Abstract
The limited availability of phosphate (Pi) in most soils results in the manifestation of Pi starvation responses in plants. To dissect the transcriptional regulation of Pi stress-response mechanisms, we have characterized the biological role of MYB62, an R2R3-type MYB transcription factor that is induced in response to Pi deficiency. The induction of MYB62 is a specific response in the leaves during Pi deprivation. The MYB62 protein localizes to the nucleus. The overexpression of MYB62 resulted in altered root architecture, Pi uptake, and acid phosphatase activity, leading to decreased total Pi content in the shoots. The expression of several Pi starvation-induced (PSI) genes was also suppressed in the MYB62 overexpressing plants. Overexpression of MYB62 resulted in a characteristic gibberellic acid (GA)-deficient phenotype that could be partially reversed by exogenous application of GA. In addition, the expression of SOC1 and SUPERMAN, molecular regulators of flowering, was suppressed in the MYB62 overexpressing plants. Interestingly, the expression of these genes was also reduced during Pi deprivation in wild-type plants, suggesting a role for GA biosynthetic and floral regulatory genes in Pi starvation responses. Thus, this study highlights the role of MYB62 in the regulation of phosphate starvation responses via changes in GA metabolism and signaling. Such cross-talk between Pi homeostasis and GA might have broader implications on flowering, root development and adaptive mechanisms during nutrient stress.
Keywords: Abiotic/environmental stress, nutrition, signal transduction
INTRODUCTION
Phosphate (Pi) is an essential macronutrient that is not readily available to plants in most soils. Plants have evolved numerous mechanisms to survive during Pi starvation-induced stress (Marschner, 1995). These adaptive mechanisms include a host of changes in the morphology and physiology of the plant mediated by various molecular components. Morphological changes such as altered root architecture, decreased shoot biomass, and increased accumulation of anthocyanin pigments as well as physiological changes such as increased acid phosphatase activity during Pi deprivation have been extensively characterized (for reviews, see Raghothama, 2000; Jain et al., 2007b). These changes appear to be the consequence of altered expression of numerous genes during Pi starvation (Misson et al., 2005). Further, many of the morphological changes observed during Pi stress such as altered root system architecture and root/shoot ratio are coordinated by phytohormones such as auxin, ethylene, and cytokinin (for review, see Zorilla et al., 2004). A complex interaction involving cytokinins and sugars during Pi starvation signaling has been proposed (Zorilla et al., 2005). The differential effects of auxin and sugars on localized Pi deficiency-induced changes in root system architecture have also been characterized (Jain et al., 2007a). There are indications that some aspects of auxin-mediated regulation of root architecture could involve GA (Fu and Harberd, 2003). The DELLA proteins, core components of GA signaling, have been shown to contribute to anthocyanin accumulation and root architecture changes but not Pi uptake or expression of PSI genes during Pi deprivation (Jiang et al., 2007). These studies are helping to establish a tangible link between gene expression and the role of phytohormones in Pi starvation responses.
Global analysis of genes expressed during Pi deprivation have identified hundreds of PSI genes in Arabidopsis, rice, white lupin, and common bean (Hammond et al., 2003; Wasaki et al., 2003; Wu et al., 2003; Misson et al., 2005; Hernández et al., 2007). Information regarding the transcriptional regulators of these genes is of particular interest, as coordinated spatio-temporal regulation of Pi-responsive genes is vital for maintenance of Pi homeostasis in plants (Hammond et al., 2004). A few transcription factors involved in regulating Pi stress responses have been identified and characterized thus far. These include PHR1, a MYB transcription factor (Rubio et al., 2001), and WRKY75, a WRKY family transcription factor (Devaiah et al., 2007a) in Arabidopsis thaliana L. A bHLH transcription factor in Oryza sativa (rice) named as OsPTF1 has also been reported to play a role in providing tolerance against Pi starvation stress (Yi et al., 2005). These transcription factors have been shown to regulate several subsets of Pi starvation stress responses positively or negatively. We have also characterized the role of the Pi stress-responsive zinc finger transcription factor ZAT6 as a repressor of primary root growth and PSI genes (Devaiah et al., 2007b). Despite the identification of these transcription factors, additional components need to be analyzed in order to decipher the transcriptional networks and signaling pathways involved in regulating adaptation of plants to Pi starvation. In this context, we identified MYB62, a Pi-responsive R2R3 MYB transcription factor based on its induction observed in an earlier microarray study of Pi-responsive genes (Misson et al., 2005).
MYB proteins represent one of the largest transcription factor families in Arabidopsis (Stracke et al., 2001). They generally contain three repeats of 52-amino acid residues called R1, R2, and R3 MYB domains, which binds DNA in a sequence-specific manner. The majority of the plant MYB proteins are of the R2R3 type. The first R2R3 MYB reported in plants was the Zea mays C1 gene, which is required for anthocyanin biosynthesis in kernels (Paz-Ares et al., 1986). Since then, R2R3 MYBs have been implicated in the regulation of morphogenesis, disease resistance, cell division, hormone signaling, and phenylpropanoid metabolism in several plant species (Jin and Martin, 1999). They have also been implicated in regulating abiotic stresses as indicated by the HOS10 R2R3 MYB transcription factor that regulates cold, dehydration and salt stress by controlling ABA biosynthesis (Zhu et al., 2005). PHR1 (Rubio et al., 2001) and PSR1 (Wykoff et al., 1999) are two well characterized R2R3 MYB transcription factors that have been implicated in the regulation of Pi stress responses. Besides this, three Arabidopsis R2R3 MYB genes, MYB33, MYB65, and MYB101, are known to mediate GA signaling during growth and flowering responses (Gocal et al., 2001).
In the present study, we have characterized MYB62, a PSI gene. We show that MYB62 is localized to the nucleus and that MYB62 overexpressing plants exhibit altered Pi starvation responses such as modified root system architecture and Pi uptake. Overexpression of MYB62 also resulted in a GA-deficient phenotype that could be partially rescued through the exogenous application of GA. The increased expression of MYB62 suppressed the expression of early GA biosynthetic genes as well as a host of PSI genes. Together, these results suggest that Pi stress-induced MYB62 negatively regulates PSI genes as well as GA biosynthetic genes. Further, the results also imply that the phytohormone GA plays an important role in regulating Pi stress responses. To our knowledge, this is the first report of a PSI transcriptional repressor of early GA biosynthetic genes.
RESULTS
MYB62 Is Responsive to Pi Stress and Is Localized to the Nucleus
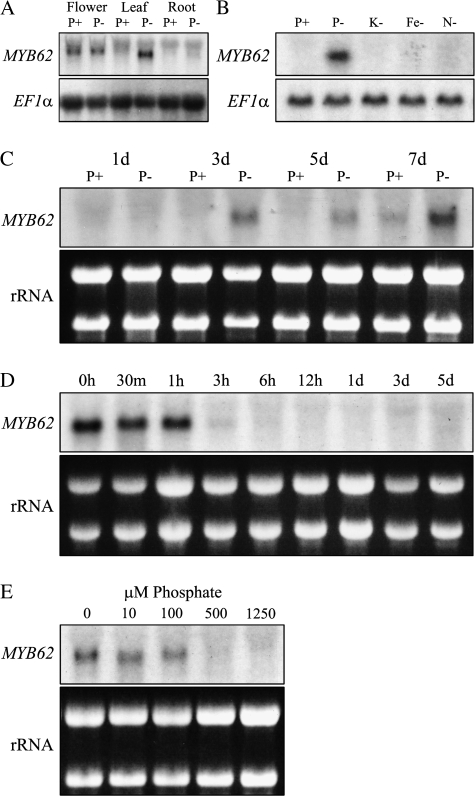
The relative abundance of MYB62 transcripts in plants grown under Pi-sufficient (P+) or Pi-deficient (P–) conditions was evaluated to characterize its response to Pi deprivation (Figure 1A). Induction of MYB62 transcripts was observed only in the leaves during Pi starvation. The expression of MYB62 was also observed in flowers, irrespective of the Pi status of the plants. This highlights the spatial and Pi stress-specific accumulation of MYB62 transcripts. To test the specificity of MYB62 response to Pi stress, the induction of MYB62 transcripts was evaluated in young seedlings grown in medium lacking P, K, Fe, and N (Figure 1B). MYB62 transcripts were induced only during Pi deprivation and not during other nutrient deficiency, suggesting that the induction of MYB62 is a specific response to Pi stress. The rapidity of MYB62 response to Pi deprivation was tested and MYB62 transcripts could be detected after 3 d and its abundance increased at 7 d of Pi deprivation (Figure 1C). When plants deprived of Pi for 7 d were transferred to medium with sufficient Pi, the MYB62 transcript level was suppressed within 1–3 h (Figure 1D). Although the effect of this decrease needs to be confirmed by estimating a corresponding decrease in protein levels, this highlights the rapid response of MYB62 to changes in Pi availability. Further, the effect of increasing availability of Pi on the expression of MYB62 was monitored by transferring plants to medium containing 0, 10, 100, 500, and 1250 μM of Pi. MYB62 transcript levels decreased with increasing availability of Pi to the plant (Figure 1E). Absence of MYB62 transcripts was observed when the Pi concentration in the medium was 500 μM or higher.
