Abstract
Dominant gain-of-function alleles of Arabidopsis phytochrome B were recently shown to confer light-independent, constitutive photomorphogenic (cop) phenotypes to transgenic plants (Su and Lagarias, 2007). In the present study, comparative transcription profiling experiments were performed to assess whether the pattern of gene expression regulated by these alleles accurately reflects the process of photomorphogenesis in wild-type Arabidopsis. Whole-genome transcription profiles of dark-grown phyAphyB seedlings expressing the Y276H mutant of phyB (YHB) revealed that YHB reprograms about 13% of the Arabidopsis transcriptome in a light-independent manner. The YHB-regulated transcriptome proved qualitatively similar to but quantitatively greater than those of wild-type seedlings grown under 15 or 50 μmol m−2 m−1 continuous red light (Rc). Among the 2977 genes statistically significant two-fold (SSTF) regulated by YHB in the absence of light include those encoding components of the photosynthetic apparatus, tetrapyrrole/pigment biosynthetic pathways, and early light-responsive signaling factors. Approximately 80% of genes SSTF regulated by Rc were also YHB-regulated. Expression of a notable subset of 346 YHB-regulated genes proved to be strongly attenuated by Rc, indicating compensating regulation by phyC-E and/or other Rc-dependent processes. Since the majority of these 346 genes are regulated by the circadian clock, these results suggest that phyA- and phyB-independent light signaling pathway(s) strongly influence clock output. Together with the unique plastid morphology of dark-grown YHB seedlings, these analyses indicate that the YHB mutant induces constitutive photomorphogenesis via faithful reconstruction of phyB signaling pathways in a light-independent fashion.
Keywords: light signaling, signal transduction, transcriptome analysis, photomorphogenesis, Arabidopsis, phytochrome
INTRODUCTION
Light sensors perform essential roles throughout the lifecycle of plants to mediate adaptive responses to the changing quality, quantity, duration, and direction of light in the natural environment (Chen et al., 2004; Franklin et al., 2005; Schäfer and Nagy, 2005). Arguably amongst the most important of these are the phytochromes, a family of biliprotein photoreceptors optimized for sensing red and far-red light (Nagy and Schäfer, 2002; Quail, 2002; Rockwell et al., 2006; Bae and Choi, 2008). The five Arabidopsis phytochrome genes (PHYA-E) encode highly related apoproteins, all of which bind the same linear tetrapyrrole (bilin) chromophore (Sharrock and Quail, 1989). Despite their similar molecular architectures, the modes of photosensory perception by the phyA-E holoproteins are distinct. The phyA photoreceptor is primarily responsible for both very low fluence responses (VLFR) and high irradiance responses to far-red light (FR–HIR), while the phyB–E photoreceptors function as red/far-red (R/FR) photoreversible sensors in the low fluence range (Shinomura et al., 1996; Whitelam and Devlin, 1997; Shinomura et al., 2000). This photosensory diversity enables long-term adaptation to FR-enriched shade environments and confers an adaptive advantage to shade-avoiding plant species that can effectively compete for limited photosynthetically active radiation with their neighbors (Mathews, 2006; Franklin, 2008).
Although distinct in their mode of light sensing, the five Arabidopsis phytochromes share a similar mechanism of action, namely to regulate expression of a distinct and overlapping set of genes following light-activated translocation from the cytoplasm to the nucleus (Nagatani, 2004; Jiao et al., 2007; Kevei et al., 2007). Whole-genome microarray analyses have established that a significant percentage of the Arabidopsis genome is regulated by light. To determine which genes are specifically phytochrome-regulated, comparative R- and FR-dependent transcription profiling of wild type and phytochrome-null mutants has been undertaken by a number of laboratories. Quail and colleagues (2007) focused on gene expression during seedling de-etiolation following exposure to R or FR light. Their studies indicated that phyA is wholly responsible for the rapid transcriptional response to FR, that phyA and phyB together regulate nearly all of the early R-responsive genes, and that phyC–E further contribute to sustained regulation of a subclass of R-responsive genes (Tepperman et al., 2001, 2004, 2006). By contrast with seedling de-etiolation analyses, Deng and colleagues examined the effect of sustained light treatment on both Arabidopsis and rice transcriptomes (Ma et al., 2001; Wang et al., 2002; Jiao et al., 2005; Ma et al., 2005). Their studies not only fingerprint the process of seedling photomorphogenesis at the transcriptional level, but also represent a useful approach to elucidate the regulatory roles of individual phytochromes and of potential signaling components following prolonged illumination.
Whole-genome profiling analyses have established that phytochromes reprogram the plant transcriptome primarily through a rapid light-dependent regulation of a transcription factor cascade (Jiao et al., 2007; Quail, 2007). Although the precise mechanism of this reprogramming process has not been fully elucidated, it is clear that phytochromes do not effect gene regulation via direct DNA binding. Instead, photoactivated phytochromes interact with a diverse array of signaling molecules to alter gene transcription (Bae and Choi, 2008; Josse et al., 2008). Recent studies indicate that phytochromes target many of these factors for degradation by the 26S proteosome—a process that is preceded by their phosphorylation (Shen et al., 2005; Al-Sady et al., 2006; Shen et al., 2007, 2008). Genetic approaches have identified two major transcriptional networks mediated by phytochromes. One network involves the PIFs, members of the PIF3 family of bHLH transcription factors that regulate genes involved in hormone biosynthesis/perception pathways impacting seed germination, elongation growth, cell division, and photosynthetic pigment biogenesis (Khanna et al., 2004; Al-Sady et al., 2006). The direct interaction between photoactivated phytochromes and PIFs initiates this signaling cascade apparently by targeting both PIFs and phytochrome for degradation (Al-Sady et al., 2008; Leivar et al., 2008). The second network entails reprogramming of protein degradation through suppression of the activity of COP/DET/FUS complexes that target key transcription factors HY5, HFR1, and LAF1 in darkness (Ma et al., 2003). The molecular mechanism of phytochrome-mediated inactivation of COP/DET/FUS factors is not fully understood; however, it appears to involve both direct and indirect pathways (Chen et al., 2004; Jiao et al., 2007).
The process of plant photomorphogenesis involves a complex interplay between multiple light-sensing systems that include multiple regulatory photoreceptors, photosynthetic pigments, and other photoprotective or photodynamic pigments (Schäfer and Nagy, 2005). In order to understand the contribution of specific photosensors to this process, investigators typically use monochromatic irradiation regimes to selectively activate photoreceptors. While monochromatic R irradiation can distinguish between B/UV-absorbing photoreceptors and phytochromes, it is difficult to distinguish the effect of R absorbed by each of the five phytochromes, by porphyrin/chlorin precursors and/or by the photosynthetic apparatus itself. Our recent discovery of a new class of gain-of-function missense alleles of phytochromes, which confer their light-independent activation, represents a valuable tool to assess the regulatory roles of individual phytochromes without exciting other photoreceptor systems (Su and Lagarias, 2007). In the present study, we examine the influence of the constitutively active Y276H missense allele of Arabidopsis PHYB (designated as YHB throughout) on the Arabidopsis transcriptome using Affymetrix ATH1 microarrays. Comparative transcription profiling of wild-type and YHB-expressing transgenic Arabidopsis seedlings grown in darkness or in continuous red light (Rc) reveals that YHB faithfully regulates the process of Rc-dependent photomorphogenesis at the genome level. Our investigations also indicate that a significant subset of the YHB-regulated gene complement is suppressed by other Rc-sensing photoreceptor systems, thereby documenting the utility of gain-of-function phytochrome alleles to elucidate the interplay between phytochrome-dependent and phytochrome-independent photomorphogenetic pathways in plants.
