Abstract
Thellungiella salsuginea (halophila) is a close relative of Arabidopsis thaliana but, unlike A. thaliana, it grows well in extreme conditions of cold, salt, and drought as well as nitrogen limitation. Over the last decade, many laboratories have started to use Thellungiella to investigate the physiological, metabolic, and molecular mechanisms of abiotic stress tolerance in plants, and new knowledge has been gained in particular with respect to ion transport and gene expression. The advantage of Thellungiella over other extremophile model plants is that it can be directly compared with Arabidopsis, and therefore generate information on both essential and critical components of stress tolerance. Thellungiella research is supported by a growing body of technical resources comprising physiological and molecular protocols, ecotype collections, expressed sequence tags, cDNA-libraries, microarrays, and a pending genome sequence. This review summarizes the current state of knowledge on Thellungiella and re-evaluates its usefulness as a model for research into plant stress tolerance.
Keywords: abiotic/environmental stress, ion channels, adaptation - evolutionary, comparative genomics, gene expression, Brassica
INTRODUCTION
The sensitivity of crops to harsh climates and soil conditions is a major limitation for worldwide food production (Cakmak, 2002). However, tolerance to cold, drought, and salinity has evolved in many wild plant species, and these so-called extremophiles represent an important genetic reservoir that can be exploited for improving crop performance on marginal land.
In September 2004, a group of researchers from around the globe gathered in Paris to discuss the prospects of a small cruciferous plant to become a new model for molecular research into plant stress tolerance (Bressan et al., 2001; Zhu, 2001; Amtmann et al., 2005). The plant under question was Thellungiella, a member of the Brassica family and a close relative of the queen of plant models, Arabidopsis thaliana (Bressan et al., 2001; Zhu, 2001; Warwick et al., 2006). Thellungiella, or ‘salt cress’ as it is also referred to, resembles its famous cousin in many assets that make this plant so popular with geneticists and molecular biologists, including short lifecycle, small genome (approximately twice the size of the Arabidopsis genome), and copious seed production. In addition, Thellungiella is a real ‘tough cookie’, able to grow and reproduce under conditions of extreme cold, drought, and salinity.
Based on the experience gathered with Thellungiella in the individual laboratories, the Paris workshop participants decided to recommend Thellungiella as an extremophile plant model system and to promote its usage by sharing protocols, seed stocks, and BAC and cDNA libraries (www.thellungiella.org). Four years on, with some 40 experimental papers published and the full genome sequence pending, Thellungiella research is well underway. In this review, I will summarize the most important results obtained to date and reassess the usefulness of Thellungiella as a model species.
PHYLOGENY AND ECOTYPES
The exact phylogenetic position of Thellungiella is still uncertain. Based on comparison of nuclear and chloroplast sequences, Thellungiella and Arabidopsis are considered to belong to different clades, despite their similar appearance and, within its clade, Thellungiella seems to be most closely related to Eutrema (Al-Shehbaz et al., 1999; Warwick et al., 2006; Sun et al., 2007). The Thellungiella genus contains four closely related species: T. salsuginea (Pallas) O.E. Schulz, T. halophila (C.A. Meyer) O.E. Schultz, T. parvula (Schrenk) Al-Shehbaz and O'Kane, and a recently described new species, T. botschantzevii (German, 2008). The Shandong ecotype from the North Eastern coast of China, which is most commonly investigated in the laboratories, has been cited as T. halophila but close inspection indicates that it belongs in fact to T. salsuginea and not to T. halophila (Figure 1). To avoid confusion, we will generally allude to the Shandong ecotype when talking about ‘Thellungiella’ or otherwise specify the ecotype. In addition to the well characterized T. salsuginea ecotypes Shandong (from China) and Yukon (from Canada), several ecotypes from Russia and Kazakhstan belonging to T. salsuginea (Altai, Buriatia, Tuva, and Yakutsk), T. halophila (Bayanaul), and T. botschantzevii (Alei and Saratov) have been collected by Bert de Boer, Alexei Babakov and colleagues (Bert de Boer, personal communication). Other Thellungiella ecotypes have been found in the USA (Ray Bressan, personal communication). Phenotyping and genotyping of these collections will provide an essential resource for gene identification by QTL analysis and mutant mapping.
Figure 1.
The ‘Real’ Thellungiella halophila (Bayanaul) Growing in the Glasshouse of the Vrije University of Amsterdam.
Seeds supplied by D.A. German (Botanical Institute Barnaul, Russia). Photo by A.H. de Boer.
DEVELOPMENT AND MORPHOLOGY
Thellungiella differs from A. thaliana in basic developmental programmes. In accordance with its extremophilic lifestyle, Thellungiella germinates and sets seeds over extended periods of time, which, in the laboratory, can create problems for comparative phenotypic characterization and needs to be taken into account in transformation protocols (e.g. through repetitive flower-dipping, see protocol by Bressan and colleagues, www.thellungiella.org). It also flowers later and requires vernalization to promote flowering. Fang and colleagues (2006, 2008) showed that ectopic expression of the Thellungiella FLC gene, a key regulator of the vernalization response pathway, caused a late-flowering phenotype in A. thaliana (Ler). Conversely, an RNAi construct silencing the endogenous ThFLC in Thellungiella produced an early flowering phenotype while retaining the same salt tolerance as wild-type. The authors propose that this transgenic line provides a better research model for plant salt tolerance studies than wild-type. Another way to obtain plants that are similar in growth rate and development (Figure 2) is to subject Arabidopsis and Thellungiella to different light regimes (10 h light at 200 μE m−2 s−1 for A. thaliana and 14 h light at 250 μE m−2 s−1 for Thellungiella deliver plants with large rosettes; Wang et al., 2006).
Figure 2.
Typical Rosette of (A) Arabidopsis thaliana (Col0) and (B) Thellungiella salsuginea (Shandong) Grown in Low-Salt Hydroponic Culture with 10 and 14 h Daylight, Respectively.