Figure 1.
Pi Deprivation Induces the Expression of MYB62.
(A) RNA blot analysis of MYB62 gene expression. Wild-type plants were grown either hydroponically or in liquid culture conditions for 7 d and then transferred to medium containing Pi (P+) or lacking Pi (P–), where they were grown for an additional 7 d. The expression of MYB62 in these samples was detected through RNA blot analysis. An elongation factor (EF1α) gene probe was used as a loading control.
(B) Induction of MYB62 expression is specific to Pi starvation stress. Seven-day-old plants were transferred to medium lacking Pi (P–), potassium (K–), iron (Fe–) or nitrogen (N–) and grown for 7 d before RNA blot analysis.
(C) Expression of MYB62 is induced after 3 d of Pi deprivation. Seven-day-old plants transferred to P+ (1 mM) or P– medium. The expression of MYB62 was monitored at 1, 3, 5, and 7 d after transfer through RNA blot analysis.
(D) Decrease in MYB62 transcripts as Pi-deprived plants are replenished with Pi. Plants that were Pi-deprived for 7 d were moved into Pi-sufficient media. The expression of MYB62 was monitored at 0, 30 min, 1, 2, 3, 6, 12 h, 1, 3, and 5 d through RNA blot analysis.
(E) Expression of MYB62 is suppressed upon increased availability of Pi. Seven-day-old plants grown on half-strength MS media were moved into media containing 0, 10, 100, 500, and 1250 μM Pi and grown for 7 d. The expression of MYB62 was monitored in these samples using RNA blot analysis. EtBr-stained ribosomal RNA prior to transfer is shown to indicate loading and integrity of the RNA.
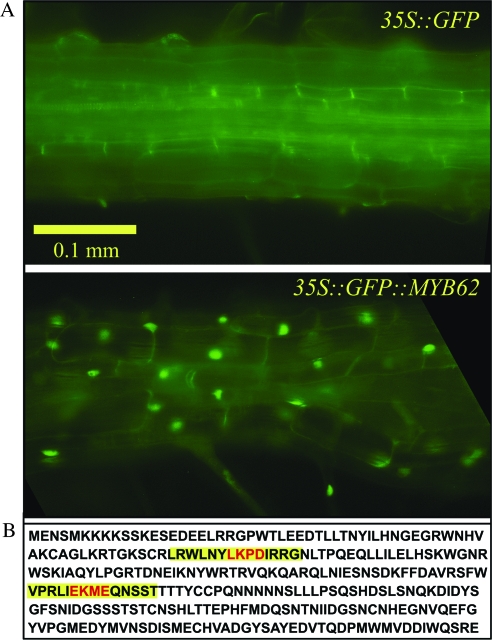
In order to identify the sub-cellular localization of MYB62, its coding region was fused with the 3′ end of an ENHANCED GREEN FLUORESCENT PROTEIN (EGFP) reporter gene. This was expressed constitutively under the control of a cauliflower mosaic virus (CaMV) 35S promoter. Transgenic Arabidopsis plants expressing the control EGFP gene and the chimeric MYB62::GFP gene were analyzed for GFP fluorescence under both P+ and P– conditions. In the control transgenic plant, GFP fluorescence was uniformly distributed all over the cell, while in plants with the MYB62–GFP protein, fluorescence was localized to the nucleus (Figure 2A). These results were confirmed through DAPI staining of the nucleus (Supplemental Figure 1). Exactly similar results were observed under both P+ and P– conditions, suggesting that MYB62 is nuclear localized irrespective of the Pi status of the plant. In addition, two conserved sumoylation target domains were identified on the deduced amino acid sequence of MYB62 (Figure 2B). Together, these results suggest that MYB62 is a nuclear localized transcription factor that is spatially and temporally regulated in plants during Pi deprivation.
Figure 2.
MYB62 is nuclear localised and has sumoylation domains.
(A) Sub-cellular localization of MYB62. Nuclear localization of a GFP::MYB62 fusion protein. The panels show microscopic images of root cells from Arabidopsis plants transformed with a control gene 35S::GFP (upper panel) or a 35S::GFP::MYB62 fusion gene (lower panel).
(B) Deduced amino acid sequence of MYB62. Two predicted sumoylation domains are highlighted in yellow on the sequence.
Overexpression of MYB62 Results in Gibberellic Acid Deficiency Symptoms
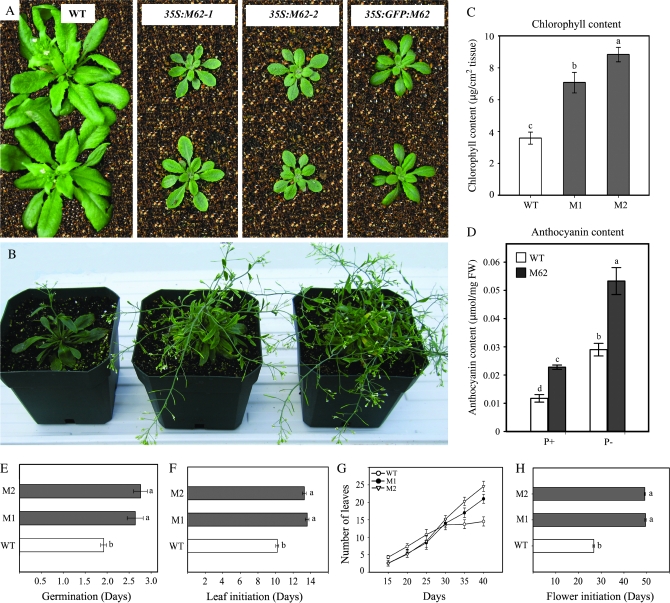
Our repeated attempts to identify a homozygous T-DNA insertion mutant for MYB62 were not successful. On the other hand, antisense and RNAi-mediated silencing of MYB62 did not suppress the expression of MYB62 for unknown reasons. Therefore, plants that overexpressed MYB62 were generated as an alternate tool to define the role of MYB62 during Pi stress. Transgenic plants with the full-length MYB62 cDNA under the control of a CaMV 35S promoter were developed and three independent transgenic lines were screened through RNA blot analysis (Supplemental Figure 2). Two representative transgenic lines with increased MYB62 expression were selected for further experiments. The MYB62 overexpressing and wild-type plants were germinated and grown on a peat–vermiculite mixture for 3 weeks under greenhouse conditions (Figure 3A). The MYB62 overexpressing seedlings were small and dark green with relatively thicker leaves as compared to the wild-type plants. The 35S::GFP::MYB62 transgenic line demonstrated an identical phenotype when used as an internal control, thus supporting the validity of the phenotype. As MYB62 overexpressing plants grew older, their inflorescence stalks lost apical dominance and produced numerous lateral branches with short internodes, bending downwards at every node (Figure 3B). The chlorophyll content of the transgenic plants was relatively higher than the wild-type plants (Figure 3C) and could have contributed to the dark green color of these plants. A significant accumulation of anthocyanin was observed in the MYB62 overexpressing plants relative to the wild-type plants grown hydroponically under P+ and P– conditions (Figure 3D). Significant delay in germination (Figure 3E) and rosette leaf initiation (Figure 3F) as well as increased number of rosette leaves (Figure 3G) were observed in MYB62 overexpressing plants as compared to the wild-type plants. In addition, bolting of the inflorescence stalk was delayed by almost 3 weeks in the MYB62 overexpressing plants relative to the wild-type (Figure 3H). The transgenic plants also exhibited delayed senescence and remained green at 12 weeks after germination as compared to the wild-type plants, which senesced and completed their lifecycle at 6 weeks. The effects of MYB62 overexpression described above are similar to those described in GA-deficient (e.g. ga1–ga5) and GA-insensitive mutants (e.g. gai) by earlier workers (Koornneef and van der Veen, 1980; Koornneef et al., 1985). Therefore, these results suggest that the overexpression of MYB62 could impair GA biosynthesis.
