Abstract
Objectives
This study is aimed to determine the role of capsular polysaccharide (CPS) and lipooligosaccharide (LOS) in modulating antimicrobial resistance and natural transformation of Campylobacter jejuni, an important food-borne human pathogen.
Methods
A series of C. jejuni mutants, which are defective in either CPS or LOS or both, were constructed. The antimicrobial susceptibility, bacterial surface hydrophobicity, natural transformation frequency and DNA binding and uptake were measured and compared between the mutants and the wild-type strain.
Results
Truncation of LOS greatly reduced (8-fold) the intrinsic resistance of C. jejuni to erythromycin, a key antibiotic used for treating human campylobacteriosis, while the loss of CPS did not result in significant changes in the susceptibility to antimicrobial agents. Notably, mutation of LOS also significantly increased (>16-fold) the susceptibility to erythromycin in C. jejuni mutants carrying the A2074G mutation in 23S rRNA. The increased susceptibility to erythromycin in the LOS mutant was probably due to enhanced permeability to this antibiotic, because the LOS mutation rendered the surface of C. jejuni more hydrophobic. Loss of CPS and truncation of LOS increased the transformation frequency by 4- and 25-fold, respectively, and mutation of both CPS and LOS resulted in a 97-fold increase in the transformation frequency. Consistent with the increased transformation frequencies, the CPS and LOS mutants showed enhanced rates of DNA uptake.
Conclusions
These results demonstrate that the surface polysaccharides in C. jejuni contribute to the resistance to erythromycin, a clinically important antibiotic, but restrict natural transformation.
Keywords: food safety, macrolides, competence, drug susceptibility
Introduction
Gram-negative bacteria possess surface polysaccharides such as capsular polysaccharide (CPS) and lipopolysaccharide (LPS) on the outer membrane. These surface polysaccharides are attached to bacterial cells by covalent linkages to their lipid moieties.1,2 Since the surface polysaccharides constitute the outermost layer of the Gram-negative bacterial cell, they directly interact with the surrounding environments, providing a permeability barrier to noxious antimicrobial agents and conferring resistance to host immunity.1,3 The LPS structure embedded in the outer leaflet of the outer membrane considerably decreases the permeability of hydrophobic compounds through the outer membrane and contributes to the general resistance of Gram-negative bacteria to hydrophobic antibiotics such as macrolides.1,4
Campylobacter jejuni is a Gram-negative bacterium and a major food-borne pathogen frequently associated with human gastroenteritis worldwide.5 More than 2 million cases of Campylobacter infections occur in the USA each year.6 Campylobacter infection is characterized by diarrhoea, fever and abdominal cramps and occasionally causes Guillain–Barré syndrome as a post-infection complication.7 When clinical treatments are warranted, macrolides and fluoroquinolones are the drugs of choice to treat Campylobacter infections.7 However, the resistance of Campylobacter to both classes of antibiotics has been on the rise and is considered a threat to public health.8 Several general mechanisms involved in bacterial resistance to antibiotics have been described, including antibiotic inactivation, target modification, active efflux and reduced cell permeability.9 The first three general mechanisms have been well studied in Campylobacter,10 but the influence of cell surface structures on the susceptibility to clinically important antibiotics by altering cell permeability is still poorly understood in this bacterium.
One of the unique features of C. jejuni is its ability to take up DNA by natural transformation.11 Our recent work showed that natural transformation plays an important role in horizontal transfer of antibiotic resistance determinants in Campylobacter.12 As a first step during the process of natural transformation in Gram-negative bacteria including C. jejuni, free DNA in the environment must interact with the surface polysaccharide layer prior to contacting the competence proteins in the cell membrane. Thus, it is conceivable that alteration of the surface polysaccharide structures would affect the efficiency of natural transformation.
C. jejuni possesses both CPS and lipooligosaccharide (LOS).13 The LOS differs from LPS in that LOS does not possess an O-antigen polysaccharide chain.13 These surface polysaccharide structures are highly variable among different Campylobacter strains14,15 and are related to the virulence of this bacterium.16,17 Molecular mimicry between C. jejuni LOS and human gangliosides is also implicated in Guillain–Barré syndrome, a paralytic neuropathy.13 Despite the advances in understanding the structures and functions of CPS and LOS, their roles in the natural transformation and antibiotic resistance in Campylobacter remain to be determined. In a previous study, Kanipes et al.16 demonstrated that C. jejuni LOS is associated with resistance to fusidic acid, novobiocin and SDS, but its contribution to clinically important antibiotics, such as fluoroquinolones and macrolides, was not determined in that work. In this study, we demonstrated that LOS, but not CPS, plays an important role in Campylobacter resistance to erythromycin and that surface polysaccharides restrict the natural transformation frequency of C. jejuni NCTC 11168.