RESULTS
YHB-Expressing Seedlings Exhibit Constitutive Photomorphogenesis
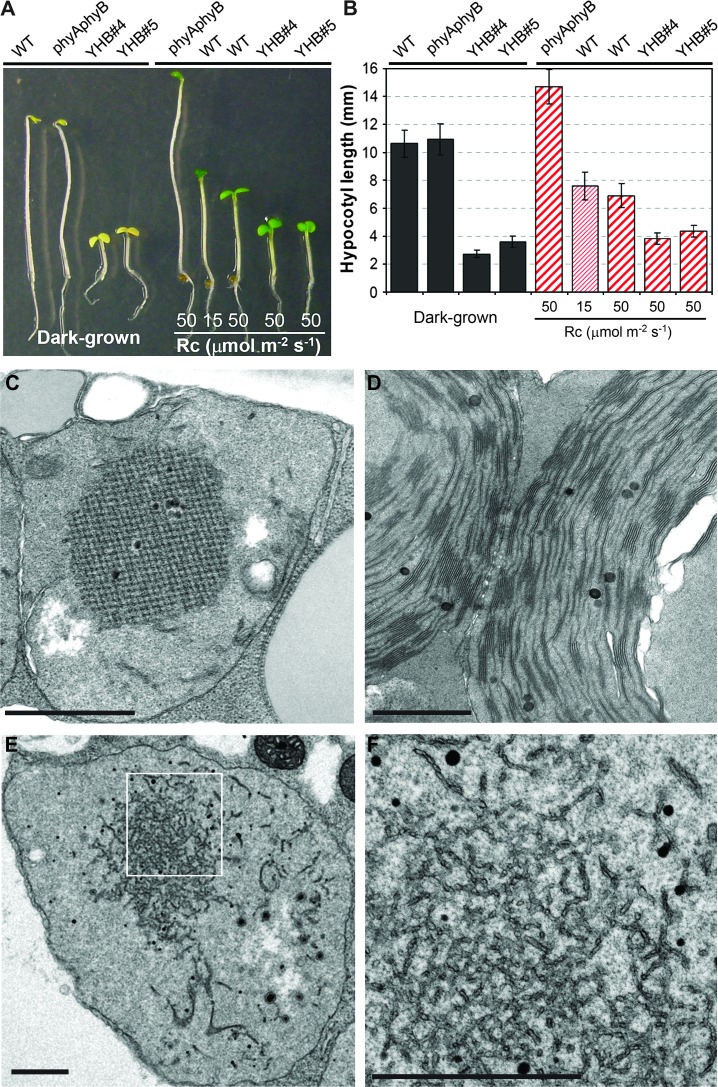
We previously reported that expression of YHB alleles, either as the CaMV 35S-promoter-driven cDNA or as a native promoter-driven genomic construct, confers a dominant, constitutive photomorphogenic (cop) phenotype to dark-grown Arabidopsis seedlings (Su and Lagarias, 2007). As shown in Figure 1A and 1B, two independent genomic YHB/phyAphyB transgenic lines grown in darkness possess short hypocotyls, open apical hook, and expanded cotyledons, in striking contrast with the etiolated phenotype of both Ler wild type and the phyAphyB parent. When grown under continuous red light (Rc), hypocotyl growth inhibition and cotyledon expansion of the phyAphyB double mutant are absent, while wild type exhibits hypocotyl elongation inhibition in a fluence rate-dependent manner. Moreover, YHB seedlings grown under a high fluence rate of Rc (50 μmol m−2 s−1) are significantly shorter than wild type grown under the same light conditions, indicating that YHB confers an even stronger photomorphogenic phenotype than that observed for light-grown wild-type seedlings.
Figure 1.
Constitutive Photomorphogenic Phenotype of YHB Seedlings.
(A) Morphology comparison of 4-day-old seedlings grown in darkness or under different fluence rates of continuous red light (Rc).
(B) Hypocotyl lengths (mean ± S.D.) of 4-day-old seedlings of different genotypes (n = 40∼60).
(C–F) TEM micrographs of an etioplast of dark-grown wild type (C), of a chloroplast of light-grown wild type (D), and of an etioplast from dark-grown YHB seedlings ((E) and (F); (F) is a close-up view of (E)). Scale bars = 1.0 μm.
Since dark-grown YHB seedlings exhibit a cop mutant phenotype, we examined plastid development in dark-grown YHB by electron microscopy. Etioplasts of dark-grown wild-type seedlings are small and possess prominent crystalline prolamellar bodies, while chloroplasts of light-grown seedlings possess stacked granal as well as stromal lamellar membranes (Figure 1C and 1D). By comparison, etioplasts of dark-grown det1 and cop1 mutants are larger and possess thylakoid membranes—a sign of bona fide chloroplast development (Chory et al., 1989; Deng et al., 1991). While also larger than wild-type etioplasts, plastids of dark-grown YHB seedlings lack recognizable thylakoid membranes and prolamellar bodies. Instead of thylakoids, YHB etioplasts possess masses of irregular, disrupted membrane fragments throughout (Figure 1E and 1F). Considerably smaller prolamellar bodies surrounded by membrane fragments were occasionally observed in some YHB etioplasts. The distinct morphology of dark-grown YHB etioplasts suggests that the cop phenotype of YHB is distinct from those of det1 and cop1 mutants.
YHB Misregulates the Vast Majority of Rc-Responsive Genes in Darkness
Owing to the potential complication of transgene-mediated (in)activation of genes near the site of insertion, two independent genetically ‘single-insertion’ YHB transgenic lines (genomic YHB lines #4 and #5 in a phyAphyB double mutant background) were used for microarray analysis (Su and Lagarias, 2007). Accurate mapping analysis revealed that both lines possess two physically linked copies of the genomic YHB transgene (Supplemental Figure 1A). Line #4 was shown to contain two YHB transgenes inserted within the AtPHYB locus on chromosome 2, while line #5 possesses an inverted repeat of two YHB transgenes inserted within a small intergenic region on chromosome 3. These mapping results were confirmed by PCR using primers specific to the flanking genomic sequences and a left-border primer of the binary transformation vector pJM63 (data not shown). Since the YHB transgene was constructed by introducing the Y276H mutation into the PHYB gene (Su and Lagarias, 2007), an elevated level of YHB expression was consistent with the two copies present in both transgenic lines (Supplemental Figure 1B). Despite the enhanced YHB transcript levels, however, the YHB protein accumulation in the two transgenic lines was similar to that of phyB in wild type (Su and Lagarias, 2007). In view of the distinct sites of YHB transgene insertion in the two lines, comparative transcriptome analysis of both lines provides an internal control for insertion site effects.
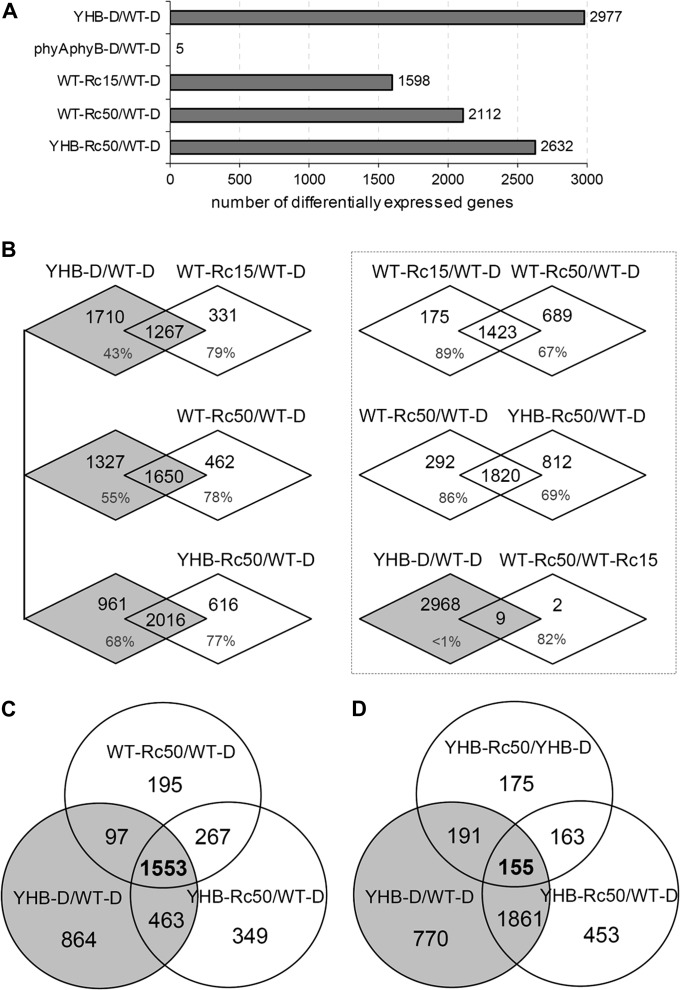
To assess whether the cop phenotype of YHB plants accurately reflects red light-induced photomorphogenesis of wild type, expression profiles of dark- and Rc-grown wild type, the phyAphyB mutant, and two YHB transgenic lines were determined using Affymetrix ATH1 microarrays. Transcript profiles of dark-grown wild type (WT–D) and phyA-201phyB-5 double mutant (phyAphyB–D) were first compared to ensure that the phyAphyB mutant background did not significantly influence the transcriptome. Only five genes with statistically significant two-fold (SSTF) expression alteration were observed by this comparison (Figure 2A)—a result consistent with the indistinguishable morphologies of the two dark-grown genotypes (Figure 1A). By contrast, 2977 genes (13.1% of the genome represented by the chip) showed SSTF misregulation in dark-grown YHB lines (YHB–D) compared with WT–D (Supplemental Table 1). In addition, both YHB lines possessed highly similar expression profiles (correlation coefficient R ≥ 0.9875), indicating that the site of transgene insertion did not significantly influence global gene expression. Comparison of YHB–D and phyAphyB–D profiles also revealed that 86–90% of the SSTF YHB-regulated genes were shared for the pairwise YHB–D/WT–D and YHB–D/phyAphyB–D comparison. Based on these results, we hereafter report SSTF differential expression for pairwise comparisons between each profile of a specifically defined experimental treatment and that of WT–D.