Figure 3.
Plants Overexpressing MYB62 Have Altered Morphology, Physiology, and Development.
(A) Phenotype of MYB62 overexpressing (35S:M62-1, 35SM62-2) and GFP::MYB62 overexpressing (35S::GFP::M62) plants relative to wild-type (WT) plants. Plants were germinated and grown on soil for 3 weeks.
(B) Phenotype of older MYB62 overexpressing plants at various stages of bolting. Plants grown on soil are shown left to right in the panel.
(C) Total chlorophyll content in two independent MYB62 overexpressing transgenic lines (M1 and M2) relative to wild-type (WT). Chlorophyll content was estimated in leaf discs from 3-week-old soil-grown plants. Values are mean ± SE (n = 4).
(D) Total anthocyanin content in wild-type (WT) and MYB62 overexpressing plants (M62) grown hydroponically under Pi-sufficient (P+) and Pi-deprived (P–) conditions. Values are mean ± SE (n = 6).
(E) Germination is delayed in MYB62 overexpressing (M1 and M2) transgenic lines as compared to wild-type (WT). The germination rate was scored after the seeds were sown on peat and vernalized for 4 d.
(F) Leaf initiation is delayed in MYB62 overexpressing (M1 and M2) transgenic lines as compared to wild-type (WT). The emergence of the first rosette leaf was recorded.
(G) MYB62 overexpressing (M1 and M2) transgenic lines have significantly more rosette leaves. Number of rosette leaves was recorded after 15, 20, 25, 30, 35, and 40 d of growth on soil medium.
(H) Bolting is delayed in MYB62 overexpressing (M1 and M2) transgenic lines. Time of appearance of the first inflorescence stem was recorded in plants grown on soil medium. Values in panels (E)–(H) are mean ± SE (n = 25).
The Phenotype of MYB62 Overexpressing Plants Can Be Partially Rescued by Exogenous GA Application
Since MYB62 overexpressing plants demonstrated a GA-deficient phenotype, the effect of exogenous GA application was tested on these plants. Wild-type and two independent MYB62 overexpressing transgenic lines that were grown on peat medium for 15 d were treated with 5 μM GA4 or water on three consecutive days (Figure 4A). The MYB62 overexpressing plants treated with water remained small and dark green compared to the wild-type plants. On the other hand, the transgenic plants treated with GA4 appeared to recover initially and showed robust growth similar to wild-type plants. However, the application of 5 μM GA4 did not alter the dark green color of leaves or delayed bolting of the MYB62 overexpressing plants. Consequently, the effect of higher concentrations and repeated applications of GA3 and GA4 was tested on two independent MYB62 overexpressing transgenic lines and wild-type plants. Wild-type and MYB62 overexpressing plants that were grown on peat medium for 20 d were treated with 50 and 100 μM GA3 or 25 and 50 μM GA4 on eight alternate days (Figure 4B). Plants sprayed with water at the same time intervals served as a control. The results showed that the production of numerous lateral branches and suppression of apical dominance was reduced in all the MYB62 overexpressing plants sprayed with GA as compared to untreated plants. Despite GA treatments, MYB62 overexpressing plants remained relatively smaller than the wild-type plants. The effect of GA applications described above, on initiation of the inflorescence stalk (bolting), was also evaluated in two independent MYB62 overexpressing transgenic lines and wild-type plants (Figure 4C). The delay in bolting of the MYB62 overexpressing plants was less pronounced in GA3 and GA4 treated plants as compared to water-sprayed transgenic plants. Despite this reduction in bolting time due to GA application, the time required by MYB62 overexpressing plants to bolt remained significantly longer relative to wild-type plants treated similarly. These results indicate that exogenous application of GA partially rescues the MYB62 overexpressing plants with respect to their height and bolting time. This is similar to earlier observations that many GA-deficient mutants were partially responsive to exogenous GA application (Koornneef and van der Veen, 1980). Thus, the similarity of MYB62 overexpressing plants and GA-deficient mutants in their response to exogenous GA suggests a possible role for MYB62 in regulating GA biosynthesis.
Figure 4.
Exogenous Application of GA Partially Reverts the Effects of MYB62 Overexpression.
(A) Initial effect of GA4 on young plants. Five μM GA4 was applied on the stems of 15-day-old plants grown on soil medium on three consecutive days. Pictures were recorded 1 week later.
(B) Effect of higher doses of various GAs on flowering and stem elongation. Plants grown on soil for 20 d were sprayed with 50 and 100 μM GA3 as well as 25 and 50 μM GA4 on eight alternate days over 2 weeks. Control plants were sprayed with water.
(C) Effect of exogenous GA application on flowering time of MYB62 overexpressing plants. Time of bolting was scored in plants treated with different GAs as described above. White bars represent wild-type (WT) and shaded bars represent two independent MYB62 overexpressing transgenic lines (M1 and M2). Values are mean ± SE (n = 10).
GA Biosynthetic and Floral Regulatory Genes Are Suppressed in MYB62 Overexpressing Plants
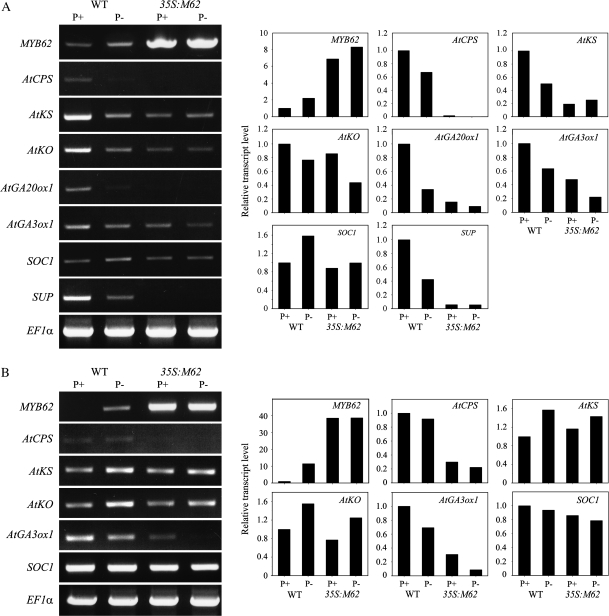
The GA-deficient phenotype observed in MYB62 overexpressing plants (Figure 3) and its partial reversal by exogenous GA application (Figure 4) strongly suggested a role for MYB62 in regulating GA biosynthetic genes. Therefore, the relative transcript abundance of five genes encoding the primary enzymes required for GA biosynthesis was evaluated in the inflorescence (Figure 5A) and leaves (Figure 5B) of wild-type and MYB62 overexpressing plants grown under P+ and P– conditions. The transcript level of each gene was expressed relative to its level in wild-type plants under P+ conditions, which was normalized to one. The five genes tested included AtCPS, encoding ent–copalyl diphosphate synthase (Koornneef et al., 1983); AtKS, encoding ent–kaurene synthase (Zeevaart and Talon, 1992); AtKO, encoding ent–kaurene oxidase (Zeevaart and Talon, 1992); AtGA20ox1, encoding GA 20–oxidase (Talon et al., 1990); and AtGA3ox1, encoding GA 3β–hydroxylase (Talon et al., 1990). The results showed that, in inflorescence, transcripts of all the GA biosynthetic genes were significantly suppressed in MYB62 overexpressing plants relative to wild-type plants under both P+ and P– conditions (Figure 5A). Interestingly, the transcripts of GA biosynthetic genes were suppressed under P– conditions in inflorescence of wild-type plants, confirming the observations of Jiang et al. (2007). In contrast to the global suppression of GA biosynthetic genes in inflorescence, a differential pattern of suppression was observed in the leaves of MYB62 overexpressing plants (Figure 5B). AtCPS and AtKO were suppressed to a lesser extent in leaves relative to their suppression in inflorescence of MYB62 overexpressing plants. In contrast, AtGA3ox1 was suppressed to a greater extent in leaves relative to that in inflorescence of MYB62 overexpressing plants. AtKS was slightly suppressed under P– conditions in the leaves of the MYB62 overexpressing plants. The expression of AtGA20ox1 was not detected in the leaves of either wild-type or transgenic plants. In contrast to the suppression of AtKS and AtKO during P– conditions in inflorescence, these genes were induced under P– conditions in leaves of wild-type plants. The differential expression pattern of these genes indicates that Pi deficiency is a part of the complex regulatory mechanism associated with GA biosynthesis and signaling. Preliminary estimation of bioactive GA4 through LC–MS analysis also indicated substantial changes in the quantum of GA4 present in wild-type plants during Pi stress, while no GA4 could be detected in MYB62 overexpressing plants (data not shown). These results suggest a complex interaction between GA biosynthesis and PSI responses in Arabidopsis.
