiki | 28 | 10485 | Wataribune 3 |
| 5 | 12536 | Suweon 258 | 29 | 13503 | Kaohsiung 139 |
| 6 | 13070 | Guizhao 2 | 30 | 15910 | Saturn |
| 7 | 15539 | Dunghan Shali | 31 | 15884 | Dawn |
| 8 | 37853 | Tannemochi | 32 | 13953 | Nga Cheik |
| 9 | 60201 | India Dular | 33 | 15963 | Toro |
| 10 | 77001 | Fukei 158 | 34 | 12031 | Shinriki |
| 11 | 88629 | Kantou Mochi 157 | 35 | 10919 | Funakiomachi |
| 12 | 88632 | Kantou Mochi 160 | 36 | 7507 | Shigawatarifune 6 |
| 13 | 88636 | Kantou Mochi 164 | 37 | 8223 | Kiryouyoshi |
| 14 | 95756 | Fukei 161 | 38 | 42267 | Habiganj Boro 2 |
| 15 | 95758 | Fukei 163 | 39 | 16089 | Dourado Agulha |
| 16 | 81339 | Babutong | 40 | 6956 | Nan-Ei |
| 17 | 119182 | Taichung Native 1 | 41 | 13592 | Azucena |
| 18 | 13043 | Guangluai 4 | 42 | 13580 | BPI 76 |
| 19 | 13076 | Erjiuqing | 43 | 13524 | Raminad Strain 3 |
| 20 | 12930 | Nanjing 11 | 44 | 94590 | Siam 29 |
| 21 | 16082 | Dourado | 45 | 15594 | Bomba |
| 22 | 13622 | IAC25 | 46 | 15595 | Bomba |
| 23 | 16085 | Pratao Precoce | 47 | 15920 | H501 |
| 24 | 129373 | Niaw Sampa Tong | 48 | 15337 | H501 |
Lanes in Fig. 1 (Lane no.) and stock numbers in Genebank of NIAS (Stock no.) are indicated.
Seed protein extraction and Western blotting
Maturing or mature seeds were ground to a fine powder with a Multi-Beads Shocker (Yasui kikai, Osaka, Japan). Seed proteins were extracted with protein extraction buffer (50 mM TRIS-HCl (pH 6.8), 4% SDS, 8 M urea, 5% 2-mercaptoethanol, and 20% glycerol) by shaking vigorously for 2 h at room temperature. Extracts were then centrifuged at 15 000 rpm for 5 min to remove debris. The anti-glutelin antibody was kindly provided by Dr S Utsumi (Kyoto University). Anti-GluD peptide antibody was produced against the synthetic peptide RQEEHRQYQQVQYREGQY, which is in the C-terminal region of the acidic subunit between positions 267 and 284. Antibodies against GluA, GluB, and GluC were prepared as described previously (Takagi et al., 2006). Proteins were separated by SDS-PAGE and transferred to PVDF membranes (Millipore). The blots were blocked in TRIS-buffered saline plus Tween 20 TBST; 25 mM TRIS-HCl (pH 7.5), 150 mM NaCl, and 0.05% Tween 20 containing 5% skimmed milk, and then reacted with antibodies in blocking buffer. The blots were washed three times in TBST and incubated with anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody in blocking buffer. Signals were detected with an ECL kit (GE Healthcare).
Expression of recombinant GluD-1 acidic subunits in E. coli
The acidic subunit regions of GluD-1 were amplified from Nipponbare and Wataribune 2 cDNA by PCR with forward and reverse primers containing overhanging NcoI and BamI recognition sites, respectively. After NcoI and BamI digestion, PCR fragments were cloned into pET15b (Novagen). The expression of recombinant GluD-1 acidic subunits was induced using OvernightExpress (Novagen) according to the manufacturer's protocol.
RNA extraction and expression analysis
Total RNA from roots, shoot apices, and leaf blades was extracted with Trizol reagent (Invitrogen) according to the manufacturer's protocol. Total RNA from seeds at 5, 10, 15, 20, and 30 d after flowering (DAF) were extracted as described previously (Takaiwa et al., 1987). After RNase-free DNase (Takara) treatment, 2.0 μg of total RNA was reverse-transcribed using the oligo(dT) primer and SuperScript III (Invitrogen). The primer sets used were 5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCAGGCATCTG-3′ for ACTIN, and 5′-GGATTGACTTTTCCTGGTTGCC-3′ and 5′-TTACTCTTGCAGCACCCATTCC-3′ for GluD-1.
To produce PGluD-1:GUS constructs, the 5’ flanking regions of GluD-1 (–1679/–29, –1225/–29, –589/–29, –429/–29, and –231/–29) were amplified by PCR using forward and reverse primers containing overhanging SalI and SmaI recognition sites, respectively. PCR fragments were digested with SalI and SmaI, and cloned into pGPTV-HPT (Becker et al., 1992). The PGluD-1:GUS construct was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Rice transformation was performed as described by Goto et al. (1999). For histochemical analysis, seeds at 7, 10, and 15 DAF were sectioned with a razor blade and incubated in 50 mM sodium phosphate buffer (pH 7.0) containing 0.5 mM X-Gluc (5-bromo-4-chloro-3-indolyl glucuronide) and 20% methanol at 37 °C. Quantitative analysis of promoter strength was performed as described previously (Jefferson, 1987). Maturing seeds at 17 DAF were homogenized in GUS extraction buffer (50 mM NaPO4 [pH 7.0], 10 mM 2-mercaptoethanol, 10 mM Na2-EDTA, 0.1% SDS, 0.1% Triton X-100), and centrifuged. Ten μl of the supernatant was mixed with 90 μl GUS assay buffer (GUS extraction buffer with 1 mM 4-methyl-umbelliferryl-β-D-glucuronide (4MUG)) and incubated at 37 °C for 60 min. Reactions were stopped with 900 μl 0.2 M Na2CO3. A dilution series of 4-methylumbelliferone (4MU) was used as a standard. Protein concentrations were determined by the Bradford method with a Protein assay kit (Bio-Rad), using a bovine serum albumin (BSA) dilution series as the standard. Three or four seeds from each independent transgenic plant were assayed.
Phylogenetic analysis
A BLAST search for glutelin genes was carried out against the Nipponbare genomic sequence at RAP-DB (http://rapdb.dna.affrc.go.jp/). Multiple sequence alignments were performed using ClustalW (http://clustalw.ddbj.nig.ac.jp). A phylogenetic tree was constructed using a Neighbor–Joining method (TreeView programs) (Page, 1996). The number of times that each branch topology was found during bootstrap analyses was determined and are indicated at branch points (n=1000) (see Supplementary Fig. S1 at JXB online).
Transient expression assay
The PCR product of the 5′ flanking region of GluD-1 (–1679/–29) and (–239/–29) were cloned into pBI201 to construct PGluD-1:GUS. The GLM mutagenized forms, GCN4 and mGCN4, were amplified by overlapping PCR, and cloned into pBI201. PGluA-2 (–682/+11):GUS and PGluB-1(–2290/+44):GUS were constructed as described previously (Yamamoto et al., 2006). CaMV P35S:RISBZ1 and P35S:RPBF were constructed as effecters as described by Yamamoto et al. (2006). Transient expression in rice callus protoplasts and enzyme assays were performed by electroporation as previously described (Wu et al., 1998b).