Photos by V. Martínez, University of Glasgow.
Thellungiella and Arabidopsis are very similar in form and shape. However, leaves of Thellungiella are often serrated (depending on growth conditions and ecotype) and appear slightly more succulent and waxy (Figure 2). In fact, one of the first experimental papers published on Thellungiella described a higher concentration and different composition of epicuticular waxes in leaves of T. parvula and halophila compared to A. thaliana (Teusink et al., 2002). At the tissue level, leaves have a second layer of palisade mesophyll cells while roots develop both an extra endodermis and cortex cell layer compared to Arabidopsis (Inan et al., 2004). The same authors also observed that stomata in Thellungiella are present at higher density but are less open than in Arabidopsis, and respond to salt stress by closing more tightly. However, other laboratories reported that transpiration and stomatal conductance are less affected (decreased) by salt in Thellungiella than in Arabidopsis (Volkov et al., 2004; M'rah et al., 2006, 2007).
STRESS TOLERANCE
Elizabeth Weretilnyk and colleagues from the Universities of McMaster and Waterloo in Canada are investigating the phenotypic plasticity of the Yukon ecotype in its natural environment. The alkaline salt flats in the Yukon Territory of Canada are characterized by a semiarid climate with a very short growing season. The permafrost soils are saturated with calcium carbonate, magnesium sulfate, and sodium chloride, and are deficient in essential macronutrients (Griffith et al., 2007). Depending on the climate conditions of a particular year, Thellungiella can be found as small, individual plants or as tall plants covering large areas (Figure 3). Profiling of transcriptome and metabolome from plants collected in different years is currently underway to correlate gene expression and metabolism with climatic data in the hope to reveal adaptive mechanisms.
Figure 3.
Thellungiella salsuginea (Yukon) Growing in the Laboratory and its Natural Field Site in the Yukon Territory in Canada.
Upper left: Chamber-grown plant showing terminal flowers and prominent rosette. Photo by E.A. Weretilnyk, McMaster University. Upper right: Yukon field site in May 2006. Photo by J. Dedrick, McMaster University. Bottom: Yukon field site in July 2005. Photo by J. Dedrick, McMaster University.
Tolerance of Thellungiella to high salt, drought, and cold has been confirmed in the laboratory. Unlike Arabidopsis, Thellungiella still thrives in high salt concentrations (100–500 mM; Inan et al., 2004; Taji et al., 2004; Gong et al., 2005), survives and re-generates after extended periods of drought (Arie Altman, personal communication), and withstands freezing (LT50 values of –13°C for non-acclimated plants and –19°C after acclimation (Griffith et al., 2007). Furthermore, Kant and colleagues showed that Thellungiella also grows better than Arabidopsis under moderate (1 mM nitrate) and severe (0.4 mM nitrate) nitrogen-limiting conditions (Kant et al., 2008). Higher salt tolerance of Thellungiella than Arabidopsis is reflected in lower concentrations of sodium (Na+) and chloride (Cl–) in the shoots and a higher potassium (K+) to Na+ ratio but not in a difference in leaf dehydration (Inan et al., 2004; Taji et al., 2004; Volkov et al., 2004; M'rah et al., 2007; Ghars et al., 2008). Several studies have linked superior ion homeostasis of Thellungiella under salt stress to the selectivity and regulation of individual ion transporters, and these will be described in more detail below. Probably due to the restriction of toxic Na+ in the leaves, shoot growth reduction in high salt—albeit evident in Thellungiella—is much less pronounced than in Arabidopsis, at the level of both leaf initiation and leaf expansion (M'rah et al., 2007). Drought-stressed Thellungiella plants maintain constant water contents, thereby protecting shoot meristems from desiccation, which, in turn, allows fast re-growth after re-watering (Arie Altman, personal communication). Cold-treated Thellungiella plants lack endogenous ice nucleation or anti-freeze activity, indicating a potential for supercooling (Griffith et al., 2007). Finally, higher nitrogen-usage efficiency of Thellungiella is evident in higher nitrate uptake, and a higher content in total amino acids and total soluble protein (Kant et al., 2008).
METABOLITES
Proline can act as a compatible solute and osmoprotectant in the cytoplasm (Munns and Tester, 2008). In response to salt treatment, proline levels seem to increase to higher levels in Thellungiella than in Arabidopsis, but there are conflicting data as to whether the mechanism of accumulation differs between the two species (Inan et al., 2004; Taji et al., 2004; Kant et al., 2006; Ghars et al., 2008). Proline accumulation was linked to constitutively high transcript levels of P5CS involved in proline biosynthesis (Taji et al., 2004), and to low expression of PDH required for proline catabolism (Kant et al., 2006).
Thioredoxin is a critical component of the defense system against oxidative damage and lipid peroxidation, and its abundance (CDSP32 thioredoxin) was found to be higher in Thellungiella than in A. thaliana under both control conditions and salt treatment (M'rah et al., 2007). Phytoalexins play an important, albeit not fully understood, role in pathogen defense (Rogers et al., 1996). Both Arabidopsis and Thellungiella increase phytoalexin biosynthesis also in response to abiotic stress, but differ in the type of compounds produced (e.g. camalexin by A. thaliana, wasalexins, and methoxybrassenin by Thellungiella; Pedras and Adio, 2008). Together with the above-mentioned findings on cuticular wax production, these studies emphasize the potential model function of Thellungiella not only for investigating stress tolerance, but also for deciphering specific biochemical pathways.