Figure 2.
Microarray Analysis Shows Significant Misregulation of the Arabidopsis Genome by YHB.
(A) Number of statistically significant two-fold (SSTF) regulated genes in dark-grown YHB (YHB–D), in dark-grown phyAphyB double mutants (phyAphyB–D), in Ler wild type grown under 15 μmol m−2 s−1 (WT–Rc15) or 50 μmol m−2 s−1 (WT–Rc50), and in YHB grown under 50 μmol m−2 s−1 (YHB–Rc50) compared with dark-grown Ler wild type (WT–D).
(B) Venn diagrams show pairwise comparison of SSTF-regulated genes with percentage values indicating the proportion of shared genes for each expression profile.
(C, D) Venn diagrams show comparison among three different sets of SSTF-regulated genes.
As expected from previous studies (Ma et al., 2001, 2005), Rc-grown wild-type seedlings showed significant changes in their transcriptome compared with that of WT–D seedlings. Although the number of SSTF-regulated genes proved fluence-rate dependent, namely 1598 genes for wild-type seedlings grown under 15 μmol m−2 s−1 Rc (WT–Rc15) versus 2112 genes for wild-type seedlings grown under 50 μmol m−2 s−1 Rc (WT–Rc50), both were fewer than the 2977 genes regulated by YHB in darkness (Figure 2A and Supplemental Table 1). Interestingly, YHB seedlings grown under 50 μmol m−2 s−1 red (YHB–Rc50) revealed 2632 genes to be SSTF expressed (Figure 2A). This number was notably less than that observed for YHB–D (by 345 genes), revealing that Rc perception attenuates the regulatory impact of YHB. A more detailed pairwise comparison documented that the complement of SSTF misregulated genes in YHB–D overlapped with ∼78% of those observed for Rc-grown WT and YHB-expressing lines (Figure 2B). The overwhelming majority of Rc-regulated transcripts were therefore shared with those regulated by YHB in darkness. Indeed, all Rc-grown seedlings possessed similar expression profiles, namely 89% of the light-regulated genes for WT–Rc15 overlapped with those for WT–Rc50, and 86% of the light-regulated genes for WT–Rc50 overlapped with those for YHB–Rc50 (Figure 2B). In wild-type seedlings, elevated Rc fluence rates regulated more genes than the lower Rc fluence rates, as expected from previous studies. Nevertheless, we detected only 11 genes with SSTF difference in expression between WT–Rc15 and WT–Rc50, nine of which (82%) were also SSTF misregulated in YHB–D (Figure 2B). These genes apparently require elevated Rc fluence rates to reach a level of expression sufficient to achieve the two-fold threshold.
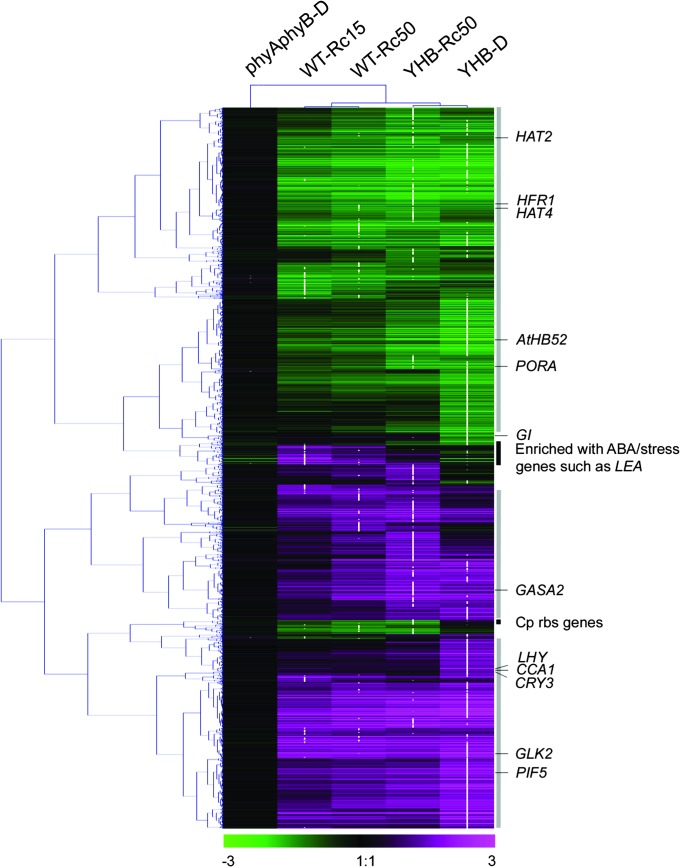
Three-way comparison of SSTF-regulated genes in WT–Rc50, YHB–Rc50, and YHB–D seedlings revealed a core set of 1553 genes shared by all three experimental treatments that are likely to reflect the strongly phyB-dependent genes (Figure 2C). This core set represents 74, 59, and 52% of the SSTF-regulated genes observed in WT–Rc50, YHB–Rc50, and YHB–D, respectively. The greater overlap of genes SSTF regulated in the Rc50-grown WT and YHB seedlings compared with those of dark-grown YHB seedlings appears to reflect other Rc-dependent processes possibly mediated by other phytochromes and/or by the photosynthetic apparatus. While YHB–Rc50 profiles revealed altered expression of 520 more genes than WT–Rc50 (Figure 2A), only 33 of these displayed SSTF difference between WT–Rc50 and YHB–Rc50. Thus, the observed difference in Rc-regulated genes appears to be quantitative in nature rather than qualitative. Indeed, cluster analysis revealed that the majority of differentially expressed genes of YHB–D and Rc-grown seedlings show a similar trend of expression alteration, with, in most cases, genes of YHB–D exhibiting more pronounced expression alteration (Figure 3, maximum regulation noted with white dots). This reinforces the interpretation that YHB–D expression profiles were qualitatively similar to, but quantitatively greater than, Rc-dependent wild-type expression profiles.
Figure 3.
Hierarchical Cluster Analysis of 3889 Core YHB- and Rc-Regulated Genes Reveals Great Overlap of Expression Pattern.
White dots indicate absolute maximum of expression change for each gene among the five experimental treatments described in Figure 2. Vertical gray bars at the right side mark gene sets with similar trend of expression change between YHB–D and Rc-grown seedlings. Solid black bars illustrate two gene sets differentially regulated by YHB and Rc. Some representative genes are indicated in the map. The numerical values for the green-to-magenta gradient bar (bottom) represent log2-fold change relative to WT–D, with induction represented in magenta and repression represented in green.
YHB-Misregulated Genes Are Involved in a Diverse Set of Physiological Processes
Numerous genes with diverse functions were misregulated by YHB in darkness. Functional categorization of these genes using GO annotations from TAIR (www.arabidopsis.org/) showed that, in terms of biological process, genes involved in stress/stimulus response and electron transport or energy pathways are significantly enriched compared to the whole-genome gene set (i.e. about 1.9-fold enrichment). We are particularly interested in the influence of YHB on the expression patterns of known light-regulated genes—especially those encoding components of the photosynthetic apparatus and genes encoding known factors of light signal transduction pathways. Genes encoding 15 chlorophyll a/b binding proteins (LHCA and LHCB/CAB families), 13 thylakoid lumen/membrane proteins, five FtsH chloroplast-localized proteases, and more than 120 chloroplast/photosystem structural and metabolic polypeptides were all significantly up-regulated, indicating that YHB misregulates expression of the photosynthetic apparatus in a light-independent manner. Many key genes encoding enzymes of the chlorophyll biosynthesis pathway were also significantly misregulated (Supplemental Figure 2 and Supplemental Table 2). The three most pronounced of the SSTF up-regulated genes in YHB–D seedlings include HEMA1 that encodes glutamate tRNA reductase (the first enzyme committed to tetrapyrrole synthesis in plants (McCormac et al., 2001; McCormac and Terry, 2002)), CAO that encodes chlorophyll a oxygenase (the last step of chlorophyll b synthesis (Harper et al., 2004)), and GUN4 (a key positive regulator of Mg-chelatase (Larkin et al., 2003)). All of these genes are strongly induced by light (Tanaka and Tanaka, 2007) and have been mapped to the same gene network (Masuda and Fujita, 2008). Consistent with previously reported expression changes during de-etiolation under red or white light, the NADPH:protochlorophyllide oxidoreductase genes PORA and PORB were both down-regulated in YHB–D seedlings while PORC was up-regulated (Oosawa et al., 2000; Su et al., 2001). With few exceptions, the expression changes of tetrapyrrole biosynthetic genes in YHB–D were in good agreement with those regulated by light during de-etiolation of Arabidopsis seedlings (Matsumoto et al., 2004; Stephenson and Terry, 2008). All six nuclear-encoded chloroplast sigma factors that support plastid transcription (Kanamaru and Tanaka, 2004) were up-regulated in YHB–D seedlings (four of them were SSTF-induced), the most notable being SIGE, which was induced by more than 23-fold. As a known blue light-regulated gene, SIGE has been implicated to be a stress-responsive sigma factor that regulates expression of plastid-encoded, photolabile components of photosystem II (Tsunoyama et al., 2004).