Prediction of cis-elements
Putative cis-elements within the GluD-1 promoter were analysed using the Plant cis-acting regulatory DNA elements (PLACE) database (Higo et al., 1999).
Gel shift assay
The coding sequences of RISBZ1 and RPBF were amplified using forward and reverse primers containing overhanging BamI recognition sites. PCR products were inserted into pET15b (Novagen). His-tagged recombinant proteins were expressed in Rossetta-gami Escherichia coli cells (Novagen), then purified on a Ni-NTA spin column (Qiagen). GluD-1 (–239/–118) and GluB-1 (–199/+23) promoter fragments were amplified by PCR. P box-mutagenized forms were amplified by overlapping PCR. To generate double-stranded oligonucleotides, complementary single-stranded oligonucleotides were annealed. Primers were 5′-TATGGTATGAATCATTTCAAC-3′ for GLM-wt, 5′-TATGGTATGAgTCATTTCAAC-3′ for GLM-GCN4, and 5′-TATGGTATctagaATTTCAAC-3′ for GLM-mGCN4. Lower case letters indicate mutations. Probes were labelled with DIG dUTP using terminal transferase (Roche). DNA binding reactions were performed as described previously (Onodera et al., 2001).
Immunocytochemical observation
Immature 15 DAF seeds were fixed in 4% paraformaldehyde, 0.2% glutaraldehyde, and 50 mM sodium phosphate buffer (pH 7.2) overnight at 4 °C. After washing in sodium phosphate buffer, the samples were dehydrated in a graded ethanol series and embedded in LR White resin (London Resin). Ultrathin sections, prepared using an ultramicrotome and mounted on nickel grids, were blocked in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2HPO4) containing 1% BSA, then incubated with anti-GluD-1 rabbit antibody. Grids were washed in washing buffer (PBS containing 0.1% BSA, 0.5 M NaCl, and 0.05% Tween 20) and incubated with gold-labelled goat anti-rabbit IgG antibody in PBS containing 1% BSA. Grids were rewashed and fixed with PBS containing 1% glutaraldehyde, rinsed twice with distilled water, and stained with uranyl acetate and lead citrate. Sections were observed with a transmission electron microscope (H-7100, Hitachi) at an accelerating voltage of 75 kV.
Results
Seed storage proteins
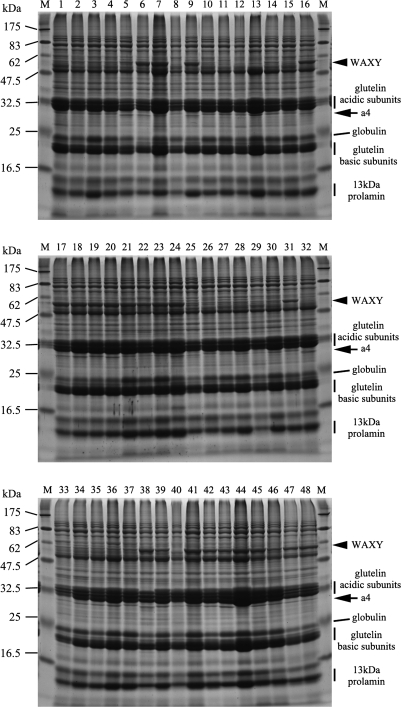
Mature seed proteins extracted from 48 japonica and indica accessions were separated by SDS-PAGE (Fig. 1). Irrespective of variations in relative protein amounts contained in a grain, the composition of total seed proteins as manifest by their SDS-PAGE banding patterns were nearly identical, except for two bands with molecular weights (MW) of approximately 60 kDa and 28 kDa. The c. 60 kDa band is thought to be the granule-bound starch synthase (GBSS) known as WAXY. High levels of WAXY expression are characteristic of indica varieties, because WAXY is absent or is expressed at very low levels in nearly all japonica varieties (Sano, 1984). Interestingly, the c. 28 kDa band runs just below the major acidic subunits of glutelin and has some heterogeneity in size among the japonica cultivars. This band has been observed previously, and tentatively assigned to the glutelin family (‘a4’; (Jahan et al., 2005)).
Fig. 1.
SDS-PAGE analysis of seed protein composition among 48 rice accessions. Analysed accessions are listed in Table 1. Glutelin acidic/basic subunits, globulin, and 13 kDa prolamins are indicated at the right of the panels. 60 kDa (WAXY) and 28 kDa (a4:GluD-1) proteins are indicated by arrows.
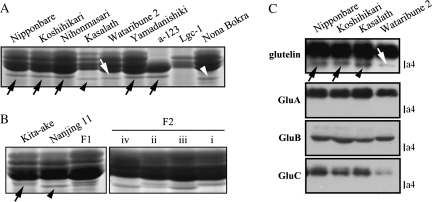
Japonica cultivars Nipponbare, Koshihikari, Nihonmasari, Yamadanishiki, and others produce a larger a4 protein (a4J) than indica cultivars such as Kasalath (a4I) (Fig. 2). Among japonica cultivars, Wataribune 2, Wataribune 3, Funakiomachi, and Shigawataribune produce an intermediate-sized a4 (a4M) (Fig. 2). Nona Bokra had the smallest a4 (a4N; Fig. 2). Thus, at least four allelic variations in size were observed for a4.
Fig. 2.
SDS-PAGE analysis and Western blot analysis of allelic polymorphisms of the a4 protein band. a4J, a4I, a4M, and a4N are indicated by black arrows, black arrowheads, white arrows, and white arrowheads, respectively. (A) Allelic polymorphisms of the a4 band among cultivars Nipponbare, Koshihikari, Nihonmasari, Kasalath, Wataribune 2, Yamadanishiki, and Nona Bokra; and the a-123 and Lgc-1 glutelin mutants. (B) Allelic interactions between a4J and a4I. F1 and F2 seeds of a cross between Kita-ake (a4J) and Nanjing 11 (a4I) were analysed. F2 seeds segregated into one of four patterns. (C) Western blot analysis of glutelins with total seed protein from Nipponbare, Koshihikari, Kasalath, and Wataribune 2 seeds.
Allelic relationship between genes encoding a4
All of the F1 progeny seed from a cross between Kita-ake (a4J) as female and Nanjing 11 (a4I) as male had both a4J and a4I, and their accumulation levels were similar (Fig. 2B). In the F2 population, a4 expression patterns fell into four classes: (i) only a4J was expressed; (ii) a4J and a4I were expressed at similar levels; (iii) a4J and a4I were both expressed, but with a4I at a higher level; and (iv) only a4I was expressed (Fig. 2B). Since endosperm is triploid tissue, the difference between (ii) and (iii) results from the dosage effect. The segregation ratio of these expression classes in F2 populations was 37, 31, 22, and 32, respectively, fitting to the expected ratio of 1:1:1:1 (χ2=3.06). Thus, the a4 protein is controlled by a single incomplete dominant gene.