Figure 5.
Genes Required for Gibberellic Acid Biosynthesis Are Suppressed by MYB62 Overexpression.
Two-week-old wild-type (WT) and MYB62 overexpressing (35S:M62) plants grown in hydroponic culture under P+ and P– conditions for 7 d were used for RT–PCR analysis as described in Methods. Densitometric analysis of the RT–PCR data is shown on the right side of the panel. Transcript levels are expressed relative to the level of transcripts in wild-type plants grown under P+ conditions, which are assumed to be one.
(A) Expression of GA biosynthetic genes AtCPS, AtKS, AtKO, AtGA3ox1, and AtGA20ox1 and two floral regulatory genes, SOC1 and SUP, in inflorescence.
(B) Expression of AtCPS, AtKS, AtKO, AtGA3ox1, and SOC1 in leaves.
MYB62 overexpressing plants exhibit delayed bolting (Figure 3), which is partially reversed by exogenous GA application (Figure 4). The effect of MYB62 overexpression on bolting led us to examine the relative transcript levels of two GA-controlled genes that regulate floral homeotic genes. The transcript levels of the two genes, SUPPRESSOR OF CONSTANS 1 (SOC1; Moon et al., 2003) and SUPERMAN (SUP; Bowman et al., 1992), were analyzed in both inflorescence and leaves of the plants under P+ and P– conditions (Figure 5). The transcript abundance of SUP was significantly reduced in the inflorescence of MYB62 overexpressing plants as compared to wild-type plants (Figure 5A). The transcript abundance of SOC1 is reduced in MYB62 overexpressing plants under P– conditions relative to wild type plants. In addition, under P– conditions, the transcripts of SOC1 were induced while the transcripts of SUP were suppressed in inflorescences of wild-type plants. These results suggest that Pi deprivation influences floral development by altered expression of SOC1 and SUP by MYB62.
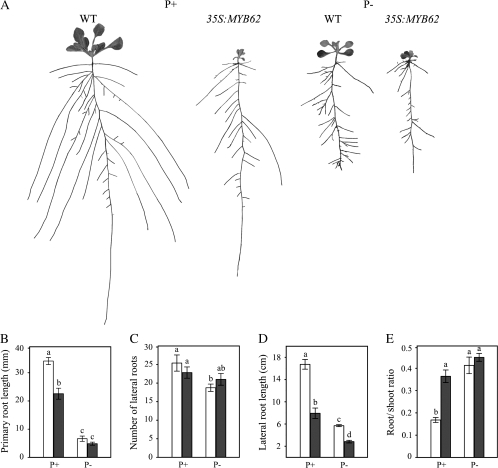
The Root System Architecture (RSA) Is Altered by MYB62 Overexpression
One of the adaptive responses to Pi starvation in Arabidopsis is the alteration of RSA (López-Bucio et al., 2003; Jain et al., 2007b). Since both GA and Pi affect RSA, it is pertinent to analyze the effect of MYB62 overexpression on RSA. We examined the RSA of 14-day-old wild-type and MYB62 overexpressing plants grown in vertically oriented agar plates under P+ and P– conditions for 7 d (Figure 6). The results indicated that the RSA of MYB62 overexpressing plants was significantly altered as compared to the wild-type plants under both P+ and P– conditions (Figure 6A). There was a significant decrease in the primary root length of the MYB62 overexpressing plants under P+ but not P– conditions as compared to the wild-type plants (Figure 6B). This result indicated that localized Pi deficiency-induced inhibition of primary root growth (Ticconi et al., 2004) is not influenced by MYB62 overexpression. Although the number of lateral roots did not vary (Figure 6C), there was a significant decrease in the length of the lateral roots in MYB62 overexpressing plants under both P+ and P– conditions as compared to the wild-type plants (Figure 6D). MYB62 overexpressing plants had a significantly larger root-to-shoot ratio than wild-type plants under P+ but not P– conditions (Figure 6E). Together, these results suggest that MYB62 plays an important role in regulating several aspects of the root architecture and could thus possibly influence Pi homeostasis.
Figure 6.
Root Architecture Is Altered by MYB62 Overexpression.
Wild-type (WT) and MYB62 overexpressing (35S:MYB62) plants were grown under Pi-sufficient (P+) and deficient (P–) conditions.
(A) Lateral roots were spread to reveal architectural details and scanned at 600 dpi. The seedlings shown are representative of 12 replicates. Panels B, C, D, and E show comparative histograms of WT (white bars), and 35S:MYB62 (black bars) with regard to various components of their root architecture under P+ or P– conditions. Different letters on the bars represent means that are statistically different (P < 0.02). Values are means ± SE (n = 12) of each genotype per treatment.
(B) Primary root length.
(C) Total number of lateral roots per plant.
(D) Length of first-order lateral roots.
(E) Root/shoot ratio on a fresh weight basis.
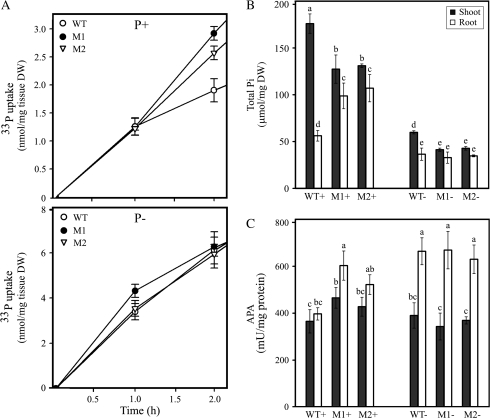
Overexpression of MYB62 Affects Pi Uptake, Pi Accumulation, and Acid Phosphatase Activity
Since changes in the RSA of MYB62 overexpressing plants (Figure 6) suggested that Pi uptake in these plants might be altered, Pi uptake measurements were done to find the effect of altered RSA caused by the overexpression of MYB62 on Pi acquisition. There was a significant increase in the Pi uptake of MYB62 overexpressing plants grown under P+ conditions (Figure 7A, upper panel) compared to wild-type plants. However, there was no difference in the Pi uptake under P– conditions between the transgenics and wild-type plants (Figure 7A, lower panel). These uptake patterns were consistent with the changes in the root architecture observed in MYB62 overexpressing plants under P+ and P– conditions, respectively (Figure 6B–6E). Thus, it appears that the changes in the RSA caused by the increased expression of MYB62 have a direct bearing on the Pi uptake of the plant.
Figure 7.
MYB62 Regulates Pi Homeostasis.