In addition to misregulation of photosynthesis-related genes, genes normally induced during the shade avoidance response were strikingly down-regulated in YHB–D seedlings, implying their insensitivity to changes in the R/FR ratio. Among these include ATHB2/HAT4, ATHB4, HAT2, PAR1, RIP, HFR1, and TAA1 (PIL1 is not present on the ATH1 chip) whose expressions are critical to the cell elongation response to low R/FR (Devlin et al., 2003; Sessa et al., 2005; Roig-Villanova et al., 2006; Stepanova et al., 2008; Tao et al., 2008). Genes contributing to phyA signal transduction were also strongly misregulated in YHB–D. These include the negative regulators SPA1 (Hoecker et al., 1999) and SPA4 (Laubinger and Hoecker, 2003) that were both up-regulated, and the positive regulators HFR1 (Fairchild et al., 2000), FHY1/PAT3 (Desnos et al., 2001), and FHL (Zhou et al., 2005) that were all down-regulated. Key morning-phased constituents of the circadian clock CCA1 and LHY (Alabadi et al., 2002) were strongly induced in YHB–D while some evening-phased circadian oscillator genes TOC1/PRR1, PRR3, GI, PCL1/LUX, and a PCL1-homolog were repressed (McClung, 2006; Para et al., 2007). A number of genes involved in blue light signaling pathways were also misregulated, such as PHOT1, PHOT2, NPH3, RPT2, CRY1, CRY3, ZTL, and MYC2 (Lin, 2002; Yadav et al., 2005). Other light signaling components showing striking expression alteration include PIF4, PIF5/PIL6, SPT, HYH, PKS3, RED1, COL1, and COL2 (Wagner et al., 1997; Sakai et al., 2000; Ledger et al., 2001; Holm et al., 2002; Hoecker et al., 2004; Lariguet and Dunand, 2005; Penfield et al., 2005; Nozue et al., 2007; Shen et al., 2007). The expression levels of a comprehensive set of genes involved in light signaling are listed in Supplemental Table 3. In addition to light signaling-related genes, a significant number of genes encoding cell wall components, as well as proteins involved in cell wall expansion and elongation processes, were differentially expressed, likely reflecting the long-term changes in hypocotyl and cotyledon growth between dark-grown wild-type and YHB seedlings (Figure 1A).
Red Light Attenuates a Significant Subset of YHB-Regulated Transcriptome
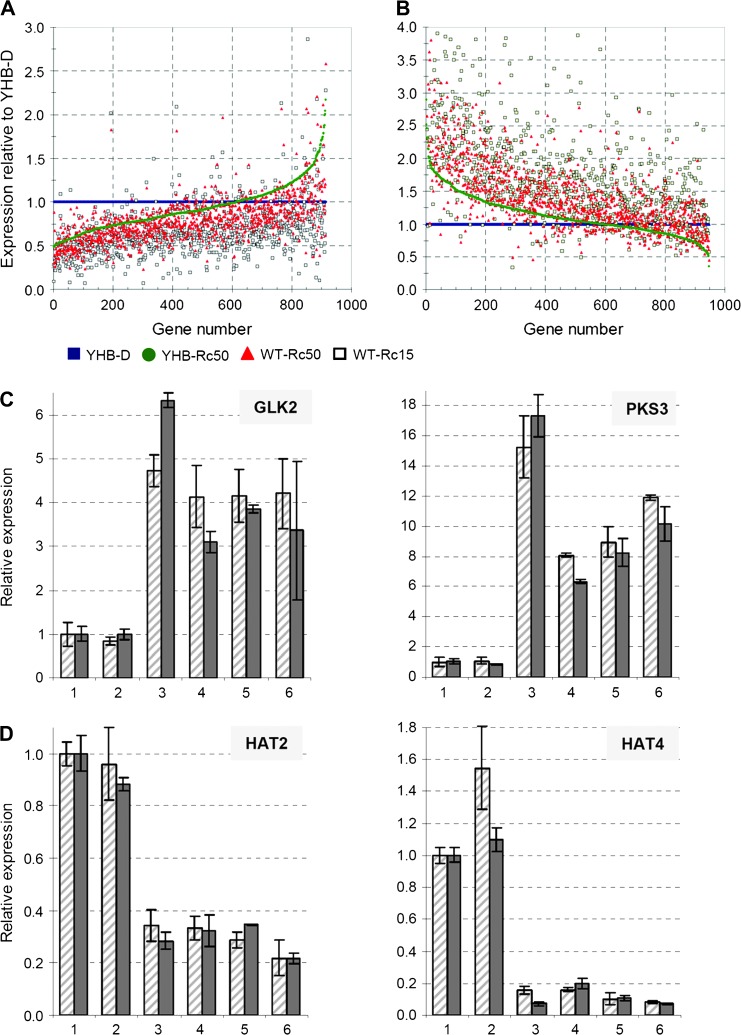
Comparative analysis of YHB–D and YHB–Rc50 profiles was performed to provide insight into the Rc-regulated YHB-independent transcriptional network and the Rc effect on YHB signal output. This analysis revealed that 92% of the SSTF-regulated genes shared by YHB–D and YHB–Rc50 (1861 out of 2016) showed no significant difference when directly compared with each other (Figure 2D). By plotting relative expression levels (normalized to those in YHB–D), however, we observed that ∼65% of the up-regulated genes exhibited quantitatively stronger misexpression in YHB–D than in YHB–Rc50 (Figure 4A). Up-regulation of these genes was also less pronounced in WT–Rc50 and WT–Rc15. A similar relative expression pattern, however, in the opposite direction, was observed for ∼65% of the down-regulated genes (Figure 4B). The expression patterns of two representative genes each for up-regulated and down-regulated gene sets were validated using real-time RT–PCR (Figure 4C and 4D). These results show that Rc attenuated expression of approximately two-thirds of the YHB-regulated genes.
Figure 4.
YHB-Dependent Misregulation of Gene Expression Is Globally Attenuated by Red Light.
Among the total of 1861 genes with comparative expression changes in both dark- and Rc-grown YHB seedlings, (A) 914 genes showed significant up-regulation, while (B) 947 genes showed significant down-regulation; expression for four experimental treatments was normalized to YHB–D and sorted by YHB–Rc50. Expression validation of two representative YHB-induced genes (C) and two representative YHB-repressed genes (D) are shown. Slashed bars = microarray measurement; solid bars = qRT–PCR measurement. Treatment codes include: 1 = WT–D, 2 = phyAphyB–D, 3 = YHB–D, 4 = WT–Rc15, 5 = WT–Rc50, and 6 = YHB–Rc50.