Identification of a new glutelin
In order to determine whether the a4 band protein is a new type of glutelin, the glu-1, glu-2, and glu-3 glutelin triple mutant (α-123), which is defective in the production of GluA-1, GluA-2, GluB-4, and GluB-5 (Iida et al., 1997), and the Low glutelin content1 (Lgc1) mutant, which is a dominant glutelin mutant due to RNA silencing (Kusaba et al., 2003), were examined for the presence of a4. α-123 produced a4J protein, indicating that it is not GluA-1, GluA-2, GluB-4 or GluB-5. There was no a4 band in Lgc1, but a4J was produced by cultivar Nihonmasari, which is the wild-type background of Lgc1 (Fig. 2A), suggesting that a4 is a glutelin. In Western blots with anti-glutelin, anti-GluA, anti-GluB, and anti-GluC antisera, anti-glutelin antibody reacted with a4J in Nipponbare and Koshihikari, a4I in Kasalath, and a4M in Wataribune 2 (Fig. 2C). By contrast, specific antibodies against GluA, GluB, and GluC did not recognize the a4 band (Fig. 2C). Taken together, a4 was identified as a different and new type of glutelin.
Mapping and molecular identification of the glutelin a4 gene
Since the size of a4 is consistently different between japonica and indica varieties, advantage was taken of chromosomal segment substituted lines (CSSLs) and back-cross inbred lines (BILs) constructed between japonica and indica cultivars. Among the 44 Koshihikari/Nona Bokra (Kos/Nona) CSSLs and their parents, Nona Bokra, SL504, SL505, and SL506 had a4N (data not shown), and only SL504, SL505, and SL506 carried a Nona Bokra segment defined by markers RM5897 and RM1313 on Chromosome 2. From a comparison of graphical genotypes and a4 phenotypes of 98 Nipponbare/Kasalath/Nipponbare (Nip/Kas/Nip) BILs (data not shown), the a4 gene mapped within a segment bounded by markers G227 and R712 (see Supplementary Fig. S2A at JXB online; http://www.gramene.org). Further genotyping indicated that the a4 gene was located between markers RM145 and G227, which spanned approximately 1.3 Mb (see Supplementary Fig. S2A at JXB online).
A BLAST search of the complete Nipponbare genome sequence indicated that there are 15 glutelin genes in the rice genome (Table 2) that can be further classified into four subfamilies, GluA, GluB, GluC, and GluD, by phylogenetic analysis (see Supplementary Fig. S2B at JXB online). GluB-1a, GluB-1b, GluB-2, GluB-6, and GluD-1 are arranged in a cluster in the 1.3 Mb region bounded by markers RM145 and G227. Since GluB-1 and GluB-2 were ruled out as possibilities for a4 as described above, the genomic sequences of GluB-6 and GluD-1 were compared between japonica cultivars Nipponbare (a4J) and Wataribune2 (a4M). There were no polymorphisms in the coding region of GluB-6 between these cultivars (data not shown). There was, however, a SNP (A173T) in the acidic subunit region of GluD-1 which results in an amino acid substitution from Asp (Nipponbare) to Val (Wataribune 2) (see Supplementary Figs S3 and S4 at JXB online), and several additional SNPs between Kasalath and Nona Bokra (see Supplementary Figs S3 and S4 at JXB online). Anti-GluD antibody reacted with both a4J and a4M, indicating that a4 represents GluD-1 (Fig. 3).
Table 2.
Rice glutelin genes
| Gene | Chr | RAP locus | Accession number |
| GluA-1 | 1 | Os01g0762500 | M17513, X05662, X05661 |
| GluA-2 | 10 | Os10g0400200 | AK107314, X05664, X06149, X05663 |
| GluA-3 | 3 | Os03g0427300 | AK107271, M28159 |
| GluA-4 | 1 | Pseudogene | – |
| GluB-1a | 2 | Os02g0249800 | AK107343, X14568 |
| GluB-1b | 2 | Os02g0249900 | X15833 |
| GluB-2 | 2 | Os02g0249600 | X54192 |
| GluB-3 | 2 | Pseudogene | X54193 |
| GluB-4 | 2 | Os02g0268300 | X14393 |
| GluB-5 | 2 | Os02g0268100 | AK107238 |
| GluB-6 | 2 | Os02g0248800 | AY429651 |
| GluB-7 | 2 | Os02g0242600 | AY196923 |
| GluC-1 | 2 | Os02g0453600 | AK064478 |
| GluC-2 | 2 | Pseudogene | – |
| GluD-1 | 2 | Os02g0249000 | AY429650 |
Located chromosomes (Chr), rice annotation project locus (RAP locus), and Accession numbers are indicated.
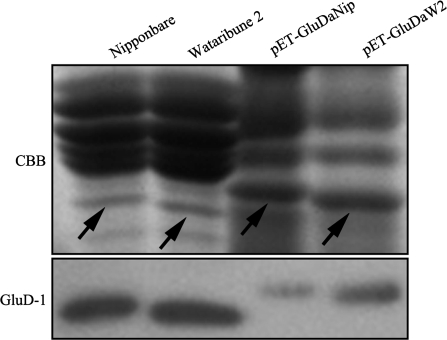
Fig. 3.
Western blot analysis and bacterial expression of the GluD-1 acidic subunit. Upper panel: SDS-PAGE of native extracts from Nipponbare and Wataribune 2 seeds, and products expressed from pET-GluDaNip and pET-GluDaW2. Lower panel: Western blot analysis with anti-GluD. Arrows indicate GluD-1 acidic subunits.
Bacterial expression of the GluD-1 acidic subunit
To examine the possibility that differences in SDS-PAGE mobility were caused by post-translational modifications, the GluD-1 acidic subunits, including signal peptides, from cvs Nipponbare (GluD1aNip) and Wataribune 2 (GluD1aW2) were expressed in E. coli cells. The expressed proteins were slightly larger than the native GluD-1 acidic subunits in rice seeds due to the presence of a signal peptide (Fig. 3). Western blots were used to confirm that these proteins were GluD1aNip and GluD1aW2 (Fig. 3, lower panel). Because a difference in gel migration between GluD1aNip and GluD1aW2 was observed in both bacterial expression products and in seed products, the difference in gel mobility is most likely not due to post-translational modification, but rather from the single substituted amino acid.
Tissue-specific expression of GluD-1
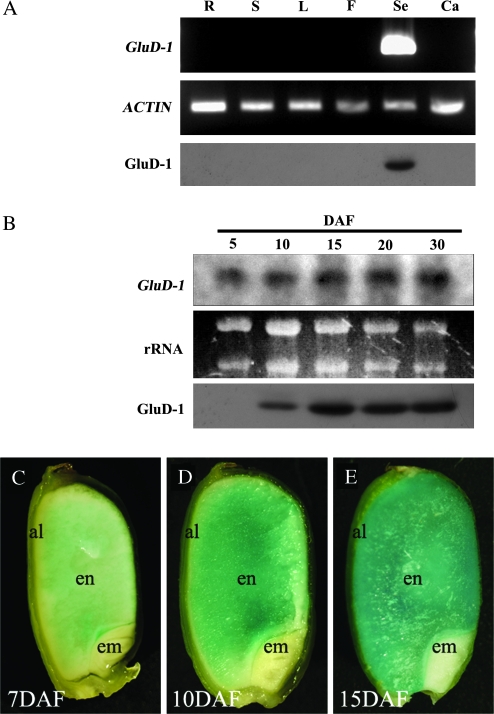
The expression of GluD-1 in plant organs was determined by RT-PCR from total RNA extracted from root (R), shoot apex (S), vegetative leaf (L), flower (F), and seed at 15 DAF (Se), and from callus (Ca). GluD-1 was specifically expressed in developing seed (Fig. 4A). GluD-1 expression was detectable by Northern blotting in seed tissue at 5 DAF with increasing intensity through 30 DAF (Fig. 4B). This spatial- and temporal-specific expression was further confirmed by Western blotting using anti-GluD-1 antibody. GluD-1 was specifically detected in mature seed, and its expression pattern was slightly different from transcription. GluD-1 was detectable by Western blotting at 10 DAF, and reached a plateau at 15 DAF (Fig. 4B).