FUNCTIONAL CHARACTERIZATION OF THELLUNGIELLA GENES
Based on high sequence identity between Thellungiella and Arabidopsis (92% on average, Inan et al., 2004), several Thellungiella genes have now been cloned and functionally characterized, including TsVP, encoding a vacuolar pyrophosphatase (Gao et al., 2006; Duan et al., 2007; Li et al., 2008), ThCBL9, encoding a calcineurin-B-like protein (Sun et al., 2008), ThHSC70, encoding a heat-shock protein (Zhang et al., 2004), ThCYP1, encoding a cyclophilin (Chen et al., 2007), and ThZF1, encoding a Cys-2/His-2-type transcription factor (Xu et al., 2007). Overexpression of TsVP improved salt and drought tolerance in maize and cotton, which was correlated with higher vacuolar H+-PPase activity (Li et al., 2008; Lv et al., 2008). Similarly, TsVP expression in tobacco improved growth on high salt and viability of msophyll protolasts under salt shock conditions (Gao et al., 2006; Duan et al., 2007). TsVP overexpressing tobacco plants accumulated more Na+ in the leaves, indicating that the reason for improved salt tolerance was indeed due to efficient compartmentalization of Na+ in the vacuoles (Gao et al., 2006). Overexpression of ThCBL9 (Sun et al., 2008) or ThHSC70 (Zhang et al., 2004) in A. thaliana enhanced tolerance to salt and osmotic stress or tolerance to high temperature and chilling, respectively. While the latter finding is in accordance with the function of HSC70 as a molecular chaperon, the role of CBL9 in salt tolerance remains to be further investigated. In Arabidopsis, this gene, in conjunction with the CBL-interacting protein kinase 23, activates the plasma membrane K+-channel AKT1 (Li et al., 2006a; Xu et al., 2006; Lee et al., 2007), and therefore salt tolerance of CBL9 overexpressing lines could be related to improved K+-uptake. Finally, heterologous expression of the ThCYP1 gene increased salt tolerance of both fission yeast and tobacco Bright Yellow 2 (BY-2) cells, suggesting that ThCYP1 may promote the appropriate folding of certain stress-related proteins (Chen et al., 2007). While all these studies provide evidence for a potential role of the respective Thellungiella genes in stress tolerance, they do not necessarily explain the difference of tolerance between Thellungiella and A. thaliana. Indeed, it has been shown that the homologous genes from Arabidopsis also endow the plant with enhanced tolerance when overexpressed (e.g. AtAVP1; Gaxiola et al., 2001). This observation not only provides evidence for an unrealized inherent potential for stress tolerance in the gene pool of A. thaliana, but raises two important questions: (1) are salt-tolerance genes differentially regulated in Thellungiella and Arabidopsis, and (2) does constitutively high expression create a disadvantage in non-stressed conditions? The above-mentioned studies report upregulation of the genes in Thellungiella, such as by heat and cold (Thhsc70), by salt, abscisic acid (ABA), H2O2 and heat shock (ThCYP1), by ABA, NaCl, and PEG (CBL9) and by salt and dought (ThZF1), but they do not provide comparative data from Arabidopsis or any other stress-sensitive species. Clearly, more detailed comparisons between Arabidopsis and Thellungiella genes are needed in the future.
GENE EXPRESSION
Several approaches have been taken to investigate stress responses of Thellungiella at the level of gene expression including comparison of EST libraries obtained from plants grown under different stress conditions, microarrays, and quantitative polymerase chain reaction (qPCR). Wong and colleagues compared 6578 ESTs representing 3628 unique genes from cDNA libraries of cold-, drought-, and salinity-stressed plants of the Yukon ecotype and found very little overlap between gene expression in different conditions, suggesting that Thellungiella tailors responses that are very specific to the individual stress (Wong et al., 2005). A similar result was obtained when microarrays spotted with the ESTs were probed with mRNA obtained from stressed plants (Wong et al., 2006). Interestingly, drought stress induced down-regulation of many genes related to pathogen defense. The authors propose that the observed expression patterns reflect the capacity of Thellungiella to be more precise and economical in its stress response than sensitive species. EST libraries and microarrays constructed from the Yukon ecotype contained genes that have no homologs in the Arabidopsis and thus provide the opportunity for discovering new genes. However, their genome coverage to date is limited (approximately 10% of the genome). Other studies have used A. thaliana microarrays representing 7000 genes as full-length cDNAs (RIKEN array; Taji et al., 2004) or more than 25 000 genes as 70mers (‘Arizona’ array; Gong et al., 2005). Unlike arrays with shorter probes (Affymetrix ATH1; Volkov et al., 2004), these arrays produce good hybridization signals when probed with Thellungiella mRNA but, in contrast to the Yukon array, they only provide information on genes that are homologous to Arabidopsis genes. Nevertheless, these studies uncovered strikingly different responses of Thellungiella and Arabidopsis to salt stress. In general, Thellungiella shows much fewer transcript changes than Arabidopsis when exposed to the same stress. In response to a very short salt shock (2 h in 250 mM NaCl), the RIKEN array identified only six transcripts as up-regulated in Thellungiella compared to 40 in Arabidopsis (Taji et al., 2004). Similarly, Gong and colleagues found that fewer transcripts responded to a 150-mM NaCl treatment (for 3 and 24 h) in Thellungiella than in Arabidopsis but the number of responsive transcripts in Thellungiella increased with stress intensity (Gong et al., 2005). Thus, many transcripts showed a similar response in Thellungiella treated with 250 mM NaCl as in Arabidopsis treated with 150 mM NaCl. In addition to two large gene clusters representing this type of expression profile (one for up and one for down-regulated genes), they also identified smaller clusters containing genes that responded in only one of the species. Both studies (Taji et al., 2004; Gong et al., 2005) pointed to the phenomenon that transcript levels of some genes that are salt-stress inducible in Arabidopsis have already a higher transcript level in Thellungiella under low-salt conditions. Hence, comparative transcriptomics between Thellungiella and Arabidopsis have highlighted three important potential adaptations to extreme conditions: (1) specificity—Thellungiella regulates a specific set of genes in each stress situation, (2) anticipation—Thellungiella is constitutively prepared for stress, and (3) sensitivity—Thellungiella requires higher stress doses to induce transcriptional responses.