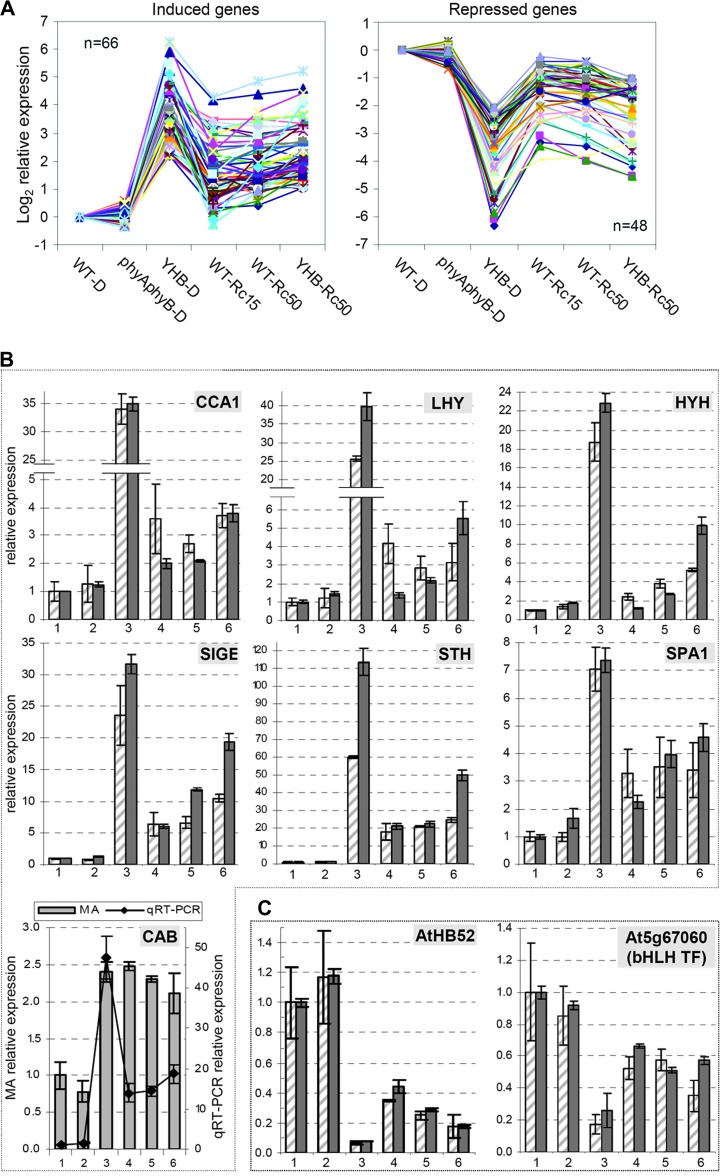
A pairwise comparison of YHB–D and YHB–Rc50 profiles revealed 684 genes to be SSTF modulated by Rc in the presence of YHB that we further categorized into four classes as described below (Figure 2D and Supplemental Table 4). Class-1 genes (155 total) were SSTF-regulated in both YHB–D and YHB–Rc50 (relative to WT–D), indicating that these YHB-misregulated genes were significantly modulated by Rc. Similar to the complement of the aforementioned 1861 YHB-regulated genes, the large majority (74%) of Class-1 genes exhibited a more significant expression change in YHB–D than in YHB–Rc50, indicating their attenuation by Rc (Figure 5A). The expression patterns of the remaining 26% Rc-modulated genes differed from the majority both quantitatively and qualitatively, suggesting that these genes may either be regulated synergistically by different Rc-sensing systems or regulated primarily by other Rc sensor systems (Supplemental Figure 3). Among the 66 Rc-induced genes in the majority population include the transcription factors CCA1, LHY, HYH, STH, SIGA, and SIGE. The phyA signaling regulator SPA1 and the HEMA1 transcript were also found in this gene set. Among the 48 Rc-repressed genes, only two transcription factors, a bHLH transcription factor (At5g67060) and the HD-Zip transcription factor AtHB52 (At5g53980), were detected. The two classes of expression patterns were validated for representative genes using real-time RT–PCR (Figure 5B and 5C). Taken together, these results indicate that Rc attenuates the misregulation by YHB in the majority of the Class-1 genes.
Figure 5.
Class-1 YHB-Regulated Genes Show a Striking Pattern of Attenuation by Red Light.
(A) Integrated expression patterns of YHB-induced and repressed genes that were attenuated by Rc (see Supplemental Table 4 for list of genes shown). Expression validation of seven representative YHB-induced genes (B) and two representative YHB-repressed genes (C) are shown. CAB gene expression is inconsistent between two measurements and thus is plotted with two axes. The legend for the relative expression in (B) and (C) is same as Figure 4C and 4D.
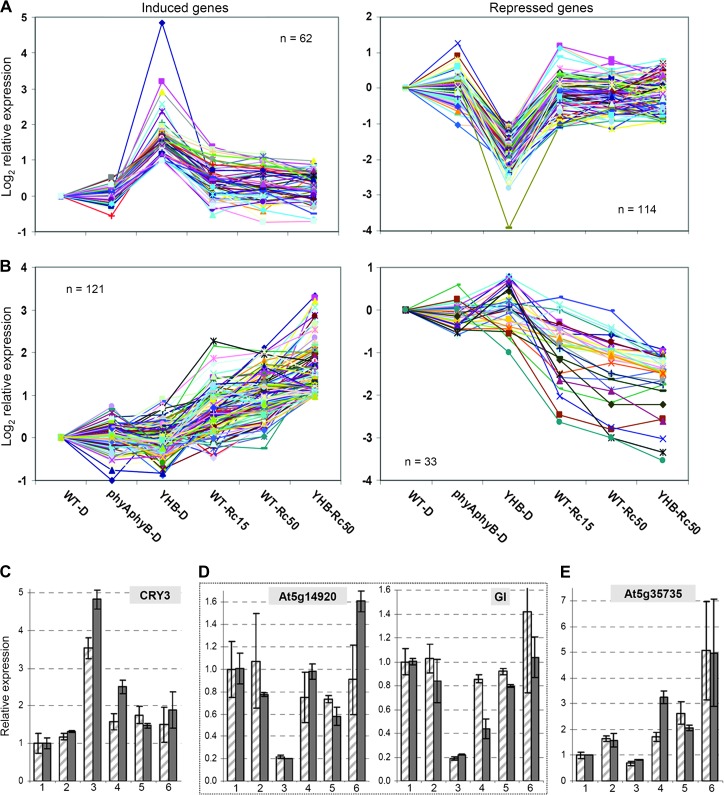
Among other genes with expression differences between YHB–D and YHB–Rc50, a second class of 191 genes showed SSTF misregulation only in YHB–D (Figure 2D and Supplemental Table 4). These Class-2 genes did not appear differentially expressed in YHB–Rc50 (relative to WT–D), indicating that Rc fully suppressed the transcriptional regulatory output of YHB. This interpretation is consistent with the pattern of the majority of genes in this category (Figure 6A). The expression patterns of representative Class-2 genes, which include the YHB-induced gene CRY3 and the YHB-repressed genes, At5g14920 (a GA-regulated gene) and GI, were also validated by real-time RT–PCR (Figure 6C and 6D).
Figure 6.
Class-2 YHB-Regulated Genes Show Almost Complete Attenuation by Red Light while Expression of Class-3 Red Light-Regulated Genes Is YHB-Independent.
(A) Integrated induced and repressed expression patterns for Class-2 genes (see Supplemental Table 4 for list of genes shown).
(B) Integrated induced and repressed expression patterns for Class-3 genes (see Supplemental Table 4 for list of genes shown).
(C–E) Expression validation of one YHB–D-induced gene (C), two YHB–D repressed genes (D), and one Rc-specific induced gene (E). The legend for the relative expression in (C)–(E) is the same as in Figure 4C and 4D.
Comparison of the Class-1 and Class-2 Rc-modulated genes with those exhibiting robust circadian fluctuation of transcript levels (Covington et al., 2008) revealed that 54% of these are clock-regulated (Supplemental Table 4). In addition, the majority of genes with elevated expression in YHB–D seedlings compared with YHB–Rc50 (i.e. genes induced relative to WT–D) exhibited peak expression at dawn, while the majority of genes with reduced expression in YHB–D (i.e. repressed genes) displayed peak expression at dusk (Supplemental Figure 4). This result suggests that Rc may globally attenuate YHB-regulated gene expression by altering output amplitude, phase, and/or period length of the circadian clock. Preliminary measurements with a CCR2::LUC reporter line into which the YHB transgene was crossed revealed that YHB plants maintain robust 24-h cycling in darkness, a phenomenon not seen in the parent reporter line (S.D. Harmer, unpublished results). While experiments to identify the Rc sensor(s) that impact on clock output are beyond the scope of the present investigation, we expect that phyC–E play key roles in the Rc-attenuation effect (Devlin and Kay, 2000).
Red Light Regulates YHB-Independent Gene Expression
In addition to Class-1 and Class-2 Rc-modulated genes described above, the other two classes of Rc-modulated genes were not strongly regulated by YHB (Figure 2D). The 163 Class-3 genes showed SSTF misregulation only in YHB–Rc50, but not in YHB–D. Most of these genes also showed greater expression alteration in YHB–Rc50 compared with WT–Rc50/15 (Figure 6B). Class-3 genes thus appear to be genes whose regulation by Rc is synergistic with the action of YHB. Many genes in this category encode factors involved in protein metabolism and stress/stimulus response. The expression pattern of the auxin-induced gene At5g35735, a representative Class-3 gene, was validated by real-time RT–PCR (Figure 6E). Finally, the fourth class of 175 Rc-modulated genes did not show SSTF regulation in either YHB–D or YHB–Rc50. Since Class-4 genes were oppositely regulated in YHB–D and YHB–Rc50, they appeared differentially expressed in the direct comparison between YHB–D and YHB–Rc50. While Class-3 and Class-4 genes did not appear to be enriched in clock-regulated transcripts (Supplemental Table 4), those that are clock-regulated show an evening phase expression pattern (Supplemental Figure 4).