Fig. 4.
Temporal and spatial expression patterns of GluD-1. (A) Organ-specific expression pattern of GluD-1 by RT-PCR, and GluD-1 by Western blot analysis. R, root; S, shoot apex; L, leaf blade; F, flower; Se, seed at 15 d after flowering (DAF); Ca, callus. (B) Temporal expression pattern during seed maturation of GluD-1 by Northern blot analysis and GluD-1 by Western blot analysis. Numerals indicate sampling stage. (C–E) GUS expression driven by the 1.6 kb GluD-1 promoter during seed maturation, at 7, 10, and 15 DAF, respectively. Al, aleurone and subaleurone layers; en, starchy endosperm; em, embryo.
Furthermore, to evaluate the spatial and temporal distribution of GluD-1 within the endosperm, transgenic rice plants harbouring the GUS reporter gene under the control of the 1.6 kb GluD-1 promoter were generated. GUS activity was observed in the inner starchy endosperm rather than in the aleurone and subaleurone layers by 7 DAF (Fig. 4C). GUS expression was less specific as the seed developed, and was found throughout the endosperm, including the aleurone and subaleurone layers by 15 DAF (Fig. 4D, E). No GUS activity was detected in either the embryos of maturing seed or in vegetative tissues (data not shown).
Regulatory domains of the GluD-1 promoter for endosperm-specific expression
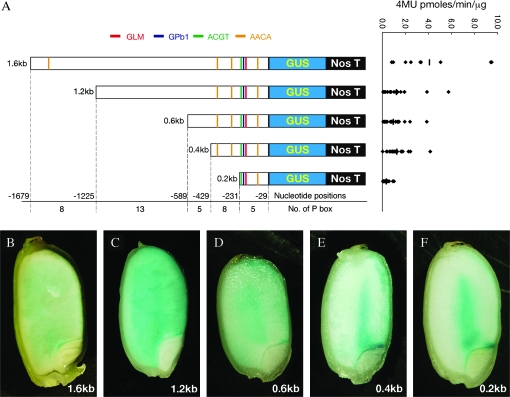
A search was made for the four SSP cis elements (GCN4, prolamin box (P box), ACGT, and AACA motifs) known to be responsible for endosperm expression using the Plant cis-acting regulatory DNA elements (PLACE) program. The 1.6 kb GluD-1 promoter contains one GCN4 like motif (GLM), 39 P boxes, one ACGT, and four AACA motifs (Fig. 5A; Table 3). It should be noted that the GCN4-like motif (GLM) around position –200 is flanked by three P boxes (Pb1, Pb2, and Pb3), constituting an ‘Endosperm box’, usually containing a GCN4 motif and a P box separated by within 10 nucleotides, as has been found in many cereal prolamin promoters such as zein, hordein, and glutenin (Hammond-Kosack et al., 1993; Muller and Knudsen, 1993; Marzabal et al., 1998).
Fig. 5.
Truncation analysis of the GluD-1 promoter. (A) Schematic representation of truncated promoter:GUS constructs for generating transgenic rice (left), and expression strength (right). Promoter lengths are indicated on the left. Negative numerals indicate the nucleotide position relative to the translational start site. Putative cis-elements predicted by the PLACE database and this study, are indicated by colour bars. Red, blue, green, and orange bars represent GCN4-like motif (GLM), Prolamin box (P box) Pb1, ACGT, and AACA motifs, respectively. Numbers of P box within truncated regions are indicated. Promoter activities at 17 DAF are expressed as 4MU pmol min−1 μg−1 protein. Diamonds represent individual transgenic lines, and bars represent mean values. (B–F) GUS expression at 7 DAF, driven by 1.6 kb, 1.2 kb, 0.6 kb, 0.4 kb, and 0.2 kb GluD-1 promoters, respectively. X-gluc staining reactions were carried out for 2 h (B), and for overnight (C–E), depending on GUS activity.
Table 3.
Predicted cis-elements in the 1.6 kb GluD-1 promoter
| cis element | Consensus | Related TF type | Positions relative to translation start site |
| GLM | TGA(G/C)TCA | bZIP | –194/–188a |
| P box | AAAG | DOF | –1640/–1637, –1620/–1617, –1496/–1493, –1490/–1487, –1426/–1423, –1393/–1390, |
| –1281/–1278, –1267/–1264, –1210/–1207, –1204/–1201, –1191/–1188, –1130/–1127, | |||
| –1118/–1115, –1056/–1053, –927/–924, –907/–904, –839/–836, –833/–830, –802/–799, | |||
| –706/–703, –684/–681, –590/–587, –583/–580, –551/–548, –486/–4832, –477/–474, | |||
| –401/–398, –369/–366, –349/–346, –343/–340, –335/–332, –328/–325, –281/–278, | |||
| –252/–249, –207/–204b, –174/–171, –157/–154, –82/–79, –75/–72 | |||
| ACGT | ACGT | bZIP | –214/–211 |
| –1500/–1495, –639/–634, –387/–382 | |||
| AACA | AACAAA | Myb | –110/–105 |
Recoginized by RISBZ1.
Recoginized by RPBF.
To characterize the GluD-1 regulatory regions required for endosperm-specific expression further, loss-of-function experiments by progressive truncations from the 5′ end of the 1.6 kb GluD-1 promoter were used to show that even when deleted to position –231, GUS activity in maturing seed was limited to the inner starchy endosperm close to the scutellum and in the peripheral aleurone and subaleurone layers of the abaxial side at an early maturating stage (Fig. 5B–F). Since the 0.2 kb upstream region is sufficient to confer GUS reporter expression, the essential regulatory elements controlling GluD-1 expression in the starchy endosperm are present within this region. The average GUS activities with upstream regions of 1.6 kb, 1.2 kb, 0.6 kb, 0.4 kb, and 0.2 kb at 17 DAP were 4.1±3.3, 1.2±1.4, 0.9±1.0, 1.3±0.9, and 0.3±0.3 pmoles 4MU min−1 mg−1, respectively (Fig. 6A, right panel), indicating that the regions –1679/–1225, and –429/–231 contain positive regulatory elements.
