Abstract
Cavernous vascular malformations occur with a frequency of 1:200 and can cause recurrent headaches, seizures and hemorrhagic stroke if located in the brain. Familial cerebral cavernous malformations (CCMs) have been associated with germline mutations in CCM1/KRIT1, CCM2 or CCM3/PDCD10. For each of the three CCM genes, we here show complete localized loss of either CCM1, CCM2 or CCM3 protein expression depending on the inherited mutation. Cavernous but not adjacent normal or reactive endothelial cells of known germline mutation carriers displayed immunohistochemical negativity only for the corresponding CCM protein but not for the two others. In addition to proving loss of function at the protein level, our data are the first to demonstrate endothelial cell mosaicism within cavernous tissues and provide clear pathogenetic evidence that the endothelial cell is the cell of disease origin.
INTRODUCTION
Cerebral cavernous malformations (CCMs) are vascular lesions causing chronic headaches, seizures and hemorrhagic stroke. Autosomal dominantly inherited forms occur with a frequency of 1:2000 to 1:10 000 and are associated with germline mutations in one of at least three genes, CCM1, CCM2 or CCM3. The occurrence of multiple CCMs in most hereditary cases but only single CCM in the majority of sporadic cases has led to the notion that biallelic mutations within the same gene are required for the formation of CCMs (1,2). Such a ‘two-hit’ disease mechanism has been supported by the observation that slowly enlarging CCMs may develop as late sequelae of cranial radiation therapy within fields of prior irradiation (3). However, a two-hit model of CCM genesis that assumes inactivation of both alleles of a given CCM gene within affected cells has been difficult to prove. Biallelic germline and somatic CCM1 mutations have thus far only been reported once. Tissue pieces from the middle part of a single cavernous lesion harbored a somatic 34 bp deletion within exon 15 of the CCM1 gene in addition to a known CCM1 germline founder mutation (4). Furthermore, two somatic CCM1 missense mutations were found in a singular lesion of an individual affected with sporadic CCM (5). While CCM1 mutations are generally truncating or result from large genomic rearrangements, the functional relevance of these two missense mutations remained unclear. It could also not be determined whether the two mutations occur in cis or in trans. Several other groups have been unable to identify somatic mutations in human and murine cavernous tissues (6–8). This may have been due to the application of insensitive genetic screening techniques given that direct sequencing revealed many CCM1 gene mutations that had been missed by denaturing high-performance liquid chromatography or SSCP in the past.
A further explanation for the difficulties to prove the two-hit model relates to the nature of the CCM tissue. Blood-filled, often thrombosed and re-organized cavities are lined by extremely thin endothelial cells that are separated by abundant collagenous connective tissue containing a mixture of different cell types (9). A putative second mutation could be envisioned to occur only in a small subset of cells and, therefore, may remain undiscovered during genetic screening.
Currently, the cell of disease origin is still unknown. In situ hybridization (5,10) and immunohistochemical studies (11) have shown CCM transcript and protein expression in endothelial and various non-endothelial cell types such as neurons and astrocytes. Notably, cavernous malformations do not only occur in the brain but occasionally also in the skin (12) and consist of endothelial cells with slight proliferative tendency (13) and lack of endothelial tight junctions (14,15) that stain positively with anti-CCM1 antibodies in cell culture (16). Such morphologically altered, extremely thin vascular endothelial cells lining the caverns appear to be the primary disease compartment that needs to be dissected and analyzed. This approach is supported by recent transplantation experiments in zebrafish that have shown that ccm1 cell autonomously regulates endothelial cellular morphology (17).
The present study demonstrates that a previously generated anti-human CCM1 antibody (16) and two newly generated anti-human CCM2 and CCM3 antibodies are immunohistochemical markers for the vascular endothelium. CCM tissues from individuals with known CCM1, CCM2 or CCM3 germline mutations revealed that immunohistochemical negativity of the corresponding CCM protein is the hallmark of vascular endothelial cells lining pathological caverns. Lack of immunoreactivity of the respective CCM protein in cavernous endothelial cells provides strong evidence for the two-hit hypothesis of CCM formation and underscores the concept of morphologically abnormal endothelial cells lining caverns as targets of disease origin. In addition, our studies reveal that the detection of loss-of-heterozygosity in CCM endothelial cells is complicated by both low number of mutated cells and the fact that the vascular endothelium within a single CCM tissue is mosaic for wild-type and mutant endothelial cells.