(A) MYB62 overexpressing seedlings have increased Pi uptake. Wild-type (WT, white ovals) and MYB62 overexpressing transgenic lines (M1 and M2, black ovals and open triangles) were grown on 0.5 X MS medium for 7 d and then transferred as groups of 10 seedlings into P+ or P– medium for 3 d. The Pi uptake of these 10-day-old seedlings was monitored over a 2-h period. The upper panel shows Pi uptake in plants from Pi-sufficient conditions and the lower panel Pi uptake in plants from Pi-deficient conditions. Error bars represent SE (n = 3). Wild-type (WT) and MYB62 overexpressing transgenic lines (M1 and M2) were grown on 0.5 X MS medium for 7 d and then transferred to Pi-sufficient (P+) and deficient (P–) medium for 7 d. Total Pi concentration and acid phosphatase activity in shoots (black bars) and roots (white bars) were estimated. Error bars indicate SE (n = 4) and different letters above the bars represent means that are statistically different (P < 0.05).
(B) Total Pi content is altered in older MYB62 overexpressing seedlings.
(C) Increased acid phosphatase activity in MYB62 overexpressing plants.
A change in the total root surface area of a plant affects its ability to explore greater soil volumes and acquire more phosphorus (López-Bucio et al., 2003). Therefore, 7-day-old wild-type and MYB62 overexpressing plants were transferred to P+ or P– medium and total Pi content was estimated in the shoots and roots of these plants after 7 d of treatment (Figure 7B). A significant increase (P < 0.05) in the total Pi concentration was observed in roots of the MYB62 overexpressing plants as compared to the wild-type plants under P+ while no changes were observed under P– conditions. However, a significant decrease in the total Pi content was observed in shoots of the transgenic plants relative to wild-type plants under both P+ and P– conditions. These results are in agreement with the changes in root architecture (Figure 6) and Pi uptake (Figure 7A) observed in MYB62 overexpressing plants. A possible defect in transfer of Pi from root to shoots, particularly under Pi sufficiency in the transgenic plants, could be one of the plausible explanations for the above results.
Production of acid phosphatases is a distinct indicator of plant responses to Pi starvation. A comparative analysis of the total acid phosphatase activity in the shoots and the roots of wild-type and MYB62 overexpressing plants grown under P+ or P– conditions was performed (Figure 7C). A small but significant (P < 0.05) increase in phosphatase activity was observed in the roots of the MYB62 overexpressing plants grown under P+ conditions as compared to the wild-type plants. Together, these results indicate that MYB62 regulates Pi uptake and accumulation through the control of root architecture and acid phosphatase activity.
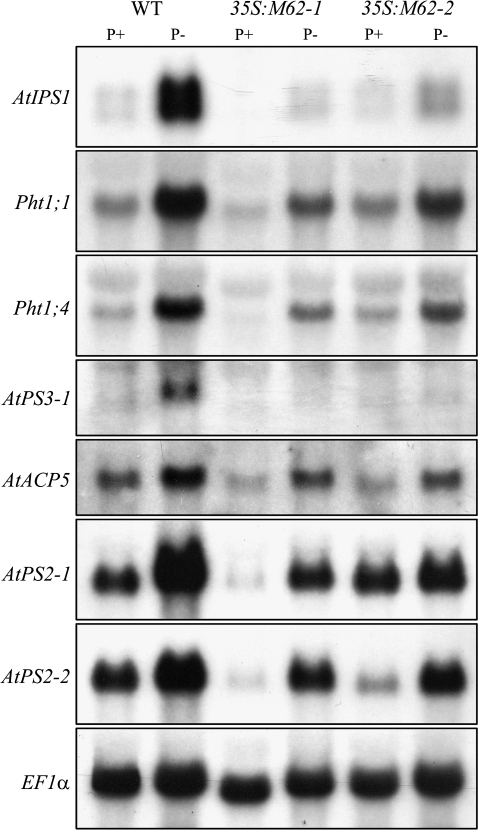
MYB62 Is a Negative Regulator of Phosphate Starvation-Induced Genes
Many genes are induced during Pi deprivation and the roles of some of these genes in Pi stress responses have been well characterized (Jain et al., 2007b). The overexpression of MYB62 was found to influence Pi homeostasis through the alteration of several Pi starvation-response mechanisms (Figures 6 and 7). Therefore, an attempt was made to find the role of MYB62 in regulating a group of PSI genes involved in maintaining Pi homeostasis. The expression of the PSI genes was evaluated in both wild-type and MYB62 overexpressing plants grown under P+ and P– conditions (Figure 8). We examined the expression profile of AtIPS1, a member of the TPSI1/Mt4 gene family that is involved in cytokinin signaling during Pi starvation (Martín et al., 2000). We also evaluated the expression of Pht1;1 and Pht1;4, which encode high-affinity phosphate transporters (Muchhal et al., 1996), as well as AtPS3-1, a member of the glycerol-3-phosphate permease family known to be responsive to Pi (Raghothama, unpublished). The expression of AtACP5, encoding an acid phosphatase involved in phosphate stress responses (del Pozo et al., 1999), was also examined. The analysis also included AtPS2-1 and AtPS2-2, encoding two members of a phosphatase family specifically induced by Pi stress (Raghothama, unpublished.), which are orthologs of the LePS2 gene family from tomato (Baldwin et al., 2001). Together, these represent a broad range of PSI genes involved in Pi stress responses. The results indicated that the transcripts of all the PSI genes tested were reduced by varying degrees in the MYB62 overexpressing plants relative to the wild-type plants during Pi deprivation (Figure 8). The overexpression of MYB62 thus suppresses an array of PSI genes involved in Pi sensing, translocation, transport and mobilization. This suggests that negative regulation of PSI genes by MYB62 is independent of the action of the phytohormone GA
Figure 8.
MYB62 Suppresses the Expression of Pi Starvation-Induced Genes.
Wild-type (WT) and two independent MYB62 overexpressing transgenic lines (35S:M62-1 and 35S:M62-2) grown in liquid culture under Pi-sufficient (P+) and deficient (P–) conditions for 7 d were used for RNA blot analysis. Total RNA (10 μg) was electrophoretically separated, blotted onto a nylon membrane and hybridized consecutively with 32P-labeled probes corresponding to: AtIPS1, Pht1;1, Pht1;4, AtPS3-1, AtACP5, AtPS2-1, and AtPS2-2. EF1α was used as the loading control.
DISCUSSION
Adaptive responses during Pi starvation are regulated by molecular determinants and coordinated through phytohormones. Recent studies on transcription factors and phytohormones are providing vital cues about intricate biological mechanisms regulating adaptation of plants to Pi stress (Devaiah et al., 2007b; Jain et al., 2007b). This study extends that line of reasoning and provides evidence for the potential involvement of GA in Pi stress responses. The involvement of GA in general stress protection has been reported earlier (Vettakkorumakankav et al., 1999) and, recently, its role has been implicated in Pi starvation response (Jiang et al., 2007). Here, we examine the potential cross-talk between Pi starvation stress and GA biosynthesis via MYB62. This potential cross-talk could lead to vital modifications in plants required for survival under Pi deficiency.
MYB62 Is a PSI Gene that Modulates Pi Homeostasis
We have shown that MYB62 is induced specifically in the leaves of young seedlings during Pi deprivation whereas it is expressed constitutively in the inflorescence (Figure 1A). It is noteworthy that the expression of any known Pi stress-responsive transcription factor does not follow the same pattern. The transcripts of Pi stress-responsive WRKY75 (Devaiah et al., 2007a) and ZAT6 (Devaiah et al., 2007b) are found in all the parts of the plant and are also induced by other nutrient stresses. On the other hand, PHR1 is not responsive to Pi starvation (Rubio et al., 2001). Therefore, the function of MYB62 is likely to be more spatially defined relative to other known Pi stress-responsive transcription factors. The specific response of MYB62 to Pi stress and the rapidity of its suppression upon Pi replenishment (Figure 1B–1E) further support this notion. Like other known Pi stress-responsive transcription factors in plants, MYB62 localizes to the nucleus irrespective of the Pi status of the plant. This suggests that unlike yeast, where nuclear localization of transcription factor is a crucial regulatory event during Pi deficiency, plants may have alternate regulatory mechanisms. One such mechanism could be post-translational regulation by sumoylation as observed in the case of PHR1, a Myb transcription factor (Miura et al., 2005). Presence of two SUMO target domains in MYB62 indicate the potential for sumoylation that is yet to be confirmed experimentally.