Among genes showing expression alteration exclusively under Rc, seven chloroplast ribosomal protein genes were all significantly down-regulated (Figure 3). The Arabidopsis chloroplast genome contains 88 genes, all of which are represented on ATH1 microarrays. Strikingly, 42 of them (48%) were included in the Rc-modulated, YHB-dependent gene set, as were 16 of the 26 chloroplast-encoded ribosomal proteins (62%). Hierarchical cluster analysis of the expression patterns of these genes suggested that these genes were co-regulated (Figure 3). Many of these genes showed an intensity-dependent repression by Rc in WT seedlings that is probably related to the completion of chloroplast biogenesis in mature photosynthetic tissue (Mullet, 1988). Remarkably, many ABA-responsive genes involved in desiccation (and possibly other stress) tolerance were up-regulated only in Rc-grown seedlings. These included the late embryogenesis abundant proteins (LEA), Em proteins, oleosins, cupin family proteins, HSPs, and other stress-responsive proteins (Gomez-Porras et al., 2007; Hundertmark and Hincha, 2008). Among all five experimental treatments, maximum expression alteration of ABA/stress-responsive genes was always observed in WT–Rc15 (Figure 3). Some of these even showed expression induction only in WT–Rc15 (Supplemental Table 1). In addition, hydroxyproline-rich glycoprotein family genes encoding cell wall structural components were also remarkably up-regulated only in Rc-grown seedlings. It is also notable that we did not find a substantial number of genes involved in photooxidative stress response to be exclusively up-regulated in Rc-grown seedlings. In this regard, one peroxiredoxin (PER1) was significantly induced. Taken together, these YHB-independent gene targets implicate other Rc-sensing systems hithertofore unrecognized.
YHB Regulates Expression of Many phyA-Dependent, Early Red Light-Responsive Genes
Quail and colleagues identified 251 early red light-responsive genes (corresponding to 256 Affymetrix probe sets) from 4-day-old, dark-grown seedlings exposed to 1 h red light irradiance (Tepperman et al., 2006). Comparative analysis showed that 67% of these genes were also SSTF-regulated in YHB–D, with most of the shared genes exhibiting the same direction of expression change. This overlap percentage was higher than the 57% or 51% overlap with genes SSTF-regulated in YHB–Rc50 and WT–Rc50, respectively (Supplemental Table 5). Considering that some early red light-responsive genes will be suppressed after photomorphogenesis is established, 67% represents a high level of overlap. A more detailed analysis revealed that 119 early red light-responsive genes were all differentially expressed in YHB–D, YHB–Rc50, and WT–Rc50, reflecting the sustained influence of phyB under Rc (Supplemental Table 5). Among these genes, 27 (23%) corresponded to Class-1/Class-2 genes differentially expressed between YHB–D and YHB–Rc50. Moreover, 25 of these 27 genes exhibited greater expression alteration in YHB–D than in YHB–Rc50 or WT–Rc50, while 10 of the 25 genes (40%) encode transcription regulators and five gene products (20%) are involved in signal transduction. Based on these results, the overlap of phyA-dependent, early red light-responsive genes in wild-type plants (Tepperman et al., 2006) and YHB-regulated genes in dark-grown phyAphyB mutant backgrounds is significant. It thus appears that YHB, and, by analogy, wild-type phyB, both regulate many of the same genes throughout photomorphogenic development of Arabidopsis.
DISCUSSION
The YHB-Mediated cop Phenotype Is Distinct from those of cop/det/fus Mutants
Our investigations fingerprint at the whole transcriptome level the process of Arabidopsis seedling photomorphogenesis mediated by the constitutively active Y276H allele of phyB (YHB) both in darkness and under continuous red light (Rc). Similar to the cop/det/fus mutants, YHB-expressing seedlings develop in darkness as if they were grown in light by exhibiting short hypocotyls, open hooks, and expanded cotyledons. However, etioplasts of dark-grown YHB seedlings are structurally distinct from those of cop/det/fus mutants whose stromal lamellae contrast with a provesicular network in YHB plastids. YHB significantly misregulates 13% of the Arabidopsis genome in darkness—the vast majority of which are Rc-responsive genes including key components of the photosynthetic apparatus normally repressed in darkness. The strong overlap of YHB-misregulated genes with known phyA-dependent, early red light-responsive genes also provides compelling support for the conclusion that phyA and phyB regulate a similar transcriptional network(s). Taken together, these results indicate that the YHB allele encodes a constitutively active phyB that regulates the same transcriptional network operating during photomorphogenesis under Rc.
COP1 are DET1 are central integrators of light signals downstream of multiple photoreceptors (Wei and Deng, 1996; Lin and Wang, 2007). A comparison between the YHB–D expression profile and those of dark-grown cop1 and det1 mutants (datasets provided by Drs Xing-Wang Deng and Joanne Chory) shows that ∼30% of COP1 or DET1-regulated genes overlap with the YHB–D expression profile. Since this overlap is much lower than that with Rc-grown wild type, YHB's cop phenotype cannot be wholly attributable to the loss of COP1 or DET1 activities. The cop phenotype of YHB–D seedlings is also distinct from plants expressing the C-terminal domain (CCT1) of CRY1 whose cop phenotype has been ascribed to CCT1-dependent inactivation of COP1 (Yang et al., 2001). HY5 and HYH are two homologous positive regulators of photomorphogenesis that are both negatively regulated by COP1 (Holm et al., 2002); however, YHB dramatically induces HYH expression, but does not alter HY5 expression in darkness. Plastid development of dark-grown YHB plants also appears different from those of dark-grown cop1, det1, or CCT1 transgenics. The circadian period length alteration seen in cop1 and det1 seedlings (Millar et al., 1995; Song and Carre, 2005) was not detected in YHB plants (S.L. Harmer, unpublished results). Taken together, we conclude that YHB reconstructs Rc-signaling by recruiting a subset of COP1/DET1-dependent pathways to achieve photomorphogenesis—a program of development that requires both gene products.
What Processes Are Responsible for the Attenuation of YHB Regulation by Rc?
YHB's light-independent activity represents a unique opportunity to evaluate the modulating influence of red light on the many signaling outputs of phyB. We have shown that the number of SSTF misregulated genes in dark-grown YHB plants is reduced under Rc, implicating reduction of YHB levels by Rc and/or compensating regulation by other photoreceptor systems. Although phyB is widely accepted to be photostable, detailed analytical studies have revealed that Rc exposure triggers a modest reduction of phyB protein levels (Sharrock and Clack, 2002; Al-Sady et al., 2008). Rc-grown YHB plants might thus maintain a sustained reduction in YHB protein abundance compared with dark-grown seedlings, which could be responsible for global suppression of its transcriptome. This explanation, however, does not account for genes with more than five-fold expression change between YHB–D and YHB–Rc50 (e.g. CCA1, LHY, and COL2). Since YHB seedlings used in this study lack both phyA and phyB photoreceptors, Rc perception by phyC–E and/or the photosynthetic apparatus itself are more likely to be responsible for this suppression. Indeed, other phytochromes besides phyB are known to contribute substantially to gene regulation during Rc-induced de-etiolation response (Tepperman et al., 2004). The hypothesis that phyC–E photoreceptors are responsible for Rc suppression of YHB-regulated transcription raises the possibility that other phytochromes attenuate the output of phyB signaling at elevated fluence rates of light. Indeed, it makes good sense for plants to possess high fluence rate sensors to prevent over-accumulation of the photosynthetic apparatus. Although plants possess ample sensory systems that respond to oxidative damage and excess reductant, among other photosynthesis-related metabolic sensors, the built-in dark reversion properties of phytochromes make them excellent candidates for high fluence rate light sensors (Furuya and Schäfer, 1996). In view of the observation that phyC–E heterodimerize with phyB (Sharrock and Clack, 2004), it is reasonable that YHB will also heterodimerize with phyC–E, since the point mutation is buried within its bilin binding GAF domain and would not be expected to interfere with dimerization (Su and Lagarias, 2007). Photoactivation of YHB:phyC–E heterodimers could therefore interfere with the activity of YHB:YHB homodimers to effect suppression of gene expression. On the other hand, while YHB mutants are poorly photoactive, being mostly locked into a constitutively active fluorescent species (Fischer et al., 2005), they possess partial photoactivity that could be responsible for the Rc suppression response. Resolution of the potential Rc-regulation of YHB levels and the role of other phytochromes to this attenuation response are beyond the scope of the present study. The former is complicated by the present lack of methods to quantify the amount of chromophore bound to the phytochrome apoprotein(s) while the latter will require introgression of the YHB transgene into phyC–E-deficient genetic backgrounds.