Sensitivity of the transcriptional responses could be linked to and should be separated from the degree of stress experience. In a recent study (B. Wang and A. Amtmann, unpublished results), we recorded transcriptional responses of membrane transporter-encoding genes of Arabidopsis and Thellungiella to salt stress using the AMT oligonucleotide array (Maathuis et al., 2003). Unlike other studies, this analysis was carried out separately for roots and shoots. We found that considerably fewer Thellungiella than Arabidopsis transcripts responded to the treatment (100 mM NaCl for 24 h) in the shoots but a similar number of transcripts (albeit different ones) responded in the roots of the two species. Considering that under identical salt treatment leaf cells are exposed to less Na+ in Thellungiella than in Arabidopsis (see above) while root cells of both species experience the same concentration, this finding suggests that the number of stress-responsive transcripts is correlated to stress experience rather than stress sensitivity. However, two observations challenge this paradigm. First, differences in shoot Na+ content will be very small after the short exposure times applied in some studies (e.g. 2 and 3 h), and, secondly, Thellungiella showed fewer transcript changes also in response to stresses that are directly sensed by the leaves (e.g. cold Wong et al., 2006) and ozone (Li et al., 2006b)). It will be interesting to test whether differential responsiveness of the two species to these stresses is already evident at the level of hormones and secondary messengers upstream of the transcriptional response.
While all studies agreed with respect to general trends in transcript regulation under stress, responses by individual genes are still under debate. For example, Taji and colleagues (2004) reported that the level of transcript encoding the plasma membrane Na+/H+ antiporter SOS1 was higher in Thellungiella than in Arabidopsis plants in control conditions and not further induced by salt. Based on qPCR experiments, Kant and colleagues (2006) measured a different and more detailed profile in which SOS1 was more strongly induced by salt in Thellungiella than in Arabidopsis as far as the shoots were concerned but constitutively higher in the roots (in control conditions). ThSOS1 induction in Thellungiella leaves during salt stress was confirmed at the protein level using an antibody against AtSOS1 (Vera-Estrella et al., 2005). The case of SOS1 exemplifies the need for detailed profiling of transcripts and proteins under a range of conditions covering a wide spectrum of stress doses and exposure times before species-specific regulation of genes that are potentially important for salt tolerance can be assessed.
ION TRANSPORT
The fact that, under salt stress, Thellungiella accumulates less Na+ in its shoot than Arabidopsis has been confirmed by many studies (see above). This observation suggests that transport processes are important for salt tolerance in Thellungiella. Lower net Na+ uptake into the shoot can be due to lower net Na+ uptake into the root (either through lower unidirectional influx or through higher unidirectional efflux), lower root–shoot transport of Na+ (either through lower xylem loading or higher recovery from the xylem), and higher Na+ recycling from the shoots into the roots (higher phloem loading). Our study with radioactive 22Na (Wang et al., 2006) showed that in 100 mM NaCl, the steady-state unidirectional Na+ influx into the roots was significantly smaller in Thellungiella than in A. thaliana (approx 50%). Surprisingly, unidirectional efflux was also slightly smaller in Thellungiella. Na+ influx was inhibited in both species by micromolar external Ca2+, which inhibits voltage-independent, non-selective channels in other species (Davenport and Tester, 2000), but not by Cs+ and TEA+, two well known inhibitors of voltage-gated K+-selective channels (Véry and Sentenac, 2002). In a subsequent electrophysiological study (Volkov and Amtmann, 2006), we confirmed that Na+ uptake across the plasma membrane of root cells generates an electric current that changes instantaneously when a voltage step is applied, indicating that the open probability of the underlying channel is not voltage-dependent. Unfortunately, the genes encoding this type of voltage-independent channel (VIC) have not yet been identified. Cyclic nucleotide-gated channels have been suggested as one candidate system mediating instantaneous current into A. thaliana roots (Maathuis and Sanders, 2001; Demidchik and Maathuis, 2007) but the Na+ current recorded in Thellungiella roots was not altered by addition of cyclic nucleotides. In accordance with the radiotracer data, Na+ inward current was significantly smaller in Thellungiella than in Arabidopsis, and this difference could be attributed to a higher selectivity of the Thellungiella VIC for K+ over Na+ (Volkov and Amtmann, 2006). The difference in Na+ permeability between Thellungiella and Arabidopsis was reflected in the membrane potential. Whereas Arabidopsis showed a large depolarization after addition of Na+ to the external medium, Thellungiella showed only a small and transient depolarization (Volkov and Amtmann, 2006). This is an important observation because membrane depolarization decreases K+ uptake through inward-rectifying K+-selective channels (Amtmann et al., 1999; Amtmann and Sanders, 1999). Our findings therefore explain not only the lower Na+ content of Thellungiella, but also the relatively higher K+ content compared with Arabidopsis (Wang et al., 2006). Furthermore, it was recently shown that expression of the high-affinity uptake system HAK5 depends on the membrane potential with HAK5-transcript levels increasing at more negative membrane potentials (Nieves-Cordones et al., 2008). Such regulation provides the basis for HAK5-induction under K+-starvation (which hyperpolarizes the membrane). More importantly, considering the difference in membrane potential between the two species (Volkov et al., 2006), it could explain higher HAK5 expression (and high-affinity K+-uptake) under salt stress in Thellungiella compared to Arabidopsis (Alemán et al., 2008). The combined evidence exemplifies how a structural feature of one transporter (K+/Na+-selectivity of VIC) can have a number of knock-on effects on the regulation of other transporters. Another important consequence of decreased Na+ permeability is related to energy conservation. Quantitative comparison of Na+ inward current at the respective resting membrane potentials with net Na+ accumulation in the shoots showed that the observed difference in Na+ influx into root cells was sufficient to explain the difference in net shoot uptake between Thellungiella and Arabidopsis. The calculations even predicted a reduced necessity for Na+ efflux in Thellungiella. Since Na+ efflux is an energy-consuming process, this finding raises the possibility that salt tolerance in Thellungiella is linked to its overall energy balance in high salt conditions.