RESULTS
Variable expression of CCM proteins in normal brain vascular endothelium
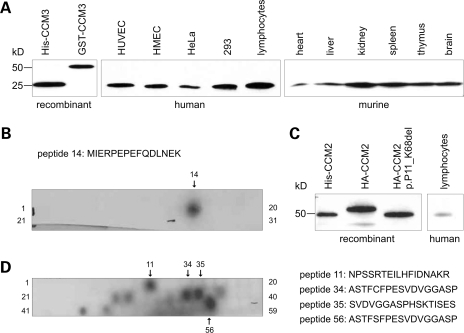
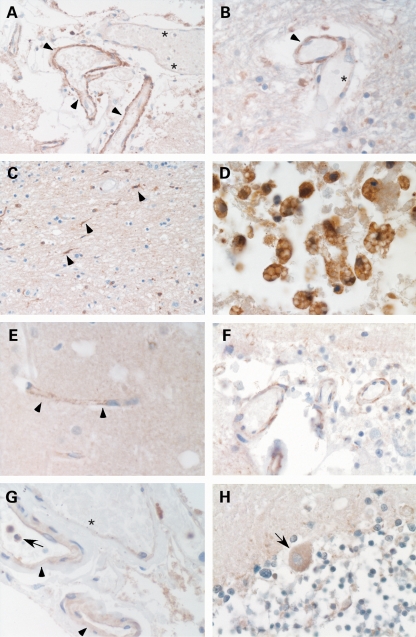
The newly generated anti-human CCM3 antibody reacts specifically with a fragment of the size of 25 kDa recombinant human CCM3 in various human cell and murine tissue lysates (Fig. 1A) and with CCM3 peptide MIERPEPEFQDLNEK (Fig. 1B). Anti-human CCM2 recognizes overexpressed CCM2 (Fig. 1C) as well as several CCM2 epitopes (Fig. 1D) but could only be used in high concentrations that yielded strong background in most western blots with cell and tissue lysates. Immunohistochemistry demonstrated that both antibodies and a polyclonal but not a monoclonal anti-human CCM1 antibody (16) stained vascular endothelium of venous, arterial and capillary blood vessels (Fig. 2). The CCM1 antibody demonstrated immunoreactivity in both meningeal (Fig. 2A) and parenchymal vessels (Fig. 2B). We observed large differences of the staining intensity for CCM1 among the endothelia of different normal brain vessels and within the meninges. Parenchymal capillary endothelia with broadened walls were often stained, whereas vessels with narrow walls regularly showed no immunoreactivity. Moreover, brain tissue cells resembling microglia revealed strong immunoreactivity (Fig. 2C). This correlated well to the strong staining of foamy macrophages in the vicinity of a cavernoma from a CCM2 germline mutation carrier (Fig. 2D). The antibody against CCM2 revealed granular staining of capillary vessels throughout the brain parenchyma and in the meninges (Fig. 2E and F). The antibody against CCM3 showed a staining pattern similar to CCM1. Most vessel endothelia were labeled by the antibody (Fig. 2G). However, there were thin-walled vessels that showed merely weak or no immunoreactivity (Fig. 2F and G). In addition, the CCM3 antibody detected polymorphonuclear blood cells and Purkinje cells in the cerebellum (Fig. 2G and H).
Figure 1.
(A) Polyclonal anti-human CCM3 antibody specifically reacts with a protein of the size of recombinant human CCM3 purified from E. coli (lane 1) in the indicated human cell and murine tissue lysates (HUVEC, human umbilical vein endothelial cells; HMEC, human microvascular endothelial cells; HeLa, human epitheloid cervix carcinoma cells; 293, human embryonal kidney cell line). Ten micrograms of lysates was separated on 10–20% polyacrylamide–SDS gradient gels and analyzed by western blotting. (B) CCM3 epitope mapping using PepSpotsTM technology. (C) Polyclonal anti-human CCM2 antibody recognizes recombinant human CCM2 purified from E. coli, HA-tagged CCM2 as well as HA-tagged CCM2:p.P11_K68del (18) overexpressed in 293 cells and a protein of the size of CCM2 in lymphocyte lysates. (D) CCM2 epitope mapping using PepSpotsTM technology. Peptide 11 resides within the PTB domain making cross-reactivity likely. Peptide 56 was artificially modified and corresponds to peptide 34.