Other possible explanations for the Rc-dependent attenuation of YHB signaling include activation of the photosynthetic apparatus and/or the light-dependent synthesis of chlorophyll. The former is not unreasonable, since photosynthesis affects nearly every metabolite and energy pool within plant cells. Photosynthesis does not operate in YHB–D seedlings, even though genes encoding components of photosynthetic apparatus are strongly induced. Sugars are known to affect light-regulated gene expression (Rook et al., 2006); hence, photosynthesis-derived carbohydrates may be responsible for attenuating YHB-dependent gene expression under red light. Rc also drives the conversion of protochlorophyllide (Pchilde) to chlorophyllide (Chlide) that is essential for chlorophyll synthesis. Enhanced chlorophyll biosynthesis under Rc would be expected to consume a greater proportion of the protoporphyrin IX pool. This might in turn decrease synthesis of bilin chromophore via reduced production of heme, thereby inhibiting phytochrome holoprotein accumulation (i.e. YHB as well as phyC–E). From this viewpoint, light may feedback regulate YHB function via reduction of bilin production—an intriguing hypothesis to test in a future investigation.
YHB Strikingly Misregulates Clock Gene Expression in Dark-Grown Plants
The observation that the suppressive effect of Rc is global in nature, affecting approximately 65% of the YHB-regulated genes (Figure 4A and 4B), raises the possibility that Rc modulates the YHB input to, or output from, the circadian clock. By imbibing/sowing/harvesting tissue at the same time of day (i.e. at dawn), we sought to minimize variations in transcript profiles due to different phases of the circadian clock. The observed YHB-dependent transcriptomes were notably enriched in up-regulated morning-phased genes and in down-regulated evening-phased genes, indicative of the morning phase of the clock at the time of harvest. The suppressive effect of Rc could be due to a light-dependent alteration of the amplitude, phase, and/or period of clock signaling. In this regard, it is known that phyB mutants show a fluence-rate-sensitive period lengthening under Rc (Somers et al., 1998), while under white light phyB mutants display a phase advance (Salome et al., 2002). Two related genes encoding components of the central oscillator (CCA1 and LHY) have also been shown to be transcriptionally induced by phytochrome photoactivation (Quail, 2002; Kikis et al., 2005); however, both genes exhibit further enhanced expression in YHB–D seedlings (Class-1 gene; see Figure 5B). Other important circadian components, notably the evening-phased genes LUX, GI, TOC1, and PRR3, are all significantly repressed only in YHB–D seedlings. This indicates compensating regulation of YHB-mediated gene expression by other Rc-dependent signaling pathways. The mechanism whereby YHB alters clock output is likely to be complex, since, for example, expression of another clock gene ELF4 (Doyle et al., 2002) is synergistically enhanced by YHB and Rc (Supplemental Table 3). This suggests that phyB activation alone is insufficient for maintaining an elevated level of ELF4 expression. The specific targets of phytochrome-mediated entrainment of the circadian clock remain an important unanswered question (Millar, 2004). Preliminary studies indicate that YHB greatly enhances the amplitude of expression of a clock output gene in dark-grown plants (S.L. Harmer, unpublished results), consistent with the observed enhancement in clock amplitude by light (Salome et al., 2008). The ability to activate YHB signaling in dark-grown plants not only represents a powerful approach to distinguish phyB-specific input pathways to the clock from other light-dependent input pathways, but also to assess the role of known modulators of the light input pathways (e.g. members of the CRY, PIF3, ZTL, and PRR families) in light-mediated clock entrainment.
Future Perspective
We have previously shown that bilin chromophore is required for the constitutive activity of the YHB transgenics (Su and Lagarias, 2007). This characteristic now makes it feasible to construct a bilin-dependent ‘inducible’ system to identify primary target genes involved in phyB signaling in the absence of light. Comparison of dark- and Rc-grown YHB expression profiles have explicitly revealed that some genes are transcriptionally regulated merely by phyB and the other are under a combinatorial control of multiple phytochromes. Genetic studies indicated that phyC–E play less pronounced roles in seedling photomorphogenesis (Devlin et al., 1998, 1999; Franklin et al., 2003). This could be due to their redundant and antagonistic activities, as well as to their altered photoregulatory activities if phyB is not present. Introduction of the YH mutation into phyC–E should also constitutively activate these phytochromes as shown by the activities of the YH mutants of phyA and phyB (Su and Lagarias, 2007). Using the similar approach of expression profiling, these phyC–E YH mutants will be valuable for defining gene sets specifically regulated by each individual, and for studying the combinatory regulatory effects of two or more phys. In summary, YH alleles of phytochromes hold great potential to unravel the overlapping and distinct roles of individual phytochromes in the absence of light activation of other photoreceptor systems.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana wild type and phyA-201phyB-5 mutant were both Ler ecotype. The construct of genomic AtPHYBY276H transgene was described previously (Su and Lagarias, 2007). Two homozygous single-locus-insertion AtPHYBY276H/phyA-201phyB-5 transgenic lines, #4 and #5, referred to as YHB for simplicity (Su and Lagarias, 2007), were used for expression analyses. Seeds used were harvested within 1 year. Seeds were surface sterilized and sowed on filter papers on MS-agar medium (solidified with 0.8% agar) lacking sucrose. Seeds were stratified at 4°C in darkness for 4 d. Germination was induced by exposure to 80 μmol m−2 s−1 white light (cool white fluorescent bulbs) for 3 h. For dark growth, plates were wrapped with several layers of aluminum foil and subjected to a 12-h 22°C/12-h 18°C temperature cycle during the first 2 d of growth to synchronize the circadian clock (Stacey Harmer, personal communication); the temperature was set to 20°C during the following 2 d of growth. The SNAP-LITE LED lighting system (Quantum Devices Inc., Barneveld, WI) was used for red light (662 ± 10 nm) illumination. Seedlings were grown for 4 d at 20°C under continuous 15 or 50 μmol m−2 s−1 red light (fluence rate measured by a LI-COR quantum photometer, model LI-189). Dark-grown seedlings were harvested and immediately frozen in liquid nitrogen in darkness with the aid of Bushnell night vision goggle with a built-in infrared illuminator (model: 26-1020 (1.0 × 20)). Seed imbibition, sowing, germination induction, and seedling harvest were performed at the same time of day (late morning to noon) for independent biological samples that were grown on different dates to reduce expression fluctuation of circadian-related genes.
RNA Isolation, cRNA Synthesis, and Microarray Hybridization
Three (dark-grown) or two (Rc-grown) independent biological replicates of each treatment were grown and processed separately. Total RNA was isolated from the whole seedlings and cleaned up using QIAGEN RNeasy Plant Mini Kit (Valencia, CA). First strand cDNA synthesis, double-stranded cDNA synthesis, cRNA synthesis, and cRNA fragmentation were performed using Affymetrix GeneChip® Expression Analysis kits (One-cycle target labeling and control reagents) (Santa Clara, CA). RNA integrity and cRNA quality were determined using Agilent Bioanalyzer 2100 (Foster City, CA). Probe hybridization, washes, and scanning were performed following Affymetrix protocols by the Microarray Core Facility at UC Davis School of Medicine (www.ucdmc.ucdavis.edu/medmicro/microarray.html).
Microarray Data Analysis
Microarray dataset (cel files) were normalized using robust multiarray average (RMA) background correction method (Bioconductor Release 2.0) (Irizarry et al., 2003). The detection calls (P/M/A) were obtained using the algorithm implemented in the Affymetrix GeneChip® Operating Software (GCOS) with default settings. The LIMMA package (Smyth, 2004) was used to perform pairwise comparison of all treatments and identify significantly differentially expressed genes with the following thresholds: adjusted p-value (adjusted for false discovery rate) <0.05, absolute fold change ≥2.0, and mean of gene expression levels across all treatments ≥10. In the Affymetrix ATH1 chip, one probe set may represent two or more relevant genes, and occasionally one gene may be represented by two probe sets; we treated one probe set as one gene when counting gene numbers. The 64 control probe sets beginning with ‘AFFX’ were filtered out from statistical analysis. Cluster analysis was performed using the Genesis software (Sturn et al., 2002) for all 3889 genes with significant expression alternation relative to dark-grown wild-type seedlings. Average Log2-transformed ratios were input for hierarchical clustering analysis, with options of Pearson correlation for computing similarities and the complete linkage for calculating distance between clusters.
Real-Time RT–PCR
Total RNA samples for real-time RT–PCR were isolated from seedlings grown independently at the same conditions as those for microarray. DNase I-treated total RNA (5.0 μg) was used for reverse transcription of cDNA using transcriptor first strand cDNA synthesis kit (Roche Applied Sci., Mannheim, Germany). For YHB, total RNA (2.5 μg) from each line was mixed together as one sample. 1.5 μl of 20-fold diluted cDNA was used for two-step RT–PCR using FastStart SYBR Green Master (Rox) (Roche Applied Sci.). Reactions were run in ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA). PCR primers were designed in Primer Express software (Applied Biosystems) and sequences are provided in Supplemental Table 5. Reactions were repeated two to four times for each gene. Gene expression values were presented relative to dark-grown wild-type seedlings after normalization to UBQ10 (At4g05320) (ΔΔCt method) (Choi and Roberts, 2007).