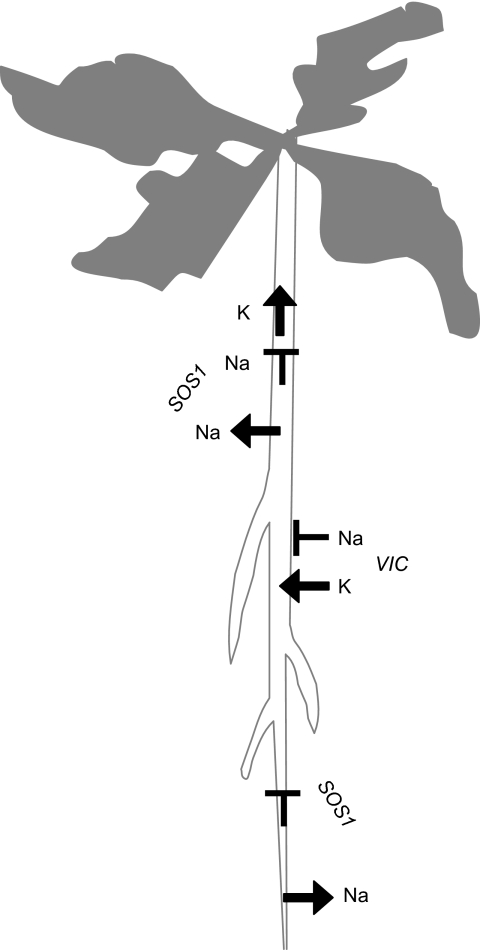
The fact that Na+ influx into root cells is sufficient to explain the difference in shoot Na+ accumulation between Thellungiella and Arabidopsis does not exclude an involvement of other transport processes in salt tolerance. A recent study by the Bohnert lab showed that knock-down of SOS1 by RNAi transforms Thellungiella into a salt-sensitive species (Oh et al., 2007, 2008). Detailed analysis of the RNAi lines provided evidence that SOS1 is required to avoid Na+ influx from the root tip into the root elongation zone during early stages of salt stress, and to recover Na+ from the xylem during prolonged salt stress. These findings prove that SOS1 is required for salt tolerance but do not necessarily explain the difference in salt sensitivity between Arabidopsis and Thellungiella. However, previous studies strongly suggest that differential regulation of this transport system exists between the two species (Taji et al., 2004; Kant et al., 2006). It has recently been shown that the stability of the Arabidopsis AtSOS1 mRNA is increased during salt stress in a ROS-dependent manner (Chung et al., 2008), raising the possibility that structural differences between ThSOS1 and AtSOS1 enhance ThSOS1 stability in the absence of salt stress and/or enhance stress-induced induction (Oh et al., 2008). This type of study should bring us closer to answering the question of whether SOS1 contributes to the difference in salt tolerance between Arabidopsis and Thellungiella. To summarize the current state of knowledge: (1) SOS1 function is essential for growth on high salt of both Thellungiella and Arabidopsis; (2) lower Na+ permeability of VICs is not sufficient for salt tolerance but critical for the difference in shoot Na+ accumulation between Thellungiella and Arabidopsis. The functions of both systems in restricting shoot Na+ accumulation are summarized in Figure 4.
Figure 4.
Effective Exclusion of Na+ from the Shoot of Thellungiella (Shandong) Is Achieved through the Combined Action of a Voltage-Independent Channel (VIC) and a Na+/H+ Antiporter (SOS1).
VIC has higher selectivity for K+ over Na+ than the respective system in Arabidopsis, thereby limiting Na+ influx into root cells and maintaining a negative membrane potential that activates K+ inward rectifying channels and high-affinity transport systems. SOS1 exports Na+ from the root tip and xylem, thereby limiting its transport into the root elongation zone and the shoot. For details and references, see text.
The observation that Thellungiella effectively restricts Na+ accumulation in the shoots suggests that Na+ compartmentalization during salt stress is less important here than in Arabidopsis or other salt-sensitive species. Nevertheless, Vera-Estrella and colleagues (2005) measured increased Na+/H+ antiport activity across the tonoplast of Thellungiella leaf cells under salt stress. Surprisingly, an antibody against the Arabidopsis vacuolar Na+/H+ antiporter AtNHX1 (also recognizing AtNHX2 and AtNHX3) did not detect the protein in Thellungiella, suggesting either low similarity of antigenicity in NHX-type transporters between the two species or the involvement of other transporters in Na+/H+ antiport.
TECHNIQUES, TOOLS, AND RESOURCES
One of the advantages of Thellungiella as a model species resides in its amenability to a wide range of physiological and molecular techniques (Figure 5). The above reviewed studies provide evidence for applicability of physiological and electrophysiological techniques (patch clamp, tracer flux analysis, and ion exchange assays), as well as transcriptomics and metabolomics. In my own lab, we have shown that micrografting techniques can be used to investigate hybrid plants between Thellungiella and Arabidopsis (G. Littlejohn and A. Amtmann, unpublished results). Efficient transformation of Thellungiella can be achieved by flower-dipping (protocol available at www.thellungiella.org) and leaf disc transformation (Li et al., 2007). On the basis of an improved transformation protocol, Bressan and colleagues have created a T-DNA knockout collection, which is available to the research community for a modest fee to cover seed divisions and handling (contact Ray Bressan at bressan@purdue.edu). A transient root transformation protocol has also been shown to work in Thellungiella (Campanoni et al., 2007). Several cDNA libraries have been constructed from Thellungiella plants grown under various stress conditions (Wong et al., 2006; Liu et al., 2007; Ni et al., 2007). One cDNA library has been used to transform fission yeast (Saccharomyces pombe) and several transformants selected on high salt are now under investigation (Liu et al., 2007). Another cDNA library has been shuttled into the binary vector pCB406 and the resulting population of A. thaliana transformants will be available for functional gene mining (Ni et al., 2007). Most importantly, K. Schumaker, R. Wing (University of Arizona) and T. Mitchell-Olds (Duke University) have launched a sequencing program at JGI, which is expected to deliver a full genome sequence of Thellungiella over the next couple of years. The sequence will provide an invaluable tool both for comparative genomics within the Brassicaceae (Schranz et al., 2006) and for the identification of novel genes.