Overexpression of OsPTF1, a rice bHLH transcription factor, was shown to increase plant tolerance to Pi stress (Yi et al., 2005). A similar observation was also made in Arabidopsis plants overexpressing the MYB PHR1 (Nilsson et al., 2007). Therefore, for the current study, we followed a similar strategy of generating plants overexpressing MYB62 to characterize its role in maintenance of Pi homeostasis. One of the characteristic responses of plants to phosphate starvation is the accumulation of anthocyanins. It has been reported that MYB proteins are known to play an important role in the control of phenylpropanoid metabolism that results in the production of anthocyanin (Martin and Paz-Ares, 1997). In this context, accumulation of anthocyanin in MYB62 overexpressing plants compared to wild-type plants (Figure 3C) is not surprising. Another interesting observation was an increase in Pi content in plants overexpressing MYB62. Presumably, the significantly altered RSA of MYB62 overexpressing plants (Figure 6) played a key role in acquiring Pi from the media. In addition to changes in RSA (Figure 6A), the observed increase in phosphatase activity (Figure 7C) in roots of MYB62 overexpressing plants may also have contributed to higher Pi uptake under P+ conditions. Higher Pi levels in roots of transgenic plants could be due to a combination of factors, including reduced transport of Pi to shoots and reduced shoot growth. It must be noted that although the overexpression of MYB62 causes significant changes in plant morphology, the transgenic plants largely retain their Pi starvation responses. This suggests that Pi homeostasis is dependent on a multiplicity of signaling molecules and pathways besides MYB62.
Changes in the expression of genes involved in Pi signaling, high-affinity Pi transport and mobilization suggested a global role for MYB62 during Pi deficiency. These results confirmed that MYB62 regulates a range of Pi starvation responses, including PSI gene expression. Suppression of PSI genes in MYB62 overexpressing plants indicates that MYB62 acts as a negative regulator of their expression. It is very likely that Pi starvation-induced MYB62 may moderate or temper the activity of other PSI genes by down-regulating their expression during Pi starvation. Thus, the role of MYB62 may be similar to that of ZAT6, another Pi-responsive transcription factor that regulates root architecture, Pi homeostasis and acts as a negative regulator of PSI genes (Devaiah et al., 2007b). On the other hand, WRKY75, which regulates root architecture independently of Pi, acts as a positive regulator of PSI genes (Devaiah et al., 2007a). Therefore, these results support the notion that both positive and negative regulators are involved in the expression of PSI genes and maintenance of Pi homeostasis.
MYB62 Regulates Pi Stress Responses through the Modulation of GA
The overexpression of MYB62 results in an archetypical GA-deficient phenotype of small dark green plants exhibiting reduced apical dominance with delayed flowering and senescence (Figure 3). GA-deficient mutants are classified as GA-responsive and GA-insensitive, based on their ability to recover upon exogenous GA application (Koornneef et al., 1985). MYB62 overexpressing plants were able to partially recover upon application of GA3 and GA4, suggesting that they are GA-deficient mutants. Besides GA, brassinosteriods (BR) also promote cell elongation and BR mutants exhibit dwarfism. Lack of response of MYB62 overexpressing plants to brassinolids (data not shown) suggested that the dwarf phenotype is not due to lack of brassinosteroids. The obvious question is how MYB62 is involved in modulating GA-induced responses. It appears that MYB62 is a regulator of GA biosynthesis. GA biosynthesis is controlled by five principal enzymes that convert geranylgeranyl diphosphate to bioactive GA1 and GA4 in a multistep process (reviewed by Hedden and Kamiya, 1997). In Arabidopsis, AtCPS, AtKS, AtKO, AtGA20ox1, and AtGA3ox1 genes encode these enzymes and their expression was suppressed to variable extents in both leaves and inflorescence by the overexpression of MYB62 (Figure 5). However, with the exception of AtGA3ox1, the transcripts of the GA biosynthetic genes were suppressed to a smaller extent in the leaves, indicating differential and spatial regulation of gene expression. These results suggest that MYB62 acts as a transcriptional repressor of GA biosynthetic genes. It is likely that MYB62 could act as an intermediary on a coregulon of both GA biosynthesis and Pi starvation response. This link between GA and Pi nutrition is further confirmed by the report that expression of AtGA20ox1 and AtGA3ox1 in Arabidopsis is suppressed under low Pi conditions (Jiang et al., 2007).
The transcriptional regulation of GA biosynthetic genes is yet to be completely understood. To date, only three transcription factors are known to regulate GA biosynthesis. The KNOX homeobox domain NTH15 in tobacco (Tanaka-Ueguchi et al., 1998) and its homolog in rice, OSH1 (Kusaba et al., 1998), repress the expression of GA20ox1. In Arabidopsis, an AP2 transcription factor named DDF1 induces the expression GA20ox1 (Magome et al., 2004). The regulation of all the GA biosynthetic genes by MYB62 described in this study is therefore significant. In addition, during Pi deprivation, the expression of the GA biosynthetic genes was suppressed particularly in inflorescence (Figure 5). However, some of them were induced in leaves, suggesting a complex organ-specific regulation of these genes by Pi stress. GA biosynthesis is known to be regulated principally by feedback inhibition, light, and temperature (Hedden and Kamiya, 1997). Our studies confirmed the data of Jiang et al. (2007) that Pi starvation causes a decrease in bioactive GA levels. These studies showing the effect of Pi stress on GA thus add a new dimension to our current understanding of GA biosynthesis.
The suppression of SOC1 (Moon et al., 2003) and SUP (Bowman et al., 1992), transcription factors associated with floral homeotic genes, in the MYB62 overexpressing plants, is interesting (Figure 5). This suggests that MYB62 not only affects PSI processes, but also impacts flowering by interfering with GA biosynthesis. Interestingly, the ectopic overexpression of Arabidopsis SUP in tobacco suppresses the expression of GA20ox1, suggesting that SUP controls cell elongation through the regulation of GA (Bereterbide et al., 2001). Studies are required to investigate whether MYB62 is epistatic to SUP or vice versa. Since Pi starvation has a significant negative impact on yield, it would be interesting to study the interaction between nutrient stress and phytohormones, not only in RSA, but also in flower development.
Many phytohormones such as auxins, cytokinins, and ethylene are known to regulate Pi stress responses, particularly the root architecture during Pi deprivation (Zorilla et al., 2005). However, the involvement of GA during Pi stress responses is just beginning to emerge (Jiang et al., 2007). In this context, it is pertinent to note that auxin is reported to regulate root growth by modulating GA repression of RGA and GAI, two transcriptional repressors of growth (Fu and Harberd, 2003). One can speculate that this interaction occurs primarily through the repressive effect of Pi stress-responsive MYB62 on the biosynthesis of GA. We therefore present the hypothesis that Pi stress-induced MYB62, a negative regulator of GA biosynthesis, controls the root architecture in a pathway that is similar to the auxin-mediated regulation of RSA. The negative regulation of PSI genes by MYB62 suggests that it may also directly impact Pi homeostasis by tempering Pi stress-induced responses. Thus, the characterization of MYB62 adds considerably to our knowledge of transcriptional regulation during Pi stress responses, and opens new avenues of research on cross-talk between abiotic stress and developmental responses. The regulatory influence of Pi stress on flowering through MYB62 also provides an important insight regarding the molecular mechanisms by which abiotic stresses affect reproduction and yield in plants.
METHODS
Plant Material and Growth Conditions
All growth studies were performed with Arabidopsis (Arabidopsis thaliana) ecotype Columbia. Different growth conditions were used to analyze the biological function of MYB62.