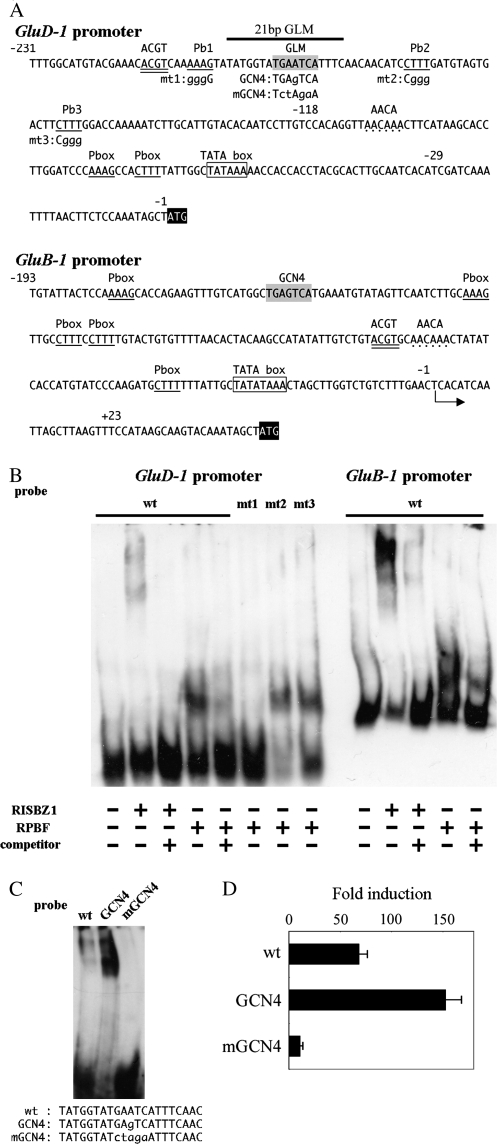
Fig. 6.
Electro-Mobility Shift Assays (EMSA) of recombinant RISBZ1 and RPBF with the GluD-1 and GluB-1 promoters, and transient expression analysis of 0.2 kb PGluD-1:GUS and GLM-mutated PGluD-1:GUS constructs. (A) Sequence of the 0.2 kb GluD-1 promoter and 0.2 kb GluB-1 promoter. Numerals indicate the nucleotide position from the translational start site for the GluD-1 promoter, and from the transcriptional start site for the GluB-1 promoter. The GCN4-like motif (GLM) is shaded in grey. Prolamin boxes (P box) are underlined. Three P boxes flanking the GLM, (Pb1, Pb2, and Pb3) are numbered. Mutant forms for loss-of-function and gain-of-function analyses are indicated by lower-case letters just below the mutated positions. The ACGT motif is double underlined. The AACA motifs are indicated by dots. A putative TATA box is framed, and the ATG start codon is shaded in black. The 21 bp oligonucleotides probe region (21 bp GLM) for EMSA is indicated. (B) EMSA of RISBZ1 and RPBF with the GluD-1 promoter, P box-mutated GluD-1 promoters, and GluB-1 promoter. Competition experiments are performed using 100-fold excess molar unlabelled wild-type probes. DIG-labelled probes are indicated above each lane. (C) EMSA of RISBZ1 with 21 bp GLM oligonucleotides and mutagenized forms within GluD-1 promoter. The oligonucleotide probes used were indicated below the panel. (D) Transient expression analysis of 0.2 kb PGluD-1:GUS and GLM-mutated PGluD-1:GUS constructs. CaMV P35S:RISBZ1 as an effecter and 0.2 kb PGluD-1:GUS (wt), its gain of function version (GCN4) and loss of function version (mGCN4) as reporters are co-transfected into callus-derived protoplasts. Trans-activation levels were represented as fold induction, relative to expression of a GUS reporter gene from each promoter without any effecter.
Trans-activation of the GluD-1 promoter by RISBZ1 and RPBF
Rice SSP genes are synergistically trans-activated by RISBZ1 and RPBF (Yamamoto et al., 2006). To determine whether GluD-1 expression is regulated by these factors, we performed transient expression assays using rice callus protoplasts. Three GUS reporter constructs fused to 0.7 kb GluA-2, 2.3 kb GluB-1, and 1.6 kb GluD-1 promoters were used to compare GUS activities upon transfection with RISBZ1 and RPBF activators. The activation of GluA-2 (328-fold) by RISBZ1 was much greater than GluD-1 (38-fold) and GluB-1 (68-fold) (Table 4). By contrast, the activation of GluB-1 (153-fold) and GluD-1 (89-fold) by RPBF was much higher than that of GluA-2 (13-fold) (Table 4). Co-transfection of RISBZ1 and RPBF resulted in synergistic activation of all three glutelin promoters. However, the synergistic effect of both effectors on the GluD-1 promoter (867-fold) was much lower than on the GluA-2 (1006-fold) and GluB-1 promoters (2549-fold) (Table 4).
Table 4.
Transient expression of glutelin promoter:GUS reporter constructs
| Reporter | Effector |
||
| (Promoter:GUS) | RISBZ1 | RPBF | RISBZ1+RPBF |
| GluA-2 | 32±48.8 | 13±0.9 | 1006±105.4 |
| GluB-1 | 62±5.9 | 153±33.8 | 2549±419.5 |
| GluD-1 | 38±3.2 | 89±6.4 | 867±60.2 |
Trans-activation levels were represented as fold induction ±SD, relative to expression of a GUS reporter gene from each promoter without any effecter.
Interactions between the GluD-1 promoter and RISBZ1 and RPBF
Gel-retardation assays were used to determine whether RISBZ1 and RPBF bind directly to the GLM and the P boxes in the GluD-1 promoter (–231/–118) (Fig. 6A). Retarded protein bands were observed after incubation with RISBZ1 or RPBF (Fig. 6B). A 100-fold molar excess of non-labelled competitor abolished these retarded bands, indicating that the GluD-1 promoter is specifically recognized by RISBZ1 and RPBF. The retardation effect associated with RISBZ1 was apparently weaker than with RPBF, and equal amounts of protein incubated with a DIG-labelled GluB-1 promoter (–199/+23) fragment gave much stronger retardation by RISBZ1 than with GluD-1. To test the specific recognition of the GLM by RISBZ1, interactions between RISBZ1 and 21 bp oligonucleotides containing the GLM within the GluD-1 promoter (wt), the typical GCN4 (gain of function; GCN4) or the altered GCN4 (loss of function; mGCN4), were analysed (Fig. 6C). Incubation of RISBZ1 and the 21 bp GLM-GCN4 containing the cognate GCN4 motif (TGAGTCA), which is present in GluB1 promoter and is known to be recognized by RISBZ1, strengthened the retardation band. By contrast, use of the 21 bp GLM-mGCN4 containing mutated GCN4 abolished retardation, indicating that RISBZ1 specifically recognized the GLM of the GluD-1 promoter, but with low affinity. This phenomenon was confirmed in vivo by transient expression assays using rice callus protoplasts. The activation of 0.2 kb PGluD-1:GUS with intact GLM by RISBZ1 (69-fold) was enhanced by gain of function (GCN4: 153-fold), and significantly decreased by loss of function (mGCN4: 16-fold) (Fig. 6D). To determine which P box was recognized by RPBF, interactions between RPBF and three fragments with altered P boxes were tested (Fig. 6B). Retardation bands were detected when RPBF was incubated with mt2 and mt3, but not with mt1, indicating that RPBF specifically recognizes P box Pb1.