Figure 5.
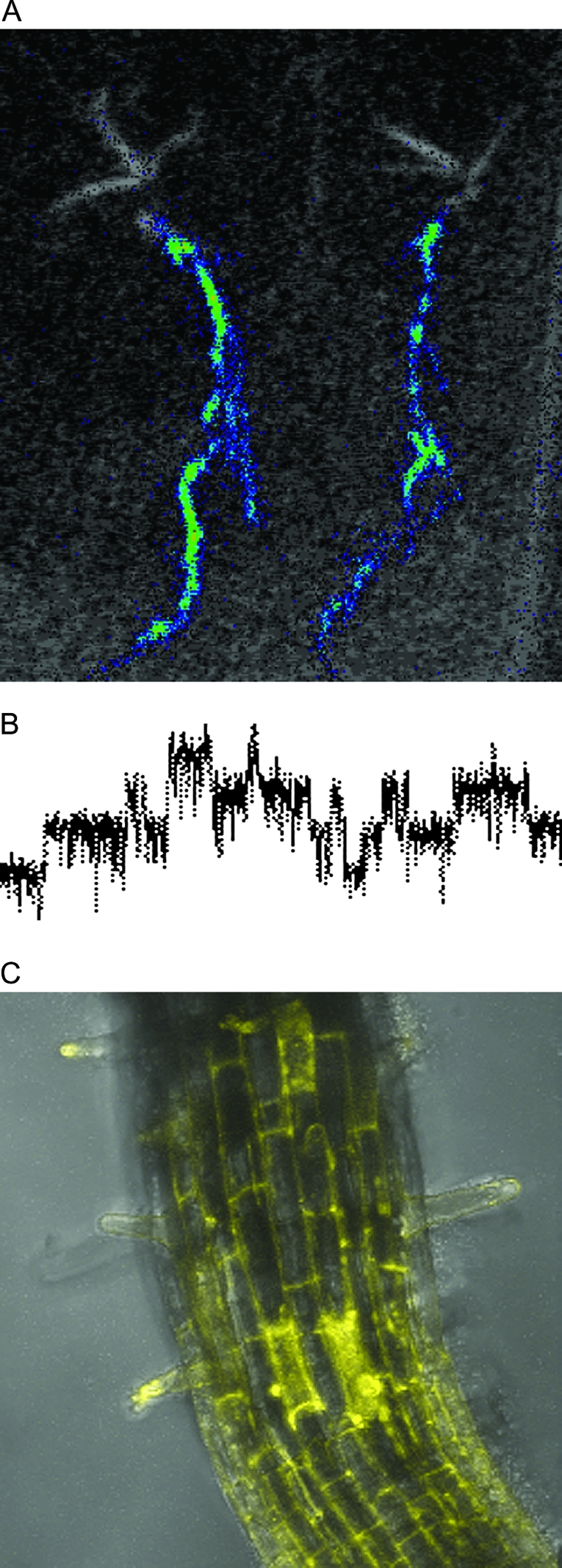
Technical Approaches to Study the Physiology and Molecular Biology of Thellungiella (Shandong).
(A) Micro-grafted hybrid plants comprising shoots of wild-type Thellungiella and roots of luciferase-transformed Arabidopsis thaliana plants (graft and photo by G. Littlejohn, University of Glasgow).
(B) Patch clamp recordings of picoAmpere-currents passing through distinctly opening and closing single ion channels in the plasma membrane of Thellungiella root protoplasts (recording by V. Volkov, University of Glasgow).
(C) Visualization of the epidermal cells of Thellungiella roots transiently expressing Yellow Fluorescent Protein (YFP) linked to the ER marker gene HDEL after transfection with Agrobacterium rhizogenes (photo by P. Camapanoni, University of Glasgow).
CONCLUSIONS
Research into Thellungiella has made enormous progress over the last few years and is likely to gain further momentum over the years to come, considering the wealth of available tools and a pending genome sequence. The studies reviewed here are evidence that Thellungiella has already yielded new information on how plants can achieve a high level of resistance against abiotic stresses. The most detailed knowledge gain to date concerns transport properties of Thellungiella, with K+/Na+ selectivity of root ion channels and function of SOS1 in Na+ exclusion from the root tip and xylem emerging as crucial mechanisms for limiting the accumulation of toxic Na+ in the shoot. The particular advantage of Thellungiella for stress tolerance research resides in its high similarity to Arabidopsis with respect to morphological appearance, metabolic pathways, and cDNA sequence and yet a striking difference in stress tolerance. Research into Thellungiella can therefore build on the enormous wealth of knowledge that has been accumulated for Arabidopsis and transfer it to comparative physiology and genomics. Direct comparison between the two species has already delivered exciting new results. The emerging picture is that Thellungiella uses similar molecular entities to Arabidopsis to deal with stress but maintains a state of stress anticipation in non-stressful conditions, and operates more specific regulation under stress. The comparative aspect is critical for future success with this species because so many of the essential components for stress tolerance already exist in Arabidopsis (and crops), and can improve stress tolerance when overexpressed. Real understanding and agricultural benefits will only be gained if we continue to investigate the species-specific regulation and the cost/benefit balance of individual stress tolerance mechanisms. Clearly, such understanding will be invaluable for avoiding unwanted side effects and ensuring sustainability of future efforts to improve crop performance on marginal and irrigated land through genetic modification. Let evolution be our teacher!