Materials and methods
Bacterial strains and culturing conditions
C. jejuni NCTC 11168 and its derivatives were used in this study. C. jejuni strains JL271, JL272 and JL273 are highly resistant to erythromycin (MIC >512 mg/L), carry the A2074G mutation in 23S rRNA and were derived from chickens fed with tylosin in a previous study.18 JB304 is a derivative of strain 11168, which was constructed in this study by transformation with the genomic DNA of strain JL271 and is highly resistant to erythromycin. The strains were grown in Mueller–Hinton (MH) medium under microaerobic (5% O2/10% CO2/85% N2) conditions at 42°C. When needed, the culture medium was supplemented with kanamycin (50 mg/L), tetracycline (5 mg/L) or chloramphenicol (10 mg/L).
Construction of CPS and LOS mutants
To construct a CPS mutant, a 1693 bp region harbouring kpsS was PCR-amplified with kpsS_F and kpsS_R primers (Table 1) and cloned into pGEM-T (Promega, Madison, WI, USA) to yield pGEM-T::kpsS. The pGEM-T::kpsS plasmid was digested with SwaI and ligated with the aphA3 gene, which was amplified from pMW1019 with Kan_F and Kan_R (Table 1). The suicide vector was introduced to C. jejuni 11168 by electroporation, and the resulting mutants were selected on MH agar plates containing kanamycin (50 mg/L). The mutation in kpsS was confirmed by PCR (data not shown) and the mutant was named JB301. For constructing an LOS mutant, a 2206 bp region containing waaF was amplified with Ex Taq™ (TaKaRa Bio Inc., Japan) using primers waaF_F and waaF_R (Table 1) and cloned into pGEM-T to yield pGEM-T::waaF. Digestion of pGEM-T::waaF with BglII removed a 649 bp internal fragment of waaF, and the construct was blunt-ended by treatment with Klenow fragment (TaKaRa Bio Inc.). The tetracycline resistance cassette (tetO) was amplified with VentR® DNA polymerase (New England Biolab, Ipswich, MA, USA) from pTet in C. jejuni 81-176,20 using TetOF and TetOR primers (Table 1). The tetO gene was inserted into the waaF on pGEM-T::waaF, and the construct was electroporated into C. jejuni 11168. The resulting transformants were selected by growth on MH agar plates containing tetracycline (5 mg/L). The waaF mutant was confirmed by PCR (data not shown) and was named JB302. The double mutant JB303, which carries mutations in both kpsS and waaF, was constructed by transforming JB301 with the genomic DNA of JB302. The double mutant was selected by growth on MH agar plates containing both tetracycline (5 mg/L) and kanamycin (50 mg/L).
Table 1.
PCR primers used in this study
| Primers | Sequence (5′ to 3′) |
|---|---|
| kpsS_F | GCT CAA GTT GAA GAT GAT GCT TCG ATG AT |
| kpsS_R | CAT ACC AAA ACA GGA TTG GGT TTA TAA GCA TGA |
| Kan_F | CTTATCAATATATCCATGGAATGGGCAAAGCAT |
| Kan_R | GATAGAACCATGGATAATGCTAAGACAATCACTAAA |
| waaF_F | CCA AAC CGA CCA GCA AAA ATG CCT TTG T |
| waaF_R | GAT GAA GAC ACG CCT TTA GAA CTT ATA AGC TT |
| TetOF | TTA TTT TTG CAT AAA CAG ATG ATT AGT G |
| TetOR | GCA AGC TGT TAA GCT AAC TTG T |
| waaF_comF | CCC AAA TTT CTA CTT GCA AAA GTG CCC AAA |
| waaF_comR | GGT ATT TCA ACG AGC GGA AAA AGC CCT AAT |
waaF and kpsS mutants were also constructed with macrolide-resistant C. jejuni strains (JB304, JL271, JL272 and JL273). The kpsS and waaF mutants of these strains were constructed by transforming these strains using the genomic DNA of JB301 (kpsS::aphA3) and JB302 (waaF::tetO), respectively, and the transformants were selected by growth on MH agar plates containing kanamycin (50 mg/L) or tetracycline (5 mg/L). The insertional mutations in kpsS and waaF of these erythromycin-resistant strains were confirmed by PCR (data not shown).