Subcellular localization of GluD-1
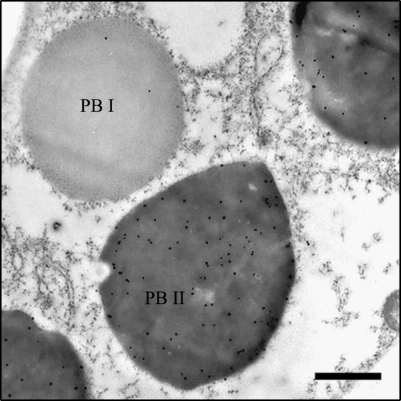
Rice SSPs are contained in ER-derived protein body I (PB-I) structures or in storage vacuoles (PB-II). The two types of PBs can be distinguished by their size, electron density, and shape. Glutelins are predominantly localized in high density, irregularly-shaped PB-II, and prolamins are found in the low density, round PB-I bodies. Gold particle-labelled anti-GluD antibodies were mainly distributed in PB-II (Fig. 7), indicating that GluD-1 is deposited in PB-II like other glutelins.
Fig. 7.
Intracellular localization of GluD-1 in the maturing seeds detected by immunoelectron microscopy. PB I and the high-density, irregularly shaped PB II are indicated. Twenty-five-nanometer (25 nm) gold particles labelled with anti-GluD-1 antibody are predominantly distributed in PB-II. Bars=1μ m.
Discussion
Rice seed endosperm is a good platform, not only for the production of recombinant proteins but also for nutriceutical delivery (Takaiwa, 2007; Takaiwa et al., 2007). Most SSP genes are specifically and highly expressed in seed, and their promoters have been used to express a number of transgenes in seed (Wu et al., 1998a; Qu and Takaiwa, 2004). Glutelin, accounting for 60–80% of rice seed protein, is synthesized as a 57 kDa precursor on rough ER, then transported into PB II storage protein vacuoles either via the Golgi apparatus, or directly, where it is processed into 37 kDa acidic and 20 kDa basic subunits (Takaiwa et al., 1999). An extensive analysis of the complete rice genome sequence indicates that there are at least 11 glutelin genes (Katsube-Tanaka et al., 2004). However, only GluA-1, GluB-4, and GluB-5 have been conclusively associated with their corresponding genes (Kusaba et al., 2003; Qu et al., 2003; Morita et al., 2007).
In this study, a glutelin with a 28 kDa acidic subunit was found to accumulate in seed. This protein, named here as GluD-1, has the smallest of the glutelin acidic subunits, and GluD-1 exhibits a few size polymorphisms among the indica and japonica varieties tested (Fig. 2). Furthermore, 28 SNPs were detected in the GluD-1 DNA sequences of four japonica and indica cultivars. The GluD-1 SNP frequency between Nipponbare and Kasalath was about 1.9%, more than double the 0.7% for the whole rice genome (Nasu et al., 2002). The amino acid (aa) sequence deduced from GluD-1 is composed of 484 aa and has c. 60% homology to GluA, c. 70% homology to GluB, and c. 50% homology to GluC. The GluD-1 aa sequence is similar to members of the GluB subfamily, but sequence similarity to GluB members was less than among GluB subfamily proteins (largely, over 80%). Thus, GluD-1 was classified into a separate group (see Supplementary Fig. S2B at JXB online).
As shown by Southern blot analysis, members of the rice GluB subfamily form a gene cluster on chromosome 2 (Okita et al., 1989; Takaiwa and Oono, 1991b). The GluB cluster is arranged in the order of GluB-6, GluD-1, GluB-2, GluB-1a, and GluB-1b. Tandem gene arrangements have been reported for several SSP genes. Prolamin-type SSP genes, encoding such as maize zein, rice 13 kDa prolamin, and wheat gliadin, are well known to form gene clusters (Reeves and Okita, 1987; Kim and Okita, 1988; Song et al., 2001).
GluD-1 was specifically expressed in maturing endosperm like other glutelin genes. However, both temporal and spatial expression patterns of GluD-1 within the endosperm were different from other glutelin genes. During seed development, mRNA levels of GluD-1 gradually increased at least until the mature stage 30 DAF, which is in contrast to the mRNA levels of many other glutelin genes, which start to increase at 5 DAF, reach a peak between 10 and 16 DAF, and then gradually decrease during seed development (Okita et al., 1989; Takaiwa and Oono, 1990; Takaiwa and Oono, 1991a; Takaiwa et al., 1991). Differential temporal expression patterns have also been observed among the soybean glycinin and maize zein families (Domoney and Casey, 1987; Kirihara et al., 1988). Furthermore, the 1.6 kb GluD-1 promoter directed GUS expression in the inner starchy endosperm at an early maturing stage, which would be complementary to other glutelin gene promoters (Qu and Takaiwa, 2004).
Because of the current initiatives to exploit plants as bioreactors for the production of various pharmaceutical and industrial recombinant proteins (Takaiwa, 2007), it has become apparent that several promoters could be used to drive expression in the starchy endosperm. The Glb-1 promoter, in particular, gives quite strong expression in the starchy endosperm (i.e. 43.5 [4MU pmol min−1 μg−1 protein]) (Wu et al., 1998a). However, expression of recombinant products by the Glb-1 promoter sometimes causes detrimental effects on growth, because Glb-1 is also expressed in vegetative tissues (Wu et al., 1998a). GluD-1, however, is specifically expressed in the endosperm, making it more suitable for expressing recombinant proteins in the starchy endosperm. The RAG-1 promoter also drives expression only in the starchy endosperm, but its expression is lower than that of the GluD-1 promoter (approximately 0.9 versus 4.1 [4MU pmol min−1 mg−1 protein]) (Wu et al., 1998a).
Several cis-elements, such as GCN4, prolamin box (P box), ACGT, and AACA motifs are conserved among various cereal SSP gene promoters and are involved in endosperm-specific expression (Zheng et al., 1993; Washida et al., 1999; Wu et al., 2000). In rice, the GCN4 motif in the proximal region of the GluB-1 promoter is recognized by RISBZ1 activator and acts as a critical cis-element for the expression in the aleurone and subaleurone layers, because multimers of the GCN4 motif can direct endosperm-specific expression (in the aleurone and subaleurone layers) in stable transgenic rice (Wu et al., 1998b; Wu et al., 2000; Onodera et al., 2001). On the other hand, multimers of the P box, ACGT, and AACA motifs cannot direct endosperm-specific expression, indicating that these elements may be involved in quantitative regulation by combinatorial interaction with the GCN4 motif in the endosperm (Wu et al., 2000). Furthermore, since mutations in individual motifs in a minimal GluB-1 promoter for the endosperm-specific expression abolished or severely suppressed GUS expression in endosperm, it has been suggested that combinatorial interactions of these four motifs are required for expression to take place in the endosperm (Wu et al., 1998b, 2000). Like these other promoters such as GluA-2 and GluB-1 promoter, the GluD-1 promoter directed endosperm-specific expression, but predominantly in the inner starchy endosperm, even though four cis-elements, essential for endosperm-specific glutelin gene expression, are present in the GluD-1 promoter. 0.2 kb of the GluD-1 promoter, which contains all four cis-elements, was sufficient to confer endosperm-specific expression (Fig. 6A).