FUNDING
Work in my laboratory is funded by The Leverhulme Trust, the Biotechnology and Biological Sciences Research Council and the Faculty of Biomedical and Life Sciences, University of Glasgow.
Acknowledgments
I am grateful to Elizabeth Weretilnyk (McMaster University, Canada), Bert de Boer (Vrije Univeristy of Amsterdam, Netherlands), Arie Altman (Hebrew University of Jerusalem, Israel), Paco Rubio (CSIC, Murcia, Spain), and Hans Bohnert (University of Illinois, USA) for sharing unpublished information, and to Ray Bressan for supplying excellent seed material over the last years. No conflict of interest declared.
References
- Alemán F, Manuel Nieves-Cordones M, Vicente Martínez V, Francisco Rubio F. Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Env. Exp. Bot. 2008 in press. [Google Scholar]
- Al-Shehbaz IA, O'Kane SL, Price RA. Generic placement of species excluded from Arabidopsis (Brassicaceae) Novon. 1999;9:296–307. [Google Scholar]
- Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 1999;29:75–112. [Google Scholar]
- Amtmann A, Bohnert HJ, Bressan RA. Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 2005;138:127–130. doi: 10.1104/pp.105.059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Jelitto TC, Sanders D. K+-selective inward-rectifying channels and apoplastic pH in barley roots. Plant Physiol. 1999;120:331–338. doi: 10.1104/pp.120.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK. Learning from the Arabidopsis experience: the next gene search paradigm. Plant Physiol. 2001;127:1354–1360. [PMC free article] [PubMed] [Google Scholar]
- Cakmak I. Plant nutrition research: priorities to meet human needs for food in sustainable ways. Plant and Soil. 2002;247:3–24. [Google Scholar]
- Campanoni P, Sutter JU, Davis CS, Littlejohn GR, Blatt MR. A generalized method for transfecting root epidermis uncovers endosomal dynamics in Arabidopsis root hairs. Plant J. 2007;51:322–330. doi: 10.1111/j.1365-313X.2007.03139.x. [DOI] [PubMed] [Google Scholar]
- Chen AP, Wang GL, Qu ZL, Lu CX, Liu N, Wang F, Xia GX. Ectopic expression of ThCYP1, a stress-responsive cyclophilin gene from Thellungiella halophila, confers salt tolerance in fission yeast and tobacco cells. Plant Cell Reports. 2007;26:237–245. doi: 10.1007/s00299-006-0238-y. [DOI] [PubMed] [Google Scholar]
- Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi HZ. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 2008;55:554–565. doi: 10.1111/j.1365-313X.2007.03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M. A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol. 2000;122:823–834. doi: 10.1104/pp.122.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Maathuis FJ. Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 2007;175:387–404. doi: 10.1111/j.1469-8137.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- Duan XG, Yang AF, Gao F, Zhang SL, Zhang JR. Heterologous expression of vacuolar H+-PPase enhances the electrochemical gradient across the vacuolar membrane and improves tobacco cell salt tolerance. Protoplasma. 2007;232:87–95. doi: 10.1007/s00709-007-0268-5. [DOI] [PubMed] [Google Scholar]
- Fang QY, Liu J, Xu ZK, Song RT. Cloning and characterization of a flowering time gene from Thellungiella halophila. Acta. Bioch. Bioph. Sin. 2008;40:747–753. [PubMed] [Google Scholar]
- Fang QY, Xu ZK, Song RT. Cloning, characterization and genetic engineering of FLC homolog in Thellungiella halophila. Biochemical and Biophysical Research Communications. 2006;347:707–714. doi: 10.1016/j.bbrc.2006.06.165. [DOI] [PubMed] [Google Scholar]
- Gao F, Gao Q, Duan XG, Yue G, Yang AF, Zhang JR. Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J. Exper. Bot. 2006;57:3259–3270. doi: 10.1093/jxb/erl090. [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl Acad. Sci. U S A. 2001;98:11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German DA. Genus Thellungiella (Cruciferae). Europe // Bot. Zhurn. (Moscow and St-Petersburg) 2008;93:1273–1280. [Google Scholar]
- Ghars MA, Parre E, Debez A, Bordenave M, Richard L, Leport L, Bouchereau A, Savoure A, Abdelly C. Comparative salt tolerance analysis between Arabidopsis thaliana and Thellungiella halophila, with special emphasis on K+/Na+ selectivity and proline accumulation. J. Plant Physiol. 2008;165:588–599. doi: 10.1016/j.jplph.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Gong QQ, Li PH, Ma SS, Rupassara SI, Bohnert HJ. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Griffith M, Timonin M, Wong ACE, Gray GR, Akhter SR, Saldanha M, Rogers MA, Weretilnyk EA, Moffatt B. Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Env. 2007;30:529–538. doi: 10.1111/j.1365-3040.2007.01653.x. [DOI] [PubMed] [Google Scholar]
- Inan G, et al. Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004;135:1718–1737. doi: 10.1104/pp.104.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Weretilnyk E, Barak S, Rothstein SJ. The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol. 2008;147:1168–1180. doi: 10.1104/pp.108.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S. Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T-halophila. Plant Cell Env. 2006;29:1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li LG, Cheong YH, Pandey GK, Lu GH, Buchanan BB, Luan S. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl Acad. Aci. U S A. 2007;104:15959–15964. doi: 10.1073/pnas.0707912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wei AY, Song CX, Li N, Zhang JR. Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol. J. 2008;6:146–159. doi: 10.1111/j.1467-7652.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- Li HQ, Xu J, Chen L, Li MR. Establishment of an efficient Agrobacterium tumefaciens-mediated leaf disc transformation of Thellungiella halophila. Plant Cell Reports. 2007;26:1785–1789. doi: 10.1007/s00299-007-0391-y. [DOI] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2006a;103:12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PH, Mane SP, Sioson AA, Robinet CV, Heath LS, Bohnert HJ, Grene R. Effects of chronic ozone exposure on gene expression in Arabidopsis thaliana ecotypes and in Thellungielia halophila. Plant Cell Env. 2006b;29:854–868. doi: 10.1111/j.1365-3040.2005.01465.x. [DOI] [PubMed] [Google Scholar]