Detection of LOS and CPS by Alcian Blue staining
CPS and LOS preparation and Alcian Blue staining were performed according to the previously reported method.21 Briefly, a loopful of 1-day-old C. jejuni culture grown on MH agar plates was resuspended in lysis buffer containing 62.5 mM Tris–HCl, pH 6.8, 2% SDS, 10% glycerol and 0.05% Bromophenol Blue. Samples were boiled for 10 min and centrifuged at 13 000 rpm for 5 min. Aliquots of the supernatant were taken and mixed with proteinase K (Sigma, St Louis, MO, USA) to a final concentration of 1 mg/mL. The samples were incubated at 50ºC for 1 h and then fractionated by SDS–PAGE. LOS and CPS were visualized with Alcian Blue staining (0.1% Alcian Blue dissolved in 40% ethanol/5% acetic acid).
Complementation of the waaF mutant in trans
Construction of the complementing plasmid for the waaF mutant was based on a previously published study.16 A 1450 bp region containing waaF and its upstream intergenic region was amplified with VentR® DNA polymerase, using waaF_comF and waaF_comR primers (Table 1). The PCR product was cloned into the SmaI site of pRY112. The constructed plasmid was introduced into the waaF mutant of C. jejuni strains 11168 (JB302), JL271 and JL273 by conjugation, as described previously.22 The waaF-complemented strain JB302 was named JB302C.
Susceptibility tests
The MICs of various antibiotics, SDS and choleate were determined with a microtitre broth dilution method as described previously.23 Antimicrobial agents were purchased from Sigma (erythromycin, cefotaxime, gentamicin, rifampicin, polymyxin B, streptomycin, SDS and choleate) and MP Biomedicals, Irvine, CA, USA (ciprofloxacin).
Hydrophobicity assay
Hydrophobicity of C. jejuni strains was measured by a salting-out method as described by Misawa and Blaser.24 Briefly, overnight cultures of C. jejuni strains were resuspended in 2 mM sodium phosphate (Sigma) to an optical density at 600 nm (OD600) of 1.0. Ammonium sulphate (4 M) was dissolved in 2 mM sodium phosphate and 2-fold serially diluted in U-bottomed 96-well plates. An equal volume (25 µL) of bacterial suspension was added to each well containing ammonium sulphate. The plates were incubated at room temperature overnight. The minimum concentration of ammonium sulphate forming bacterial aggregation was determined and used for the hydrophobicity index. Hydrophobicity is inversely correlated with the concentration of ammonium sulphate causing bacterial aggregation.
Natural transformation
Natural transformation was performed as described previously11 with some modifications. C. jejuni cultures grown overnight on MH agar plates were collected and resuspended in MH broth to OD600 of 0.05. Bacterial suspensions (0.5 mL) were transferred to sterilized tubes, incubated at 37°C with shaking (200 rpm) under microaerobic conditions for 3 h and then 1 µg of genomic DNA of JB201 or JB202,12 which carry the chloramphenicol resistance gene (cat) and the kanamycin resistance gene (aphA3), respectively, was added to each tube. After additional incubation for 4 h, bacterial cultures were serially diluted and plated on MH agar plates with or without antibiotics (10 mg/L chloramphenicol or 50 mg/L kanamycin) to count the numbers of transformants and the total bacterial number, respectively. Transformation frequency was calculated by dividing the number of transformants from 1 µg of donor DNA by the total number of bacteria.
DNA binding and uptake assay
Genomic DNA from C. jejuni NCTC 11168 was extracted using the Wizard Genomic DNA Purification Kit (Promega). Up to 400 ng of chromosomal DNA was nick translated using the Nick Translation Kit (Roche, Indianapolis, IN, USA) in the presence of 100 µCi of [α-32P]dCTP (Perkin Elmer, Waltham, MA, USA) as recommended by the manufacturer. Unincorporated nucleotides were removed by ethanol precipitation and the DNA was resuspended in 82 µL of double distilled water. The CPS and LOS mutants and the wild-type 11168, grown for 18 h on MH agar, were resuspended in 10 mL MH broth to an OD600 of 0.1. The culture tubes were incubated microaerobically with shaking (250 rpm) until the optical density had doubled (∼3 h). Cells were harvested by centrifugation at 4000 g for 20 min and resuspended in 3.3 mL MH broth. Each culture was adjusted to the same optical density at OD600 to ensure an equivalent number of cells were used in the DNA binding and uptake assay for each strain. To each culture (1 mL), 100 ng of 32P-labelled DNA was added, and the culture was incubated microaerobically with labelled DNA for 2 h and divided into two 500 µL aliquots. DNaseI was added to the aliquots for DNA uptake assay to a final concentration of 100 mg/L to digest extracellular DNA. After DNase I treatment for 10 min at room temperature, cells were washed three times with MH broth and resuspended in 100 µL of ice-cold water prior to liquid scintillation (Beckman Coulter). Since the intracellular DNA is protected from DNaseI treatment, the amount of radioactivity from the samples treated with DNaseI represents the level of DNA uptake. The radioactivity of the sample without DNaseI treatment was considered the DNA-binding level. Three independent experiments were performed.