Gel retardation assays and trans-activation experiments clearly demonstrate that the GCN4-like motif and P box in the GluD-1 promoter region –231/–29 were specifically recognized by RISBZ1 and RPBF (Fig. 6B, C; Table 3). It is important to note that activation of the GluD-1 promoter by RISBZ1 was lower than activation of the GluA-2 and GluB-1 promoters, a difference which can be accounted for by low affinity binding due to deviations in the GCN4 motif sequence from the cognate GCN4 (i.e. TGA(G/C)TCA to TGAATCA), resulting in poor RISBZ1 recognition of the GluD-1 promoter GLM (Fig. 6). The expression site also changed to the inner starchy endosperm when the mutation was introduced into the GCN4 motif in the 197 bp GluB-1 promoter to include aleurone and sub-aleurone layer-specific expression (Wu et al., 1998). It has recently been shown that co-suppression of RPBF in maturing seed also decreased GluD-1 expression, whereas co-suppression of RISBZ1 had little effect on GluD-1 expression, indicating that RPBF contributes significantly to GluD-1 expression in planta (MP Yamamoto et al., unpublished results). Notably, knockout/knockdown mutants of transcription factors often exhibit only subtle phenotypes, due to functional redundancy of transcription factors that belong to the same family (Hiratsu et al., 2003; Zhang, 2003). It has been reported that RISBZ2-5, related to RISBZ1, are also expressed in the endosperm (Onodera et al., 2001). Although these other bZIP proteins trans-activate the expression from GCN4 much less than RISBZ1, it is possible that they participate in the regulation of GluD-1 expression and compensate for RISBZ1 absence. Furthermore, rice allergenic RAG-1 and Glb-1 promoters are expressed in the inner starchy endosperm. There is no cognate GCN4 motif in these promoters, although truncated GCN4 motifs in the Glb-1 promoter are recognized by RISBZ1 in vitro. Furthermore, these promoters are not activated by RISBZ1, or the expression level is relatively low (Yamamoto et al., 2006). Taken together, the absence or truncation of the GCN4 motif from the minimal proximal promoter, which is required for endosperm-specific expression, may be associated with the observed changes in expression pattern from the aleurone and sub-aleurone to inner starchy endosperm. Another possible explanation is that an alternative motif may act as a substitute to confer expression in the starchy endosperm by interacting with other essential cis-elements, since combination among P box, AACA, and ACGT motifs was not sufficient to confer endosperm-specific expression.
On the other hand, 5′ deletion analysis of the GluD-1 promoter indicated that –1679/–1225 and –429/–231 contain enhancer domains (Fig. 5). Within these regions, one AACA (–1500/–1495) and eight P boxes, including one that is well-conserved within the endosperm box (TGCAAAG: –1270/–1264); and two AACA motifs (–639/–634 and –387/–382) are present as candidate motifs. These cis-elements may be responsible for quantitative regulation of GluD-1.
Further work will be required to determine whether the GCN4 motif acts as a critical element for determining aleurone and subaleurone-specific expression. Ideally, the relationship between the GCN4 motif and tissue-specific expression would be examined by using transgenic rice plants containing minimal endosperm-specific promoters with mutagenized GCN4 motifs.
Supplementary data
Supplementary data can be found at JXB online.
Fig. S1. Alignment of rice glutelin genes used for drawing a phylogenetic tree.
Fig. S2. Mapping of glutelin a4 gene and a phyllogenetic tree of rice glutelins.
Fig. S3. Alignment of GluD-1 cDNAs among cultivars Nipponbare, Wataribune 2, Kasalath and Nona Bokra.
Fig. S4. Alignment of GluD-1 among cultivars Nipponbare, Wataribune 2, Kasalath, and Nona Bokra.
Supplementary Material
Acknowledgments
We thank Dr S Utsumi for kindly providing anti-glutelin antibody. We also thank Ms M Utsuno, Ms Y Ikemoto, Ms H Yajima, Ms Y Suzuki, and Ms X Wang for technical assistance. This work was supported in part by a Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (19780011 to TK) and the Development of Fundamental Technologies for the Production of High-Value Materials using Transgenic Plants from the Ministry of Economy, Trade and Industry (to FT).
References
- Albani D, Hammond-Kosack MC, Smith C, Conlan S, Colot V, Holdsworth M, Bevan MW. The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. The Plant Cell. 1997;9:171–184. doi: 10.1105/tpc.9.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Molecular Biology. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. The Plant Journal. 2002;29:453–464. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- Domoney C, Casey R. Changes in legumin messenger RNAs throughout seed development in Pisum sativum L. Planta. 1987;170:562–566. doi: 10.1007/BF00402992. [DOI] [PubMed] [Google Scholar]
- Furuta M, Yamagata H, Tanaka K, Kasai Z, Fujii S. Cell-free synthesis of the rice glutelin precursor. Plant and Cell Physiology. 1986;27:1201–1204. [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. Iron fortification of rice seed by the soybean ferritin gene. Nature Biotechnology. 1999;17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack MC, Holdsworth MJ, Bevan MW. In vivo footprinting of a low molecular weight glutenin gene (LMWG-1D1) in wheat endosperm. EMBO Journal. 1993;12:545–554. doi: 10.1002/j.1460-2075.1993.tb05686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Iida S, Kusaba M, Nishio T. Mutants lacking glutelin subunits in rice: mapping and combination of mutated glutelin genes. Theoretical and Applied Genetics. 1997;94:177–183. [Google Scholar]
- Jahan M, Uemura Y, Kumamaru T, Hamid A, Satoh H. Genetic variation of glutelin acidic subunit polypeptides in Bangladesh rice genetic resources. Genetic Resources and Crop Evolution. 2005;52:977–987. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reports. 1987;5:387–405. [Google Scholar]
- Katsube-Tanaka T, Duldulao JBA, Kimura Y, Iida S, Yamaguchi T, Nakano J, Utsumi S. The two subfamilies of rice glutelin differ in both primary and higher-order structures. Biochimica et Biophysica Acta-Proteins and Proteomics. 2004;1699:95–102. doi: 10.1016/j.bbapap.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Kim WT, Okita TW. Structure, expression, and heterogeneity of the rice seed prolamines. Plant Physiology. 1988;88:649–655. doi: 10.1104/pp.88.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirihara JA, Hunsperger JP, Mahoney WC, Messing JW. Differential expression of a gene for a methionine-rich storage protein in maize. Molecular and General Genetics. 1988;211:477–484. doi: 10.1007/BF00425704. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Okita TW. Structural relationship among the rice glutelin polypeptides. Plant Physiology. 1986;81:748–753. doi: 10.1104/pp.81.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T. Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. The Plant Cell. 2003;15:1455–1467. doi: 10.1105/tpc.011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Sasaki T, Yano M. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theoretical and Applied Genetics. 1998;96:997–1003. [Google Scholar]
- Marzabal P, Busk PK, Ludevid MD, Torrent M. The bifactorial endosperm box of gamma-zein gene: characterisation and function of the Pb3 and GZM cis-acting elements. The Plant Journal. 1998;16:41–52. doi: 10.1046/j.1365-313x.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. The Plant Journal. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Morita R, Kusaba M, Lida S, Nishio T, Nishimura M. Knockout of glutelin genes which form a tandem array with a high level of homology in rice by gamma irradiation. Genes and Genetic Systems. 2007;82:321–327. doi: 10.1266/ggs.82.321. [DOI] [PubMed] [Google Scholar]