- Liu N, Chen AP, Zhong NQ, Wang F, Wang HY, Xia GX. Functional screening of salt stress-related genes from Thellungiella halophila using fission yeast system. Physiol. Plant. 2007;129:671–678. [Google Scholar]
- Lv S, Zhang KW, Gao Q, Lian LJ, Song YJ, Zhang JR. Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol. 2008;49:1150–1164. doi: 10.1093/pcp/pcn090. [DOI] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001;127:1617–1625. [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, et al. Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J. 2003;35:675–692. doi: 10.1046/j.1365-313x.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- M'rah S, Ouerghi Z, Berthomieu C, Havaux M, Jungas C, Hajji M, Grignon C, Lachaal M. Effects of NaCl on the growth, ion accumulation and photosynthetic parameters of Thellungiella halophila. J. Plant Physiol. 2006;163:1022–1031. doi: 10.1016/j.jplph.2005.07.015. [DOI] [PubMed] [Google Scholar]
- M'rah S, Ouerghi Z, Eymery F, Rey P, Hajji M, Grignon C, Lachaal M. Efficiency of biochemical protection against toxic effects of accumulated salt differentiates Thellungiella halophila from Arabidopsis thaliana. J. Plant Physiol. 2007;164:375–384. doi: 10.1016/j.jplph.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Ni WS, Lei ZY, Chen X, Oliver DJ, Xiang CB. Construction of a plant transformation-ready expression cDNA library for Thellungiella halophila using recombination cloning. J. Integr. Plant Biol. 2007;49:1313–1319. [Google Scholar]
- Nieves-Cordones M, Miller AJ, Alemán F, Martínez V, Rubio F. A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant. Mol. Biol. 2008;68:521–532. doi: 10.1007/s11103-008-9388-3. [DOI] [PubMed] [Google Scholar]
- Oh D-H, et al. Interference with Thellungiella halophila SOS1 generates plants sensitive to salinity by altered routing and deposition of sodium. Submitted. 2008 [Google Scholar]
- Oh D-H, Gong QQ, Ulanov A, Zhang Q, Li YZ, Ma WY, Yun DJ, Bressan RA, Bohnert HJ. Sodium stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNA interference line. J. Integr. Plant Biol. 2007;49:1484–1496. [Google Scholar]
- Pedras MSC, Adio AM. Phytoalexins and phytoanticipins from the wild crucifers Thellungiella halophila and Arabidopsis thaliana: Rapalexin A, wasalexins and camalexin. Phytochem. 2008;69:889–893. doi: 10.1016/j.phytochem.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FN. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol. Plant Microbe. In. 1996;9:748–757. doi: 10.1094/mpmi-9-0748. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Sun ZB, Qi XY, Li PH, Wu CX, Zhao YX, Zhang H, Wang ZL. Overexpression of a Thellungiella halophila CBL9 homolog, ThCBL9, confers salt and osmotic tolerances in transgenic Arabidopsis thaliana. J. Plant Biol. 2008;51:25–34. [Google Scholar]
- Sun ZY, Zhang XJ, Li FZ. Phylogenetic relationship between Arabidopsis and Thellungiella (Cruciferae): evidence from leaf epidermal features and chloroplast sequence analysis. Acta. Botanica Yunnanica. 2007;29:632–638. [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink RS, Rahman M, Bressan RA, Jenks MA. Cuticular waxes on Arabidopsis thaliana close relatives Thellungiella halophila and Thellungiella parvula. Int. J. Plant Sci. 2002;163:309–315. [Google Scholar]
- Vera-Estrella R, Barkla BJ, Garcia-Ramirez L, Pantoja O. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 2005;139:1507–1517. doi: 10.1104/pp.105.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. Cation channels in the Arabidopsis plasma membrane. Trends Plant Sci. 2002;7:168–175. doi: 10.1016/s1360-1385(02)02262-8. [DOI] [PubMed] [Google Scholar]
- Volkov V, Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J. 2006;48:342–353. doi: 10.1111/j.1365-313X.2006.02876.x. [DOI] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Env. 2004;27:1–14. [Google Scholar]
- Wang B, Davenport RJ, Volkov V, Amtmann A. Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J. Exper. Bot. 2006;57:1161–1170. doi: 10.1093/jxb/erj116. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Al-Shehbaz IA, Sauder CA. Phylogenetic position of Arabis arenicola and generic limits of Aphragmus and Eutrema (Brassicaceae) based on sequences of nuclear ribosomal DNA. Canadian Journal of Botany-Revue Canadienne De Botanique. 2006;84:269–281. [Google Scholar]
- Wong CE, et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006;140:1437–1450. doi: 10.1104/pp.105.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Diaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M. Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol. Biol. 2005;58:561–574. doi: 10.1007/s11103-005-6163-6. [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Xu SM, Wang XC, Chen J. Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Reports. 2007;26:497–506. doi: 10.1007/s00299-006-0248-9. [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo SL, Yin HB, Xiong DJ, Zhang H, Zhao YX. Molecular cloning and identification of a heat shock cognate protein 70 gene, Thhsc70, in Thellungiella halophila. Acta. Bot. Sin. 2004;46:1212–1219. [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]