Statistical analysis
Normality and variance of each dataset were assessed by D’Agostino Omnibus25 and Modified-Levene test,26 respectively. Differences between the wild-type and each mutant strain in hydrophobicity were tested using the Mann–Whitney U-test, and the Student t-test was used for the transformation frequency dataset. For the analysis of the results of DNA binding and uptake, ANOVA was used with the strain and DNaseI treatment as fixed variables, and the Bonferroni corrected multiple comparison t-test was applied to variables with significant main or interaction effects.
Results
Contribution of LOS, but not CPS, to the intrinsic resistance to erythromycin
Multiple genes clustered in two separate loci are involved in LOS and CPS biosynthesis in C. jejuni. Based on their importance in LOS and CPS production,27,28 we selected waaF and kpsS as target genes to disrupt LOS and CPS, respectively. The waaF gene encodes a heptosyltransferase, while kpsS encodes a putative CPS transporter. Truncation of LOS and loss of CPS in these mutants were confirmed by Alcian Blue staining [Figure S1, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. The LOS-complemented strain (JB302C) partially restored the production of intact LOS (Figure S1a). Thus, both the truncated form and the intact form of LOS were seen in JB302C. The growth rate of the CPS and LOS mutants in MH broth was comparable to that of the wild-type (data not shown).
The antimicrobial susceptibilities of the CPS and LOS mutants were compared with that of the wild-type. As shown in Table 2, the mutations had different effects on the sensitivity to various antibiotics. Although JB301 (the CPS mutant) and JB302 (the LOS mutant) did not exhibit any changes in the susceptibility to ciprofloxacin (a hydrophilic fluoroquinolone), JB302 showed a significant increase (8-fold) in the susceptibility to erythromycin. JB302C (JB302 complemented with a plasmid-carried copy of waaF) had MIC levels similar to those of the wild-type (Table 2). The LOS mutation also moderately (2-fold) increased the susceptibility of Campylobacter to polymyxin B and SDS. These changes were reproducible in four independent experiments. In contrast to LOS, the mutation of CPS did not result in an increase in the susceptibilities of the tested antimicrobials except a minor change (2-fold) in SDS. Instead, the CPS mutant showed slightly decreased sensitivity to some hydrophilic antibiotics, such as cefotaxime, gentamicin, polymyxin B and streptomycin. Compared with JB302, the MICs of erythromycin, cefotaxime and SDS were further reduced in JB303 (a CPS and LOS double mutant), but the reduction was modest (Table 2), suggesting that the MIC changes in the double mutant were mainly due to the LOS mutation.
Table 2.
Antimicrobial susceptibility of C. jejuni 11168 and its derivatives
| MIC (mg/L)a |
|||||
|---|---|---|---|---|---|
| Antimicrobials | 11168 | JB301 | JB302 | JB303 | JB302C |
| Ciprofloxacin | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Erythromycin | 0.5 | 0.5 | 0.0625 (8) | 0.0313 (16) | 0.25 |
| Cefotaxime | 1 | 2 (0.5) | 1 | 0.5 (2) | 1 |
| Gentamicin | 1 | 2 (0.5) | 1 | 1 | 1 |
| Rifampicin | >128 | >128 | >128 | >128 | >128 |
| Polymyxin B | 1 | 2 (0.5) | 0.5 (2) | 0.5 (2) | 1 |
| Streptomycin | 0.5 | 1 (0.5) | 0.5 | 0.5 | 0.5 |
| SDS | 128 | 64 (2) | 64 (2) | 32 (4) | 128 |
| Choleate | 32 768 | 32 768 | 32 768 | 32 768 | 32 768 |
aThe numbers in parentheses indicate the differences (n-fold) between wild-type 11168 and the mutant strains. JB301, CPS mutant; JB302, LOS mutant; JB303, CPS and LOS double mutant; JB302C, JB302 carrying pRY112::waaF.
