Abstract
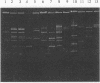
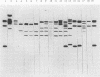
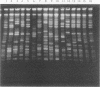
A patient with chronic myelogenous leukemia became colonized with a Staphylococcus haemolyticus strain and experienced a septic episode caused by this strain during a cytostatic course. The strain was multiply resistant to antibiotics; the MIC and MBC of vancomycin were 2 and 4 mg/liter, and the MIC and MBC of teicoplanin were 4 and 16 mg/liter, respectively. We performed a surveillance study on the carriage of S. haemolyticus in medical and nursing staff of the hospital ward where the patient was treated. S. haemolyticus was isolated from 18 sites on 12 of the 39 people tested. A number of typing methods were performed in order to investigate the possible relationships among the isolates. Methods used were immunoblotting of staphylococcal peptides, plasmid analysis, restriction fragment length polymorphism of chromosomal DNA, and pulsed-field gel electrophoresis of total DNA. Compared with the immunoblotting technique, the molecular methods were more discriminative. The strain colonizing the patient showed a consistent pattern by all typing methods during isolation. When the immunoblot technique was used, similar patterns were found with isolates from hospital staff and isolates from unrelated sources. With the molecular techniques, no evidence of a local spread of the patient's strain was found. However, plasmid profiles and restriction fragment length polymorphism and pulsed-field gel electrophoresis patterns showed that S. haemolyticus isolates collected from hospital ward personnel were related, which was not the case with isolates collected from unrelated sources. Restriction fragment length polymorphism analysis was more discriminative when IS431 was used as a DNA probe instead of a probe based on the 16S rRNA gene. S. haemolyticus, as in this case, may develop resistance to vancomycin and teicoplanin. These antibiotics are considered the last-resort drugs for the therapy of nosocomial gram-positive infections. Thus, local spread of staphylococci resistant to these drugs is an important problem, which should be prevented by strict hygienic measures and antibiotic policy.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert G., Passot S., Lucht F., Dorche G. Selection of vancomycin- and teicoplanin-resistant Staphylococcus haemolyticus during teicoplanin treatment of S. epidermidis infection. J Antimicrob Chemother. 1990 Mar;25(3):491–493. doi: 10.1093/jac/25.3.491. [DOI] [PubMed] [Google Scholar]
- Barberis-Maino L., Berger-Bächi B., Weber H., Beck W. D., Kayser F. H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59(1):107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Degener J. E., Naidoo J. L., Noble W. C., Phillips I., Marples R. R. Carriage of gentamicin-resistant coagulase-negative staphylococci in patients on continuous ambulatory peritoneal dialysis. J Antimicrob Chemother. 1987 Apr;19(4):505–512. doi: 10.1093/jac/19.4.505. [DOI] [PubMed] [Google Scholar]
- Froggatt J. W., Johnston J. L., Galetto D. W., Archer G. L. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1989 Apr;33(4):460–466. doi: 10.1128/aac.33.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R. V., Ruff E. A. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1983 Sep;24(3):450–452. doi: 10.1128/aac.24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P. W., van Soolingen D., Dale J. W., Schuitema A. R., McAdam R. A., Catty D., van Embden J. D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990 Sep;28(9):2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W. T., Armstrong D., Bodey G. P., Feld R., Mandell G. L., Meyers J. D., Pizzo P. A., Schimpff S. C., Shenep J. L., Wade J. C. From the Infectious Diseases Society of America. Guidelines for the use of antimicrobial agents in neutropenic patients with unexplained fever. J Infect Dis. 1990 Mar;161(3):381–396. doi: 10.1093/infdis/161.3.381. [DOI] [PubMed] [Google Scholar]
- Kappers-Klunne M. C., Degener J. E., Stijnen T., Abels J. Complications from long-term indwelling central venous catheters in hematologic patients with special reference to infection. Cancer. 1989 Oct 15;64(8):1747–1752. doi: 10.1002/1097-0142(19891015)64:8<1747::aid-cncr2820640832>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee W., Burnie J. P. Fingerprinting methicillin-resistant Staphylococcus aureus by the immunoblot technique. J Med Microbiol. 1988 Apr;25(4):261–268. doi: 10.1099/00222615-25-4-261. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton R. P., Hermans J., Simoons-Smit A. M., Hoogkamp-Korstanje J. A., Degener J. E., van Klingeren B. Correlations between consumption of antibiotics and methicillin resistance in coagulase negative staphylococci. J Antimicrob Chemother. 1990 Oct;26(4):573–583. doi: 10.1093/jac/26.4.573. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schwalbe R. S., Stapleton J. T., Gilligan P. H. Emergence of vancomycin resistance in coagulase-negative staphylococci. N Engl J Med. 1987 Apr 9;316(15):927–931. doi: 10.1056/NEJM198704093161507. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach L. A., Pfaller M. A., Barrett M., Koontz F. P., Wenzel R. P. Vancomycin resistance in Staphylococcus haemolyticus causing colonization and bloodstream infection. J Clin Microbiol. 1990 Sep;28(9):2064–2068. doi: 10.1128/jcm.28.9.2064-2068.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]