Figure 2.
Expression of CCM proteins in human brain tissues. (A) Polyclonal anti-human CCM1 antibody stains vascular endothelia (arrowheads) in normal meningeal tissues. The asterisks indicate an adjacent blood vessel with immunonegative endothelial cells. (B) Stronger (arrowhead) and weaker (asterisk) staining intensities with the anti-CCM1 antibody in parenchymal vessels. The CCM1 antibody also stains (C) microglia and (D) foamy macrophages in the vicinity of a cavernous lesion. (E) Granular staining of capillary vessels in the brain parenchyma (E) and the meninges (F) incubated with the polyclonal anti-human CCM2 antibody. (G and H) Positive immunoreactivity of vessel endothelia with the polyclonal anti-human CCM3 antibody which also detected (G) polymorphonuclear blood cells (arrow) and (H) Purkinje cells (arrow) in the cerebellum. (G) Similar to anti-CCM1, anti-CCM3 shows different staining intensities for endothelia of vessels with broadened walls (arrow head) when compared with adjacent thin-walled vessels (asterisk).
Absence of CCM immunoreactivity in endothelial cells lining pathological caverns correlates with the respective CCM germline mutation
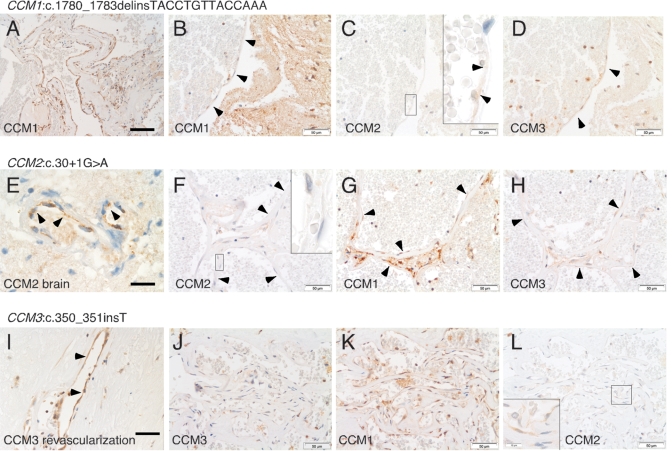
Markedly reduced to absent CCM1 immunostaining in endothelial cells lining pathological caverns was observed for cavernous endothelia analyzed from one patient carrying a CCM1 germline mutation (No. 1, Table 1, Supplementary Material, Fig. S1A). Within the cavernous lesion of a second individual with a CCM1 germline mutation (No. 2, Table 1), a subset of presumably neoangiogenic vessels was labeled with the anti-CCM antibody (Fig. 3A) while endothelia lining large pristine caverns were found to be immunonegative for CCM1 (Fig. 3B). For this familial case 2, adjacent brain tissue was available, which demonstrated prominent staining of normal arterial endothelium with the anti-human CCM1 antibody on the same tissue section (data not shown). Caverns analyzed from both CCM1 germline mutation carriers (Table 1, Nos 1 and 2) showed positive immunostaining for CCM2 and CCM3 (Supplementary Material, Figs S1B and C and Fig. 3C, D). Consistent with our recent finding that expression of CCM1:p.A715VfsX14 is reduced but not absent in vitro (18), even pristine caverns from sporadic case No. 3 (Table 1) demonstrated weak immunoreactivity (Supplementary Material, Fig. S1D).
Table 1.
Overview of CCM endothelial cell immunohistochemistry and respective germline mutations
| No. | Presentation | Gene | Exon | Nucleotide change | Amino acid change | Type of mutation | Reference | CCM1-IH | CCM2-IH | CCM3-IH |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Familial | CCM1 | 16 | c.1683_1684insA | p.V562SfsX6 | Frameshift | (22) | − | + | + |
| 2 | Familial | CCM1 | 17 | c.1780_1783delinsTA CCTGTTACCAAA | p.A594YfsX14 | Frameshift | (23) | −a | + | + |
| 3 | Sporadic | CCM1 | Intron19 | c.2143-2A>G | p.A715VfsX14 | Splice | (18) | (+)b | + | + |
| 4 | Familial | CCM2 | Intron1 | c.30+1G>A | p.? | Splice | (18) | + | − | + |
| 5 | Familial | CCM2 | 2 | c.55C>T | p.R19X | Stop | (18) | + | − | + |
| 6 | Familial | CCM2 | 2 | c.55C>T | p.R19X | Stop | (18) | + | − | + |
| 7 | Familial | CCM3 | 7 | c.350_351insT | p.D119RfsX2 | Frameshift | (18) | + | + | −a |
IH, immunohistochemistry of CCM endothelial cells. Nos 5 and 6 are siblings. GenBank accession nos are: CCM1: NM_194456.1, CCM2: NM_031443.3, CCM3: NM_007217.3. DNA mutation numbering is based on cDNA sequence with +1 corresponding to the A of the ATG translation initiation codon.