Effect of the LOS mutation on the susceptibility to erythromycin in macrolide-resistant mutants
High-level macrolide resistance in Campylobacter is often associated with point mutations in the 23S rRNA.29 To investigate if LOS affects the resistance to macrolide antibiotics in C. jejuni strains harbouring the A2074G mutation in 23S rRNA, we introduced the LOS and CPS mutations to JB304, JL271, JL272 and JL273, which are highly resistant to erythromycin (MIC >512 mg/L). While the CPS mutation did not have a measurable effect on macrolide resistance in these C. jejuni strains, the LOS mutation decreased the MIC of erythromycin from >512 to 16–32 mg/L (a >16-fold reduction). In trans complementation of the LOS mutants of JL271 and JL273 restored the high-level resistance to erythromycin (MIC >512 mg/L).
Increased hydrophobicity in CPS and LOS mutants
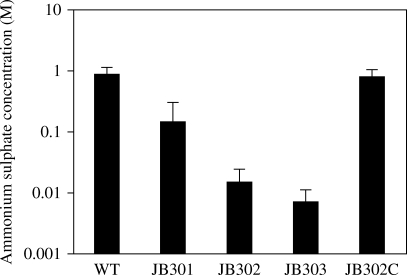
We hypothesized that the loss of hydrophilic surface polysaccharides in the CPS and LOS mutants would make the bacterial surface more hydrophobic than that of the wild-type. To test this hypothesis, we measured the hydrophobicity of the strains by the salting-out method. In this assay, a higher concentration of ammonium sulphate mediating bacterial aggregation indicates lower hydrophobicity.24 Thus, the concentration of ammonium sulphate is inversely proportional to hydrophobicity. CPS mutation was previously reported to increase the hydrophobicity of C. jejuni.30 In our study, we confirmed the previous finding regarding the C. jejuni CPS mutant and further demonstrated LOS had a much greater impact on surface hydrophobicity than CPS. Mutation of CPS and LOS increased the hydrophobicity by 3.2- and 93-fold, respectively, as reflected by the changes in the concentration of ammonium sulphate (Figure 1). The CPS and LOS double mutation resulted in a 171-fold increase in the hydrophobicity (Figure 1). The differences between the wild-type and each mutant were statistically significant (P < 0.001) by the Mann–Whitney U-test. These results demonstrated that CPS and LOS are strongly associated with the surface hydrophobicity of C. jejuni.
Figure 1.
Effect of CPS and LOS mutation on surface hydrophobicity of C. jejuni. The concentration of ammonium sulphate is inversely proportional to hydrophobicity. Each bar represents the mean ± SEM of three different experiments. WT, C. jejuni 11168; JB301, CPS mutant (kpsS::aphA3); JB302, LOS mutant (waaF::tetO); JB303, CPS and LOS double mutant; JB302C, JB302 harbouring pRY112::waaF.
Contribution of CPS and LOS to natural transformation
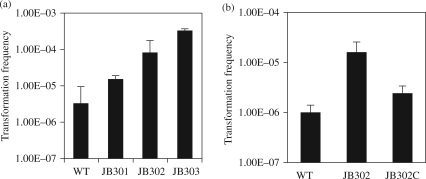
We performed natural transformation with these mutants and found that the CPS and LOS mutations increased the frequency of natural transformation by 4- and 25-fold, respectively, and the double mutation of both LOS and CPS significantly increased the transformation frequency by 97-fold (Figure 2a). The results were reproducible in four independent experiments. The difference between the wild-type and each mutant was statistically significant (P < 0.001). Since a greater change in transformation was observed with the LOS mutant than with the CPS mutant, we complemented the LOS mutant using a wild-type waaF carried on a shuttle plasmid. In trans complementation decreased the transformation frequency to a near wild-type level (Figure 2b). The differences between the wild-type and the waaF-complemented strain were statistically insignificant (P > 0.1).
Figure 2.