- Muller M, Knudsen S. The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. The Plant Journal. 1993;4:343–355. doi: 10.1046/j.1365-313x.1993.04020343.x. [DOI] [PubMed] [Google Scholar]
- Nasu S, Suzuki J, Ohta R, Hasegawa K, Yui R, Kitazawa N, Monna L, Minobe Y. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Research. 2002;9:163–171. doi: 10.1093/dnares/9.5.163. [DOI] [PubMed] [Google Scholar]
- Okita TW, Hwang YS, Hnilo J, Kim WT, Aryan AP, Larson R, Krishnan HB. Structure and expression of the rice glutelin multigene family. Journal of Biological Chemistry. 1989;264:12573–12581. [PubMed] [Google Scholar]
- Onate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P. Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. Journal of Biological Chemistry. 1999;274:9175–9182. doi: 10.1074/jbc.274.14.9175. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. Journal of Biological Chemistry. 2001;276:14139–14152. doi: 10.1074/jbc.M007405200. [DOI] [PubMed] [Google Scholar]
- Osborn T. The vegetable proteins. London, FL: Longmans, Green and Co; 1924. [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nature Biotechnolgy. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Takaiwa F. Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnology Journal. 2004;2:113–125. doi: 10.1111/j.1467-7652.2004.00055.x. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Wei XL, Satoh H, Kumamaru T, Ogawa M, Takaiwa F. Biochemical and molecular characterization of a rice glutelin allele for the GluA-1 gene. Theoretical and Applied Genetics. 2003;107:20–25. doi: 10.1007/s00122-003-1228-x. [DOI] [PubMed] [Google Scholar]
- Reeves CD, Okita TW. Analyses of α/β-type gliadin genes from diploid and hexaploid wheats. Gene. 1987;52:257–266. doi: 10.1016/0378-1119(87)90052-7. [DOI] [PubMed] [Google Scholar]
- Sano Y. Differential regulation of waxy gene-expression in rice endosperm. Theoretical and Applied Genetics. 1984;68:467–473. doi: 10.1007/BF00254822. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD-zein genes. The Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RT, Llaca V, Linton E, Messing J. Sequence, regulation, and evolution of the maize 22-kD alpha zein in gene family. Genome Research. 2001;11:1817–1825. doi: 10.1101/gr.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Utsumi S, Takaiwa F. Foreign gene products can be enhanced by introduction into low storage protein mutants. Plant Biotechnology Journal. 2003;1:411–422. doi: 10.1046/j.1467-7652.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Takagi H, Hiroi T, Yang L, Tada Y, Yuki Y, Takamura K, Ishimitsu R, Kawauchi H, Kiyono H, Takaiwa F. A rice-based edible vaccine expressing multiple T cell epitopes induces oral tolerance for inhibition of Th2-mediated IgE responses. Proceedings of the National Academy of Sciences, USA. 2005a;102:17525–17530. doi: 10.1073/pnas.0503428102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Hirose S, Yasuda H, Takaiwa F. Biochemical safety evaluation of transgenic rice seeds expressing T cell epitopes of Japanese cedar pollen allergens. Journal of Agricultural and Food Chemistry. 2006;54:9901–9905. doi: 10.1021/jf061848v. [DOI] [PubMed] [Google Scholar]
- Takagi H, Saito S, Yang L, Nagasaka S, Nishizawa N, Takaiwa F. Oral immunotherapy against a pollen allergy using a seed-based peptide vaccine. Plant Biotechnology Journal. 2005b;3:521–533. doi: 10.1111/j.1467-7652.2005.00143.x. [DOI] [PubMed] [Google Scholar]
- Takai T, Nonoue Y, Yamamoto SI, Yamanouchi U, Matsubara K, Liang ZW, Lin HX, Ono N, Uga Y, Yano M. Development of chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona bokra’ and japonica recipient cultivar ‘Koshihikari’. Breeding Science. 2007;57:257–261. [Google Scholar]
- Takaiwa F. Transgenic rice seed as a nutriceutical delivery system. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources. 2007;2:1–9. [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K. A rice glutelin gene family: a major type of glutelin messenger-rnas can be divided into 2 classes. Molecular and General Genetics. 1987;208:15–22. [Google Scholar]
- Takaiwa F, Ogawa M, Okita TW. Rice glutelins. In: Shewry P, Casey R, editors. Seed proteins. Dordrecht, FL: Kluwer Academic Publishers; 1999. pp. 401–426. [Google Scholar]
- Takaiwa F, Oono K. Interaction of an immature seed-specific trans-acting factor with the 5′ upstream region of a rice glutelin gene. Molecular and General Genetics. 1990;224:289–293. doi: 10.1007/BF00271563. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Oono K. Genomic DNA-sequences of 2 new genes for new storage protein glutelin in rice. Japanese Journal of Genetics. 1991a;66:161–171. doi: 10.1266/jjg.66.161. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Oono K. Restriction-fragment-length-polymorphism of glutelin from cultivated rice (Oryza sativa L) Japanese Journal of Genetics. 1991b;66:155–160. [Google Scholar]
- Takaiwa F, Oono K, Wing D, Kato A. Sequence of 3 members and expression of a new major subfamily of glutelin genes from rice. Plant Molecular Biology. 1991;17:875–885. doi: 10.1007/BF00037068. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Takagi H, Hirose S, Wakasa Y. Endosperm tissue is a good production platform for artificial recombinant proteins in transgenic rice. Plant Biotechnology Journal. 2007;5:84–92. doi: 10.1111/j.1467-7652.2006.00220.x. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Yamanouchi U, Yoshihara T, Washida H, Tanabe F, Kato A, Yamada K. Characterization of common cis-regulatory elements responsible for the endosperm-specific expression of members of the rice glutelin multigene family. Plant Molecular Biology. 1996;20:1207–1221. doi: 10.1007/BF00019553. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proceedings of the National Academy of Sciences, USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F. Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Molecular Biology. 1999;40:1–12. doi: 10.1023/a:1026459229671. [DOI] [PubMed] [Google Scholar]
- Wu CY, Adachi T, Hatano T, Washida H, Suzuki A, Takaiwa F. Promoters of rice seed storage protein genes direct endosperm-specific gene expression in transgenic rice. Plant and Cell Physiology. 1998a;39:885–889. [Google Scholar]
- Wu CY, Suzuki A, Washida H, Takaiwa F. The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. The Plant Journal. 1998b;14:673–683. doi: 10.1046/j.1365-313x.1998.00167.x. [DOI] [PubMed] [Google Scholar]
- Wu CY, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. The Plant Journal. 2000;23:415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K, Kasai Z. Evidence for a precursor form of rice glutelin subunits. Agricultural and Biological Chemistry. 1982;46:321–322. [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiology. 2006;141:1694–1707. doi: 10.1104/pp.106.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Zhang JZ. Overexpression analysis of plant transcription factors. Current Opinion in Plant Biology. 2003;6:430–440. doi: 10.1016/s1369-5266(03)00081-5. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Kawagoe Y, Xiao S, Li Z, Okita T, Hau TL, Lin A, Murai N. 5′ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. The Plant Journal. 1993;4:357–366. doi: 10.1046/j.1365-313x.1993.04020357.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.