aImmunonegative pristine caverns plus immunoreactive endothelial cells within same CCM lesion, the latter most likely representing neoangiogenic vessels derived from vessels with only the germline mutation.
bPartial immunoreactivity in pristine caverns corresponding to the fact that CCM1:p.A715VfsX14 was shown to be reduced but not absent in vitro (18).
Figure 3.
Serial sections from carriers of a known germline mutation in one of the three CCM genes immunostained for CCM1, CCM2 and CCM3, respectively. Immunohistochemistry with the anti-human CCM1 antibody revealed (A) CCM1-positive and (B) CCM1-negative endothelia (arrowheads) within the same cavernous malformation tissue section of a CCM1:c.1780_1783delinsTACCTGTTACCAAA germline mutation carrier (Table 1, No. 2), whereas (C) CCM2- and (D) CCM3-immunoreactivity was observed. (E) Normal brain arterial endothelium of a CCM2:c.30+1G>A germline mutation carrier (Table 1, No. 4) reacts with the anti-human CCM2 antibody while (F) cavernous endothelium in the same tissue section does not. (G and H) Immunostaining for CCM1 and CCM3 is positive in caverns from this individual. (I) Vessels in revascularized thrombosed caverns of a CCM3:c.350_351insT germline mutation carrier (Table 1, No. 7) show CCM3-immunoreactivity, whereas (J) pristine caverns do not. (K and L) Positive immunostaining for CCM1 and CCM2 in the same cavernous lesion. Scale bar in (A): 100 µm, in (E): 20 µm, all others: 50 µm. Insets in (C) and (L) demonstrate faint granular immunoreactivity for CCM2, whereas the inset in (F) shows negativity for CCM2 at five times higher magnification (original magnification 100×).
Immunohistochemical negativity of cavernous endothelial cells from three carriers of CCM2 germline mutations that are all located near the 5′ end of the CCM2 gene (Table 1, Nos 4–6) was consistent with defective CCM2 protein expression in all cavernous endothelia assessed (Fig. 3F, Supplementary Material, Fig. S1H and K). In contrast, regular brain arterial endothelium (available for individual Nos 4 and 5) showed immunoreactivity with the anti-human CCM2 antibody (Fig. 3E). As expected, cavernous endothelium from carriers of CCM2 germline mutations stained positively for anti-human CCM1 and CCM3 antibodies (Fig. 3G and H, Supplementary Material, Fig. S1G, I, J and L).
CCM3 mutations are known to account for only 10% of CCM mutations (18,19). The only individual with a germline mutation in the CCM3 gene in our panel of patients (Table 1, No. 7) showed absent CCM3 expression in cavernous endothelial cells (Fig. 3J), whereas capillary endothelial cells of vessels in revascularized thrombosed caverns within the same tissue sections reacted with the anti-human CCM3 antibody (Fig. 3I). Again, endothelial cells lining pathological caverns of the CCM3 germline mutation carrier revealed immunoreactivity for CCM1 and CCM2 (Fig. 3K and L).
Taken together, negative immunostaining of the corresponding CCM protein in endothelia from CCM tissues harboring a germline mutation suggests that the antibodies tested specifically stain the corresponding CCM1, CCM2 or CCM3 proteins. Furthermore, our data underline that CCM pathology is primarily due to defective endothelial cells. Most importantly, complete and specific lack of CCM protein in affected endothelial cells from CCM germline mutation carriers supports a genetic two-hit mechanism for CCM formation.