Effect of CPS and LOS mutation on natural transformation in C. jejuni. (a) Transformation frequencies in various mutant constructs. The result is representative of four independent experiments with triplicate transformation reactions. The donor DNA contained a chloramphenicol resistance marker (cat). Each bar represents the mean ± SEM. WT, C. jejuni 11168; JB301, CPS mutant; JB302, LOS mutant; JB303, CPS and LOS double mutant. (b) Complementation of the waaF mutant in natural transformation. JB302C is JB302 complemented with pRY112::waaF. The result is representative of three independent experiments with triplicate transformation. Since the complementing plasmid itself carried a cat gene, the genomic DNA containing the kanamycin resistance marker (aphA3) was used as the donor DNA. Each bar represents the mean ± SEM.
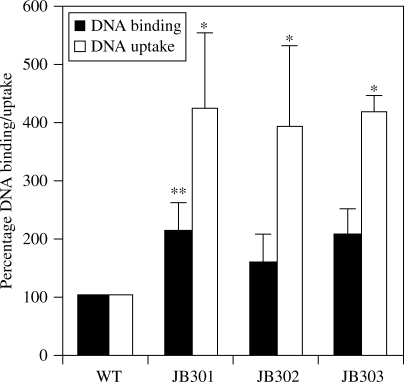
CPS and LOS affected DNA binding and uptake
DNA binding and uptake assays were performed to determine whether the LOS and CPS mutations affect DNA binding and uptake in Campylobacter. The results demonstrated that DNA binding and uptake increased 2- and 4-fold, respectively, in the mutants (Figure 3). Despite variations observed between experiments in the measured radioactivity levels, the fold differences between the wild-type and the mutant strains remained consistent. The fold changes in DNA uptake were statistically significant in all of the mutants (P < 0.01). The increase in DNA binding was statistically significant (P < 0.05) in JB301, but was marginally non-significant in JB302 and JB303 as determined by Bonferroni corrected t-test despite the trend of increased DNA binding (Figure 3). Although JB303 consistently showed a higher transformation frequency than JB301 and JB302 (Figure 2a), it did not demonstrate greater DNA binding and uptake rates than the single mutants (Figure 3).
Figure 3.
Effect of CPS and LOS mutation on DNA binding and uptake in C. jejuni. The result is representative of three independent experiments. Each bar represents the mean ± SEM of three samples in one experiment. In the repeated experiments, similar patterns of results were obtained (see the Results section). The statistically significant difference between each mutant and the wild-type strain is indicated by one asterisk (P < 0.01) or two asterisks (P < 0.05). WT, C. jejuni 11168; JB301, CPS mutant; JB302, LOS mutant; JB303, CPS and LOS double mutant.
Discussion
The role of surface polysaccharides in mediating antibiotic resistance in C. jejuni was determined in this study. Although a previous study reported that the LOS mutation sensitized C. jejuni to fusidic acid, novobiocin and SDS,16 it was unknown if CPS also contributes to antimicrobial resistance and if both CPS and LOS are important for resisting clinically relevant antibiotics, such as fluoroquinolones and macrolides. In this study, we showed that C. jejuni LOS plays a significant role in the protection against the action of erythromycin (Table 2), which is hydrophobic and a key antibiotic clinically used to treat Campylobacter infection. Compared with LOS, CPS had a limited impact on antimicrobial resistance in C. jejuni (Table 2). Instead, the CPS mutation in C. jejuni slightly decreased the susceptibility to some hydrophilic antibiotics, such as cefotaxime, gentamicin, streptomycin and polymyxin B (Table 2).
Campylobacter resistance to erythromycin is associated with mutations in 23S rRNA29 and the function of the multidrug efflux pump CmeABC.18 In this study, we showed that truncation of LOS reduced Campylobacter resistance to erythromycin (Table 2). In other bacteria, it has been shown that loss of LPS renders bacterial cells more permeable to hydrophobic agents and consequently increases the sensitivity to hydrophobic antibiotics.1,4 Many Gram-negative bacteria such as E. coli and Salmonella Typhimurium are intrinsically resistant to hydrophobic antimicrobial agents including erythromycin, and this intrinsic resistance to erythromycin is ascribed to the exclusion of hydrophobic antibiotics by the hydrophilic LPS.1,4 Since C. jejuni only has LOS and does not have the full-length LPS, it is much more susceptible to macrolides than E. coli and Salmonella, explaining why macrolide antibiotics are suitable for treating Campylobacter infections. In this study, we showed that truncation of LOS further increased the susceptibility of Campylobacter to erythromycin. Unlike hydrophilic antibiotics, which traverse the outer membrane through water-filled channels such as porins, hydrophobic macrolides diffuse through the outer membrane via hydrophobic interaction.1,4 Based on the fact that truncation of LOS significantly increased the hydrophobicity of Campylobacter (Figure 1), we speculate that the LOS mutant was more permeable to erythromycin and thus more susceptible to the antibiotic than the wild-type strain.