Mapping of Genomic Insertion Loci of the YHBg Transgene
Pure genomic DNA was extracted from T3 transgenic lines #4 and #5, respectively, using Wizard® genomic DNA purification kit (Promega, Madison, WI). The ligation and PCR-based mapping method, adaptor duplex sequences for ligation, and nested primers AP1 and AP2 were as described previously (Siebert et al., 1995; Devic et al., 1997). Nested primers anchored in the left-border region of the pJM63 vector were LB1 (5′-GTCACGTCTTGCGCACTGATTTG-3′) for primary PCR and LB2 (5′-TCGTGAACGGTGAGAAGCTCTGG-3′) for secondary PCR. Purified PCR products were sequenced with the LB2 primer. The insertion locus was determined by BLAST search of sequencing result against the Arabidopsis genome (www.arabidopsis.org/Blast/index.jsp).
Transmission Electron Microscopy
Seedlings were grown in darkness or under continuous white light (50 μmol m−2 s−1) for 13 d on 0.8% agar medium containing half-strength MS salt, half-strength Gamborg's vitamin, and 1% sucrose. Cotyledons were dissected and fixed into Karnovsky's fixative (Pelco 34700 BioWave, Ted Pella Inc., Redding, CA) by microwave under vacuum at the following power and time: 5 min at 0 watts, 10 s at 200 watts, 20 s at 155 watts, and 10 s at 250 watts. The tissues were further fixed with 1% OsO4 in 100 mM phosphate buffer for 2 h, rinsed in ddH2O, and incubated in 0.1% tannic acid for 30 min. After a brief rinse with ddH2O, tissues were treated with 1% uranyl acetate for 90 min followed by dehydration through a series of graded acetone. The samples were embedded in an epoxy resin mixture, and thin sections were cut by diamond knife (Diatome, Switzerland) and picked up on copper grids, stained with uranyl acetate and lead citrate before viewing under a Philip CM120 Biotwin transmission electron microscope (Hillsboro, OR). Photographs were taken with a Gatan MegaScan digital camera (model 794/20, Pleasanton, CA).
Accession Number and Data Deposition
The microarray data discussed in this article have been deposited in NCBI Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE8951.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the National Institutes of Health (grant GM068552 to J.C.L.).
Supplementary Material
Acknowledgments
We thank Matt Rolston (Microarray Core Facility, UC Davis) for performing Affymetrix ATH1 chip hybridization, washes, and scanning work. We also thank Patricia Kysar of the Diagnostic and Research Electron Microscopy Laboratory at UC Davis for TEM measurements. We are particularly grateful to Dr Stacey Harmer for helpful discussion and critical reading of the manuscript. No conflict of interest declared.
References
- Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl Acad. Sci. U S A. 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis . Curr. Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Ann. Rev. Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Choi WG, Roberts DM. Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem. 2007;282:24209–24218. doi: 10.1074/jbc.M700982200. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008 doi: 10.1186/gb-2008-9-8-r130. 9, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis . Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. FHY1: a phytochrome A-specific signal transducer. Genes Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M, Roscoe TJ. Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem. 1997;35:331–339. [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2509. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis . Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis . Plant Physiol. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana . Nature. 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochem. (ACS) 2005;44:15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. Shade avoidance. New Phytol. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell. 2003;15:1981–1989. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Larner VS, Whitelam GC. The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 2005;49:653–664. doi: 10.1387/ijdb.051989kf. [DOI] [PubMed] [Google Scholar]
- Furuya M, Schäfer E. Photoperception and signalling of induction reactions by different phytochromes. Tr. Plant Sci. 1996;1:301–307. [Google Scholar]
- Gomez-Porras JL, Riano-Pachon DM, Dreyer I, Mayer JE, Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AL, von Gesjen SE, Linford AS, Peterson MP, Faircloth RS, Thissen MM, Brusslan JA. Chlorophyllide a oxygenase mRNA and protein levels correlate with the chlorophyll a/b ratio in Arabidopsis thaliana . Photosynth. Res. 2004;79:149–159. doi: 10.1023/B:PRES.0000015375.40167.76. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta. 2004;219:195–200. doi: 10.1007/s00425-004-1211-z. [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis . Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana . BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat. Rev. Gen. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis . Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Foreman J, Halliday KJ. Paths through the phytochrome network. Plant Cell Environ. 2008;31:667–678. doi: 10.1111/j.1365-3040.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci. Biotechnol. Biochem. 2004;68:2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- Kevei E, Schafer E, Nagy F. Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J. Exp. Bot. 2007;58:3113–3124. doi: 10.1093/jxb/erm145. [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- Lariguet P, Dunand C. Plant photoreceptors: phylogenetic overview. J. Mol. Evol. 2005;61:559–569. doi: 10.1007/s00239-004-0294-2. [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Hoecker U. The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 2003;35:373–385. doi: 10.1046/j.1365-313x.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Blue light receptors and signal transduction. Plant Cell. 2002;(14 Suppl):S207–S225. doi: 10.1105/tpc.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RC, Wang HY. Targeting proteins for degradation by Arabidopsis COP1: teamwork is what matters. J. Integ. Plant Biol. 2007;49:35–42. [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW. Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 2005;138:80–91. doi: 10.1104/pp.104.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW. Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development. 2003;130:969–981. doi: 10.1242/dev.00281. [DOI] [PubMed] [Google Scholar]
- Masuda T, Fujita Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008;7:1131–1149. doi: 10.1039/b807210h. [DOI] [PubMed] [Google Scholar]
- Mathews S. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 2006;15:3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 2004;135:2379–2391. doi: 10.1104/pp.104.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana . Plant J. 2002;32:549–559. doi: 10.1046/j.1365-313x.2002.01443.x. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Fischer A, Kumar AM, Soll D, Terry MJ. Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana . Plant J. 2001;25:549–561. doi: 10.1046/j.1365-313x.2001.00986.x. [DOI] [PubMed] [Google Scholar]
- Millar AJ. Input signals to the plant circadian clock. J. Exp. Bot. 2004;55:277–283. doi: 10.1093/jxb/erh034. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis . Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Mullet JE. Chloroplast development and gene-expression. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1988;39:475–502. [Google Scholar]
- Nagatani A. Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Nagy F, Schäfer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Ann. Rev. Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Oosawa N, Masuda T, Awai K, Fusada N, Shimada H, Ohta H, Takamiya K. Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana . FEBS Lett. 2000;474:133–136. doi: 10.1016/s0014-5793(00)01568-4. [DOI] [PubMed] [Google Scholar]
- Para A, Farre EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Quail P. Phytochrome-regulated gene expression. J. Integ. Plant Biol. 2007;49:11–20. [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Ann. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martinez-Garcia JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis . Plant Physiol. 2006;141:85–96. doi: 10.1104/pp.105.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Hadingham SA, Li Y, Bevan MW. Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 2006;29:426–434. doi: 10.1111/j.1365-3040.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K. RPT2: a signal transducer of the phototropic response in Arabidopsis . Plant Cell. 2000;12:225–236. doi: 10.1105/tpc.12.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis . Plant Physiol. 2002;129:1674–1685. doi: 10.1104/pp.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Xie QG, McClung CR. Circadian timekeeping during early Arabidopsis development. Plant Physiol. 2008;147:1110–1125. doi: 10.1104/pp.108.117622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer E, Nagy F. Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. 3rd edn. Dordrecht, The Netherlands: Springer; 2005. [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis . Genes Dev. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Heterodimerization of type II phytochromes in Arabidopsis . Proc. Natl Acad. Sci. U S A. 2004;101:11500–11505. doi: 10.1073/pnas.0404286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Feng S, Ma L, Lin R, Qu LJ, Chen Z, Wang H, Deng XW. Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 2005;139:1234–1243. doi: 10.1104/pp.105.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Fururya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis . Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucl. Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Gen. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Song HR, Carre IA. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 2005;57:761–771. doi: 10.1007/s11103-005-3096-z. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stephenson PG, Terry MJ. Light signalling pathways regulating the Mg-chelatase branchpoint of chlorophyll synthesis during de-etiolation in Arabidopsis thaliana . Photochem. Photobiol. Sci. 2008;7:1243–1252. doi: 10.1039/b802596g. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Su Q, Frick G, Armstrong G, Apel K. POR C of Arabidopsis thaliana: a third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 2001;47:805–813. doi: 10.1023/a:1013699721301. [DOI] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Ann. Rev. Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 2004;38:725–739. doi: 10.1111/j.1365-313X.2004.02084.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl Acad. Sci. U S A. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl Acad. Sci. U S A. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis . Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma L, Habashi J, Li J, Zhao H, Deng XW. Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis . Plant J. 2002;32:723–733. doi: 10.1046/j.1365-313x.2002.01462.x. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/DET/FUS genes in light control of arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Hare PD, Yang SW, Zeidler M, Huang LF, Chua NH. FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 2005;43:356–370. doi: 10.1111/j.1365-313X.2005.02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.