Technical limitations of genetic proof of two-hit mechanism
For molecular analyses, about 200 morphologically pathological endothelial cells (as seen in Fig. 3B) from each of three small heterogeneous CCM1-3 tissues (Table 1, Nos 2, 4 and 7) were sampled by laser-assisted microdissection in order to circumvent allele drop-out problems. Whole genome amplification (WGA) of the isolated DNA was performed by improved primer extension pre-amplification PCR (I-PEP-PCR) (20,21). Subsequent amplification and direct sequencing of the CCM gene for which a germline mutation (18,22,23) and negative CCM protein expression had been found revealed that only a subset of exons could be amplified most likely due to fragmentation of the DNA in the archival paraffin-embedded tissues. These amplification products did not reveal a second somatic mutation.
DISCUSSION
Arteriovenous (ENG/ACVRL1/RASA1), venous (TIE2), glomuvenous (glomulin) and cavernous (CCM1, CCM2, CCCM3) malformations are localized defects of vascular development that have been associated with autosomal dominantly inherited heterozygous germline mutations. Apart from TIE2 mutations, all are thought to cause loss of function (24). Paradominant inheritance was first shown for glomuvenous malformations (GVMs), which are characterized by abnormally differentiated smooth muscle cells in the walls of distented venous channels. A somatic 5 bp deletion in the second glomulin allele was identified in only one out of seven resected fresh–frozen lesions from different individuals by SSCP and heteroduplex analyses followed by sequencing of the cloned amplification product. It was concluded that cellular heterogeneity of GVMs limits somatic mutation detection (25). Similarly, a subtle denaturing high-performance liquid chromatography change in only two out of three samples from one surgically excised CCM lesion led to the identification of a biallelic somatic 34 bp deletion in the CCM1 gene. Peak heights suggested that 13.5–21% of DNA derived from an unknown cell type in the middle of the lesion carried the mutation, whereas the mutation-negative sample from the lesion margin might have been largely composed of normal vessels (4). Our data define cavernous endothelial cells as the primary disease compartment and suggest that the mutation-negative vessels might correspond to immunopositive neoangiogenic vessels as seen in Figure 3I. In a recent systematic study of fresh–frozen venous malformation tissues, single and double-hit mutations in the angiopoietin-receptor gene TIE2 were found to cause both solitary and multiple sporadic forms (26).
In contrast to the venous and glomuvenous skin lesions, CCMs are less accessible and often resected at late stages after recurrent bleeding. In addition, microsurgical excision is performed by the dissection of the vessel to brain interlayer. During the excision, the cavernous lesion itself and the feeding and draining vessels are permanently shrinked by electrosurgical coagulation. Thus, the CCM tissue is often primarily thrombosed, reorganized, calcified and always artificially altered by the surgical manipulation. Furthermore, endothelial cells lining pristine caverns only represent a small subset of cells. Therefore, DNA must be purified from small amounts of laser-microdissected starting material and pre-amplified by WGA to enrich mutant cells. When implementing WGA in our laboratory, only archival paraffin-embedded tissues were available for our cohort of patients. Compared with frozen sections, the quality of cell morphology for immunohistochemistry and laser microdissection is superior. However, fixation and staining procedures as well as sampling and storage conditions are known to compromise the integrity of the DNA and present a challenge for molecular analyses. Several WGA techniques are not suitable for use with formalin-fixed, paraffin-embedded clinical samples due to fragmentation of the template genomic DNA. For paraffin-embedded tissue sections, I-PEP-PCR was previously shown to yield the most efficient downstream amplification results after pre-amplification (20) but did not lead to detection of mutations in our tissue samples. Thus, the nature of the second-hit event remains unknown and potentially includes small mutations, large inactivating gene deletions (27–30), epigenetic as well as (post-)transcriptional pathomechanisms.
Endothelial cell mosaicism within CCM tissues as seen in Figure 3A and I represents a further problem for molecular analyses. CCMs have been characterized as hamartomatous since they are benign, disorganized lesions with growth potential as a result of proliferative capacity and hemodynamic modifications. Endothelial proliferation and neoangiogenesis has been demonstrated within human CCM specimens by immunohistochemical staining for proliferating cell nuclear antigen (PCNA) and various vascular markers (13,31). Notelet et al. distinguished between areas with collagenous, thick-walled caverns and very few PCNA-stained endothelial cells and ‘territories showing irregular, dilated, closely contiguous vascular spaces separated by lacy walls often with stained endothelial cells’ as seen in Figure 3B or L. They also noted thrombi with figures of revascularization that contained PCNA-positive cells as well as staining of endothelial cells in capillaries at the periphery of these lesions and reactive glial cells, endothelial cells and pericytes in the surrounding brain parenchyma. Thus, the majority of activated proliferating endothelia within CCM tissues appears to be reactive and to correspond to the CCM-immunopositive neoangiogenic vessels seen in our study (Fig. 3A and I).