Although both CPS and LOS affected the hydrophobicity of C. jejuni (Figure 1), loss of CPS alone did not alter the bacterial susceptibility to erythromycin (Table 2). This was probably due to the possibility that LOS alone provides a sufficient barrier for diffusion of macrolides across the outer membrane in Campylobacter. WaaF is responsible for the addition of the second heptose to the inner-core region of C. jejuni LOS, and its mutation results in the formation of truncated LOS16,28 (Figure S1a). The LOS mutant containing the waaF mutation showed a slight (2-fold) reduction in the MIC level of polymyxin B (Table 2). The increased sensitivity to SDS by the LOS mutation was reported in a previous study16 and was confirmed by our result (Table 2). However, the CPS and LOS mutants did not show any changes in the susceptibility to choleate (Table 2), indicating that the surface polysaccharides do not contribute significantly to bile resistance.
Natural transformation is an important mechanism of horizontal gene transfer and significantly mediates the spread of antimicrobial resistance determinants in Campylobacter.12,31 Natural transformation requires many protein components and is distinct from other artificial DNA transformations such as heat-shock transformation and electroporation.32 Although the role of LPS in conjugation and heat-shock transformation was reported in naturally non-competent Salmonella Typhimurium,33,34 to our knowledge this is the first report documenting the role of bacterial polysaccharides in natural transformation. In this study, we showed that bacterial surface polysaccharides, especially LOS, reduced the transformation frequencies in C. jejuni (Figure 2). How the surface polysaccharides reduce natural transformation in Campylobacter may be explained by two possibilities. First, both DNA and the surface polysaccharides are negatively charged and electrostatic repulsion between them may restrict the contact of transforming DNA with the bacterial surface. Second, CPS and LOS may act as physical barriers, sterically and/or structurally hindering the binding of DNA to receptor protein(s) located in the outer membrane. In either case, the surface polysaccharide would reduce the rate of DNA uptake by bacterial cells. Indeed, our results showed that loss of CPS and LOS significantly increased DNA uptake into Campylobacter cells (Figure 3), supporting the notion that surface polysaccharides limit the interaction of transforming DNA with specialized DNA binding and uptake apparatuses in the membrane. JB303 (double mutant) showed a higher transformation frequency than the single mutants (Figure 2a), but the measured rate of DNA binding and uptake in JB303 was not higher than those of the single mutants (Figure 3). The reason for this discrepancy is unknown, but it might be explained by the low sensitivity of the DNA uptake assay, which did not allow the differentiation of DNA uptake rates among the mutants.
It was previously reported that the galE mutation in C. jejuni strain 81 116 resulted in the formation of truncated LOS and a 20-fold decrease in the natural transformation frequency, suggesting that the LOS mutation reduced the natural transformation frequency of this C. jejuni strain,35 which is in contrast to the findings obtained in this study. However, GalE is an epimerase with dual functions that converts glucose to galactose and N-acetylglucosamine to N-acetylgalactosamine36 and is required for three important pathways including CPS biosynthesis, LOS production and N-linked protein glycosylation.13,37 Thus, mutation of galE would result in pleiotropic changes and affect the production of LOS and CPS35,36 as well as the N-linked protein glycosylation.36 Since the N-linked protein glycosylation plays a significant role in the natural transformation of C. jejuni,38 it is likely that the reduced natural transformation frequency in the C. jejuni galE mutant was not caused by the truncation of LOS, but was due to an altered N-linked protein glycosylation system.
In conclusion, we demonstrated in this study that loss of LOS, but not CPS, significantly reduced Campylobacter resistance to erythromycin, an important antibiotic for treating human campylobacteriosis. It was also shown that loss of LOS and CPS increased the frequency of natural transformation in C. jejuni strain NCTC 11168, which may ultimately affect the horizontal transfer of antibiotic resistance determinants. Given that both CPS and LOS are highly diverse and phase variable in Campylobacter,13,30 the results from this study suggest that the variable production of CPS and LOS in different Campylobacter strains may contribute to their intrinsic differences in antimicrobial susceptibility and natural transformation of antibiotic resistance genes.
Funding
This study was supported by the National Institutes of Health grant R01DK063008 and the National Research Initiative competitive grant 2007-35201-18278 from the USDA Cooperative State Research, Education, and Extension Service.