Hydroponic Culture
Seeds were germinated in Premier ProMix PGX peat mix (Premier Horticulture Inc., Quakertown, PA). Plants were grown under greenhouse conditions under a 16-h light/8-h dark cycle at 1000 μmol m−2 s−1 PAR. Seedlings at the five to seven leaf stage were transferred to hydroponics after their roots were gently washed. After a recovery period of 7 d in 0.5 X modified Hoagland's solution, plants were transferred to hydroponic solutions containing 250 μM Pi (P+) or no Pi (P–) for 7 d before different parts were harvested (Devaiah et al., 2007a).
Petri Dish Culture
Seeds were surface sterilized, stratified at 4°C and germinated initially on 0.5 X MS medium. Seven-day-old seedlings were transferred to MS medium modified according to Devaiah et al. (2007) and supplemented with 1% (w/v) agar and 1.5% (w/v) sucrose. Treatments with phosphate sufficient and deficient medium were supplemented with 1 mM KH2PO4 or 0.5 mM K2SO4, respectively. The seedlings were grown under a 16-h light/8-h dark cycle at 22°C with 75 μmol m−2 s−1 PAR. The plates were inclined at a 65° angle to allow the roots to grow along the agar surface.
Liquid Culture
This method was used to generate material for all gene expression analysis except where stated otherwise. Surface sterilized seeds were dispensed into conical flasks containing half-strength MS medium without agar. The seedlings were grown under a 16-h light (125 μmol m−2 s−1 PAR)/8-h dark cycle at 22°C with constant shaking (85 rpm). Seven-day-old seedlings were rinsed thrice with distilled water and transferred into MS liquid media with Pi (1 mM) or without Pi. Plants were grown for 7 d, harvested, blot dried, frozen immediately in liquid nitrogen, and stored at –70°C until being used for RNA extraction (Karthikeyan et al., 2002).
GA Treatment of Plants
Plants were treated with GA in two ways, depending on the developmental stage and experimental requirement. GA4 and GA3 (Sigma Aldrich) were dissolved in alcohol and made up to required concentrations with water. When younger plants were treated, 5 μM of GA was placed as a droplet on the stem just below the oldest rosette leaf on three consecutive days. Higher concentrations of GAs were applied to older plants by spraying the GA on aerial parts eight alternate days. Ten replicates were maintained for each treatment and genotype; experiments were repeated twice.
Plant Transformation
Arabidopsis plants were transformed with two different gene constructs using the floral dip method (Clough and Bent, 1998).
The first construct was used to generate a GFP::MYB62 translational fusion protein. The full-length MYB62 cDNA was amplified with the primers 5′-CCCAAGCTTATGGAAAATTCGATGAAGAAGAAG-3′ and 5′-GCGGATCCTTACTCCCTAAACT GCCAAATGT-3′. The amplified fragment was digested with HindIII and BamHI and cloned into the binary pEGAD expression vector with the ENHANCED GREEN FLUORESCENT PROTEIN (EGFP) as an N-terminal translational fusion. This construct and an empty vector control were stably transformed into Arabidopsis and transgenic seedlings were selected by spraying 50 μl L−1 Basta.
The vector construct for overexpressing MYB62 was developed by modifying the pEGAD vector and cloning the MYB62 cDNA into the modified vector. The sequence encoding EGFP was excised from pEGAD using AgeI and EcoRI enzymes. The 5′ overhangs on the vector were end filled using DNA Polymerase1 (New England Biolabs Inc.) and ligated to form a new overexpression vector. The full-length MYB62 cDNA was amplified using the same primers mentioned above. The amplified fragment was cloned just after the CaMV 35S promoter into the modified vector using the HindIII and BamHI restriction enzymes. This construct was transformed into Arabidopsis and transgenic seedlings selected as described above. Each independent transgenic line was multiplied over three generations before analysis.
Visualization of GFP
Wide-field fluorescence imaging was performed using a NIKON E800 compound microscope equipped with a SPOT RT-slider digital camera (Diagnostic Instruments Inc.) interfaced to a computer. GFP excitation was performed with standard FITC filters. Images of the roots were taken through FITC filters under the 20X objective. To confirm the nuclear localization of MYB62, roots from 35S::GFP::MYB62 transgenic and wild-type control plants were stained with 4′,6-diamidino-2-phenylindole (DAPI). They were fixed in Phosphate Buffered Saline (PBS), pH 7.2, containing 4% (w/v) paraformaldehyde, 50 mM EDTA and 100 mM NaCl for 1 h and then washed thrice over 40 min with DAPI stain solution (100 mM PBS, 50 mM EDTA and 1 μg mL−1 DAPI). The nuclear specific dye DAPI co-localized with GFP fluorescence in the cells (Supplemental Figure 2).
RNA Gel Blot and Semi-Quantitative RT–PCR Analysis
Total RNA was extracted from plant samples using the TRIzol reagent (Invitrogen). Ten micrograms of total RNA was electrophoretically separated in a denaturing formaldehyde agarose gel and blotted onto nylon membranes. The nylon membranes were hybridized overnight with 32P-labeled DNA probes at 42°C, washed stringently and exposed to X-ray films.
For semi-quantitative RT–PCR analysis, two micrograms of DNAse-treated (RQ1 DNAse, promega) total RNA was used as a template for first-strand cDNA synthesis with Superscript II (Invitrogen) and an oligo(dT) primer. Semi-quantitative RT–PCR was performed as described by Devaiah et al. (2007a). The following gene-specific primers were used to detect cDNA of respective genes: MYB62, 5′-ATGGAAAATTCG ATGAAGAAGAAG-3′ and 5′-TTACTCCCTAAACTGCCAAATGT-3′; At4g02780 (AtCPS), 5′-GTGGGGTGTGCGCAGAAG-3′ and 5′-GTTTGGAGATGATCGCCAC-3′; At1g79460 (AtKS), 5′-TCTCTGGGGCTGCAACTTT-3′ and 5′-GTGAATCCATCGTCC TTCCT-3′; At5g25900 (AtKO), 5′-GGGAAAGATGTGGAATCC-3′ and 5′-GTCCTCTGGT CTCTCTC-3′; At4g25420 (AtGA20ox1), 5′-CATCTCCTGAGGAAGAAG-3′ and 5′-ACCC ATGTCCCAACGC-3′; At1g15550 (AtGA3ox1), 5′-TGGCATCGAAATTGATGTGG-3′ and 5′-CCATGTCACCGATTGGTATAG-3′; At3g23130 (SUPERMAN), 5′-ATGGAGAG ATCAAACAGC-3′ and 5′-TTAAGCGAAACCCAAACG-3′; SOC1, 5′-GGTGAGGGGCA AAAC-3′ and 5′-GAAGAACAAGGTAACCC-3′. Ten μl reactions were set up for each sample and amplified through 25 cycles. The product was seperated on an agarose gel and the results were documented. The ImageQuant 5.1 program (GE Healthcare Life sciences) was used for densitometric quantification of the relative intensity of the DNA bands. The RT–PCR analysis was repeated twice, with consistent results.
Physiological Measurements
Anthocyanin Estimation
Rosette leaves of plants raised hydroponically as described above were used for anthocyanin estimation. About 100 mg of frozen ground tissue from each treatment and line was used for the quantification of anthocyanins as described by Lange et al. (1971). The optical density was measured at A532 and A653. Subtraction of 0.24 A653 compensated for the small overlap in absorbance at 532 nm by the chlorophylls. The concentration was determined by using the corrected absorbance and the molar extinction coefficient (ϵ) of 38 000 L mol−1 cm−1 for anthocyanin.
Quantification of Chlorophyll
Chlorophyll was quantified in rosette leaves of 3-week-old plants grown on peat–vermiculite mix using the method described by Fitter et al. (2002). Leaf discs of 0.5 cm2 area were collected using a bore with an 8-mm diameter. The discs were flash frozen in liquid nitrogen, ground in a micro-centrifuge tube and then suspended in 1 ml 80% acetone. Following centrifugation to remove debris, the OD of the supernatant was measured at 645 and 663 nm. Chlorophyll concentration was calculated using the formula: chlorophyll concentration (mg mL−1) = (OD645 × 20.2) + (OD663 × 8.0); this value was then divided by 0.5 to obtain the total chlorophyll concentration per cm2 tissue.