Experimental data from zebrafish knockdown and knockout studies suggest alterations in endothelial cell morphology rather than endothelial cell proliferation as likely pathomechanism (17). We have observed that thick-walled thrombosed caverns often even lack endothelial cells at the cavernous wall (data not shown). It is conceivable that biallelic loss of a CCM-coding gene might affect other than tumor suppressor functions of CCM interactors like RAPIA, e.g. regulation of endothelial cell-cell junctions (16). Therefore, somatic inactivation of the TIE2, CCM and glomulin genes suggests that the Knudsonian two-hit mechanism may not be limited to a classic tumor suppressor model but also plays a pivotal role in the pathogenesis of these non-neoplastic vascular malformations.
MATERIALS AND METHODS
Generation of antibodies against human CCM2 and CCM3
Full-length human CCM2 and CCM3 genes were cloned into pET28b and overexpressed in Escherichia coli strain BL21 (DE3), respectively. Highly purified protein samples were obtained by IMAC capture of His-tagged fusion proteins using Ni-NTA superflow matrix (Qiagen), subsequent thrombin proteolysis of the His-tag followed by a subtractive IMAC step to remove uncleaved protein and a final gel filtration step on a preparative Superdex75 column (GE Healthcare) (CRELUX GmbH, Martinsried, Germany). The purified proteins were pure as judged by SDS–PAGE analysis and Coomassie staining. Polyclonal anti-human CCM2 and anti-human CCM3 sera were prepared through the immunization of rabbits with purified human CCM2 or CCM3 proteins coupled to keyhole limpet hemocyanin (immunoGlobe GmbH, Himmelstadt, Germany). Sera were affinity purified on columns consisting of non-tagged CCM2 or CCM3 protein covalently coupled to NHS–Sepharose 4FF beads. Western blot analyses of various tissue and cell lysates demonstrated that the anti-human CCM3 antibody reacted with a specific 25 kDa band as well as an additional 70 kDa protein representing Hsp70 and 30S ribosomal protein S1 as determined by mass spectrometry (data not shown). To get rid of these contaminations, epitope mapping of the 212 amino acid CCM3 sequence was carried out using PepSpots™ technology (JPT Peptide Technologies GmbH, Berlin). Two libraries of 15mer peptides with eight or nine amino acids overlap revealed MIERPEPEFQDLNEK as a major epitope. In an alternative approach, a peptide corresponding to this sequence was used for purification of the anti-CCM3 antiserum yielding an affinity-purified antibody (IG-626; immunoGlobe GmbH, Himmelstadt, Germany) specific to human, murine, rat, chicken and Xenopus CCM3. The respective peptide from Danio rerio is not recognized by this antibody (data not shown). The PepSpotsTM technology was also applied for CCM2 epitope mapping. However, due to the extremely low yield of the peptide-purified anti-CCM3 antibody, this approach was not pursued for the anti-CCM2 antibody that proved to work specifically in immunohistochemical applications.
Western blotting
Cell lysates were prepared as described by Voss et al. (32). Mouse tissues were homogenized in an iso-osmotic homogenization buffer (0.25 m sucrose, 10 mm Tris pH 7,2, 1 mm EDTA, Roche complete EDTA-free protease inhibitor) by five passes with 1000 U/min using a Potter S homogenizer (5). After a centrifugation step of 1000 r.p.m. for 2 min (rotor F45-30-11, eppendorf, Hamburg), the post-nuclear supernatants were frozen in liquid nitrogen. Cell lysates were prepared with lysis buffer (1% Triton-X-100, 10 mm Tris pH 8, 1 mm EDTA, Roche complete EDTA-free protease inhibitor). For western blot analysis, 10 µg of tissue and cell lysates were loaded onto 10–20% SDS–PAGE gels, electroblotted onto nitrocellulose and probed with the polyclonal rabbit anti-human CCM3 antibody at a dilution of 1 µg/ml. Secondary antibodies conjugated to peroxidase were purchased from Dianova.