Transparency declarations
None to declare.
Supplementary data
Figure S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Supplementary Material
Acknowledgements
We would like to thank Jun Lin at the University of Tennessee for providing some of the erythromycin-resistant isolates used in this study.
References
- 1.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 4.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mead PS, Slutsker L, Dietz V, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman CR, Neimann J, Wegener HC, et al. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2000. pp. 121–38. [Google Scholar]
- 7.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–6. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 8.Engberg J, Aarestrup FM, Taylor DE, et al. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001;7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–93. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Plummer PJ. Mechanisms of antibiotic resistance in Campylobacter jejuni. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2008. pp. 263–76. [Google Scholar]
- 11.Wang Y, Taylor DE. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–55. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon B, Muraoka W, Sahin O, et al. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob Agents Chemother. 2008;52:2699–708. doi: 10.1128/AAC.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlyshev AV, Ketley JM, Wren BW. The Campylobacter jejuni glycome. FEMS Microbiol Rev. 2005;29:377–90. doi: 10.1016/j.femsre.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Fouts DE, Mongodin EF, Mandrell RE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–8. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 16.Kanipes MI, Holder LC, Corcoran AT, et al. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect Immun. 2004;72:2452–5. doi: 10.1128/IAI.72.4.2452-2455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perera VN, Nachamkin I, Ung H, et al. Molecular mimicry in Campylobacter jejuni: role of the lipo-oligosaccharide core oligosaccharide in inducing anti-ganglioside antibodies. FEMS Immunol Med Microbiol. 2007;50:27–36. doi: 10.1111/j.1574-695X.2007.00225.x. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, Yan M, Sahin O, et al. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother. 2007;51:1678–86. doi: 10.1128/AAC.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wösten MM, Boeve M, Koot MG, et al. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–9. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacon DJ, Alm RA, Burr DH, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun. 2000;68:4384–90. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlyshev AV, Wren BW. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using alcian blue dye. J Clin Microbiol. 2001;39:279–84. doi: 10.1128/JCM.39.1.279-284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller WG, Bates AH, Horn ST, et al. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl Environ Microbiol. 2000;66:5426–36. doi: 10.1128/aem.66.12.5426-5436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46:2124–31. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misawa N, Blaser MJ. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect Immun. 2000;68:6168–75. doi: 10.1128/iai.68.11.6168-6175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Agostino RB, Belanger A, D’Agostino RB., Jr A suggestion for using powerful and informative tests of normality. Am Stat. 1990;44:316–21. [Google Scholar]
- 26.Levene H. Robust tests for equality of variances. In: Olkin I, editor. Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Stanford, CA: Stanford University Press; 1960. pp. 278–92. [Google Scholar]
- 27.Karlyshev AV, Linton D, Gregson NA, et al. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000;35:529–41. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- 28.Oldfield NJ, Moran AP, Millar LA, et al. Characterization of the Campylobacter jejuni heptosyltransferase II gene, waaF, provides genetic evidence that extracellular polysaccharide is lipid A core independent. J Bacteriol. 2002;184:2100–7. doi: 10.1128/JB.184.8.2100-2107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibreel A, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006;58:243–55. doi: 10.1093/jac/dkl210. [DOI] [PubMed] [Google Scholar]
- 30.Bacon DJ, Szymanski CM, Burr DH, et al. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol Microbiol. 2001;40:769–77. doi: 10.1046/j.1365-2958.2001.02431.x. [DOI] [PubMed] [Google Scholar]
- 31.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–79. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 32.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–44. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 33.Sanderson KE, Janzer J, Head J. Influence of lipopolysaccharide and protein in the cell envelope on recipient capacity in conjugation of Salmonella typhimurium. J Bacteriol. 1981;148:283–93. doi: 10.1128/jb.148.1.283-293.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLachlan PR, Sanderson KE. Transformation of Salmonella typhimurium with plasmid DNA: differences between rough and smooth strains. J Bacteriol. 1985;161:442–5. doi: 10.1128/jb.161.1.442-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fry BN, Feng S, Chen YY, et al. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect Immun. 2000;68:2594–601. doi: 10.1128/iai.68.5.2594-2601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernatchez S, Szymanski CM, Ishiyama N, et al. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem. 2005;280:4792–802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 37.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–37. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 38.Larsen JC, Szymanski C, Guerry P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J Bacteriol. 2004;186:6508–14. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.