Quantification of Total Pi
Total Pi concentration was quantified using a modification of the US Environmental Protection Agency (EPA) Method 365.2. About 50 mg of fresh sample from plants grown in Petri dish culture as described above was taken in a pre-weighed vial and oven dried. After recording their dry weight, the samples were flamed to ash and dissolved in 100 μl of concentrated HCl. Ten μl of this sample was diluted in 790 μl of water. To a reaction containing 800 μl of diluted sample, 200 μl of mixed reagent (4.8 mM NH4MoO4, 2.5 N H2SO4, and 35 mM of ascorbic acid) was added and incubated at 45°C for 20 min. Total Pi content was measured at A650 using appropriate standards and expressed as total Pi/mg tissue dry weight.
Quantification of Total Acid Phosphatase Activity (APA)
Total Acid Phosphatase was measured as described earlier using the pNPP hydrolysis assay (Richardson et al., 2001) using material from plants grown in Petri dish culture. Samples were extracted from about 30 mg of finely ground frozen tissue. The enzyme activity was measured at A405. Total protein was estimated separately using Bradford's reagent and the total acid phosphatase activity expressed as mU/mg protein.
Measurements of Roots and Root/Shoot Ratio
Seedlings were grown on Petri dishes under Pi-sufficient or deficient conditions as described earlier. After 7 d of treatment, the primary root length, lateral root number, and lateral root lengths were measured. The roots from 12 individual plants of each line per treatment were spread out carefully with a fine brush, scanned at 600 dpi and the different root traits were evaluated using the ImageJ program (Abramoff et al., 2004). For measuring root/shoot ratio, shoots from plants grown as described above were excised just below the hypocotyls. Shoots and roots from three plants of each genotype were pooled and treated as one biological sample. Samples were weighed and the root/shoot ratio was calculated by dividing the root fresh weight by shoot fresh weight. Values are the mean of seven replicates and the experiment was repeated twice.
Pi Uptake Assay
Pi uptake assay was performed using the method described earlier by Devaiah et al. (2007a). Briefly, 7-day-old wild-type and MYB62 overexpressing seedlings were grown in Pi-sufficient or deficient medium for 3 d. Groups of 10 seedlings were used as one biological sample. The roots of the seedlings were incubated in a pre-treatment solution (5 mM MES and 0.1 mM CaCl2, pH 5.7) for 20 min before moving them into 2 ml of uptake solution (5 mM MES, 0.1 mM CaCl2, 50 μM KH2PO4, pH 5.7) containing [33P]orthophosphate (0.15 μCi mL−1). Samples were moved into ice-cold de-sorption solution (5 mM MES, 0.1 mM CaCl2 and 1 mM KH2PO4, pH 5.7) at the end of 1 and 2 h, respectively. After two washes with fresh de-sorption solution for 45 min, the samples were blot dried, placed in pre-weighed scintillation vials, oven dried overnight at 65°C and their dry weight recorded. Four ml of scintillation cocktail was added into each vial and radioactivity was measured with a scintillation counter (Beckman Coulter).
Statistical Analysis
Statistical significance of difference between mean values was determined using Student's t-test and ANOVA. Different letters on the error bars of histograms were used to indicate means that were statistically different at P ≤ 0.05.
Genebank identifiers of MYB62: At1g68320; AY519568.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by grants from United States Department of Agriculture and the McKnight Foundation.
Supplementary Material
Acknowledgments
We would like to thank Mike Poling and Ajay Jain for their valuable help in preparing and editing this manuscript and Amber Jannasch of the Purdue Metabolomics facility for helping in the estimation of GA. We also thank the Arabidopsis Resource Center at the Ohio State University for providing the vectors used in this study. No conflict of interest declared.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG. LEPS2, a phosphorus starvation–induced novel acid phosphatase from tomato. Plant Physiol. 2001;125:728–737. doi: 10.1104/pp.125.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereterbide A, Hernould M, Castera S, Mouras A. Inhibition of cell proliferation, cell expansion and differentiation by the Arabidopsis SUPERMAN gene in transgenic tobacco plants. Planta. 2001;214:22–29. doi: 10.1007/s004250100584. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development. 1992;114:599–615. doi: 10.1242/dev.114.3.599. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/ oxidative stress condition. Plant J. 1999;19:579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007a;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis is synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 2007b;145:147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- Gocal GFW, et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001;127:1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ. Genetic responses to phosphorus deficiency. Ann. Bot. (Lond.) 2004;94:323–332. doi: 10.1093/aob/mch156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis, enzymes, genes and their regulation. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:431–60. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hernández G, et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 2007;144:752–767. doi: 10.1104/pp.107.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. Differential effects of sucrose and auxin on localized Pi–deficiency induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 2007a;144:232–247. doi: 10.1104/pp.106.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vasconcelos MJ, Sahi SV, Raghothama KG. Molecular mechanisms of plant adaptation to phosphate deficiency. In: Janick J, editor. Plant Breeding Reviews. Vol. 29. NJ: John Wiley & Sons; 2007b. pp. 359–399. [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. Phosphate-starvation root architecture and anthocyanin-accumulation responses are modulated by the GA–DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D'Urzo MP, Damz B, Raghothama KG. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002;130:221–233. doi: 10.1104/pp.020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Caskin P, Spray CR, Suzuki Y, Phinney BO, MacMillan J. Metabolism and biological activity of gibberellin A4 in vegetative shoots of Zea mays, Oryza sativa and Arabidopsis thaliana. Plant Physiol. 1993;102:379–386. doi: 10.1104/pp.102.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen Induction and analysis of gibberellin sensitive mutants of Arabidopsis thaliana (l.) Heynh Theor. Appl. Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Louren-Martinet EP, van Rign L, Zeevaart JAD. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 1985;65:33–39. [Google Scholar]
- Koornneef M, van Eden J, Hanhart CJ, de Jongh AMM. Genetic fine-structure of the GA-1 locus in the higher plant Arabidopsis thaliana (L.) Heynh. Genet. Res. 1983;41:57–68. [Google Scholar]
- Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M. Alteration of hormone levels in transgenic tobacco plants over–expressing the rice homeobox gene OSH1. Plant Physiol. 1998;116:471–476. doi: 10.1104/pp.116.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Shropshire W, Jr, Mohr H. An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol. 1971;47:649–655. doi: 10.1104/pp.47.5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004;37:720–729. doi: 10.1111/j.1365-313x.2003.01998.x. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition higher plants. London: Academic Press; 1995. [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:1–11. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. TIG. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl Acad. Sci. U S A. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl Acad. Sci. U S A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Sung-Suk S, Lee H, Choi K-R, Hong C, Paek N-C, Kim S-G, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003;35:613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Műller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 2007;30:1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Wienand U, Peterson PA, Saedler H. Molecular cloning of the c1 locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO J. 1986;5:829–834. doi: 10.1002/j.1460-2075.1986.tb04291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000;3:182–187. [PubMed] [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE. Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J. 2001;25:641–649. doi: 10.1046/j.1365-313x.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl Acad. Sci. U S A. 1990;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M. Over-expression of a tobacco homeobox gene NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA20-oxidase. Plant J. 1998;15:391–400. doi: 10.1046/j.1365-313x.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- Vettakkorumakankav NN, Falk D, Saxena P, Fletcher RA. A crucial role for gibberellins in stress protection of plants. Plant Cell Physiol. 1999;40:542–548. [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T. Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ. 2003;26:1515–1523. [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl Acad. Sci. U S A. 1999;96:15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Talon M. Gibberellin mutants in Arabidopsis thaliana. In: Karssen CM, van Loon LC, Vreugdenhil D, editors. In Progress in Plant Growth Regulation. Dordrecht: Kluwer; 1992. pp. 34–42. [Google Scholar]
- Zhu J, et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl Acad. Sci. U S A. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zorilla JMF, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J. The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 2004;55:285–293. doi: 10.1093/jxb/erh009. [DOI] [PubMed] [Google Scholar]
- Zorilla JMF, Martín AC, Leyva A, Paz-Ares J. Interaction between phosphate starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005;138:847–857. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.