Immunohistochemistry
Immunohistochemistry was performed on neutral-buffered formalin-fixed and paraffin-embedded tissue slices as described before (33). Briefly, tissue sections (3 µm) were deparaffinized in xylene and rehydrated in graded alcohol. Antigen retrieval was performed in citrate buffer (pH 6.0) in a standard microwave oven for 10 min followed by blocking the endogenous peroxidase with H2O2 (3%) in methanol and blocking unspecific antibody binding with goat normal serum. Polyclonal anti-human CCM1 antibody (pAB anti-KRIT-1, Rb6832), a kind gift of Drs Marc Ginsberg and Angela Glading (University of California, San Diego), anti-human CCM2 antibody and anti-human CCM3 antibody were used at a dilution of 1:1000 (16) or concentrations of 16 and 6 µg/ml for immunohistochemistry, respectively. Antigen binding was visualized with the Histostain-Plus kit (Zymed Laboratories, South San Francisco, CA, USA).
Laser-capture microdissection, WGA and mutation analyses
Laser-capture microdissection of 10 µm CCM tissue sections was performed with a Leica LMD6000 microscope. Tissue slices were deparaffinized, rehydrated in graded alcohol and stained with 4,6-diamidino-2-phenylindole dihydrochloride at a concentration of 400 ng/ml for 10 min in order to visualize the nuclei of endothelial cells for microdissection.
Genetic testing was approved by local Ethics Committees (University of Würzburg, Study 21/05; Philipps-University Marburg, Study 149/05) and performed with informed consent. Published protocols were followed for DNA isolation of about 200 endothelial cells per tissue (20) and WGA by I-PEP-PCR (21). Subsequently, coding CCM1-3 exons including adjacent splice sites were directly sequenced on a Beckmann CEQ 8800 capillary electrophoresis system according to (18).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Bavarian Genome Network and the Deutsche Forschungsgemeinschaft/Graduiertenkolleg 1048 (U.F. and S.S.). Funding to pay the Open Access charge was provided by the Bavarian Genome Network.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the affected individuals for participating in this study, Drs Marc Ginsberg and Angela Glading for the anti-human CCM1 antibody (pAB anti-KRIT-1, Rb6832), Dr Ismail Moarefi at CRELUX GmbH for comments on the manuscript, Gerhard Zimmermann at Leica microsystems for valuable support with the LMD and Heike Geißel and Sabine Gätzner for expert technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Revencu N., Vikkula M. Cerebral cavernous malformation: new molecular and clinical insights. J. Med. Genet. 2006;43:716–721. doi: 10.1136/jmg.2006.041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labauge P., Denier C., Bergametti F., Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6:237–244. doi: 10.1016/S1474-4422(07)70053-4. [DOI] [PubMed] [Google Scholar]
- 3.Strenger V., Sovinz P., Lackner H., Dornbusch H.J., Lingitz H., Eder H.G., Moser A., Urban C. Intracerebral cavernous hemangioma after cranial irradiation in childhood. Incidence and risk factors. Strahlenther. Onkol. 2008;184:276–280. doi: 10.1007/s00066-008-1817-3. [DOI] [PubMed] [Google Scholar]
- 4.Gault J., Shenkar R., Recksiek P., Awad I.A. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 5.Kehrer-Sawatzki H., Wilda M., Braun V.M., Richter H.P., Hameister H. Mutation and expression analysis of the KRIT1 gene associated with cerebral cavernous malformations (CCM1) Acta Neuropathol. (Berl.) 2002;104:231–240. doi: 10.1007/s00401-002-0552-6. [DOI] [PubMed] [Google Scholar]
- 6.Marini V., Ferrera L., Pigatto F., Origone P., Garre C., Dorcaratto A., Viale G., Alberti F., Mareni C. Search for loss of heterozygosity and mutation analysis of KRIT1 gene in CCM patients. Am. J. Med. Genet. A. 2004;130:98–101. doi: 10.1002/ajmg.a.30122. [DOI] [PubMed] [Google Scholar]
- 7.Plummer N.W., Gallione C.J., Srinivasan S., Zawistowski J.S., Louis D.N., Marchuk D.A. Loss of p53 sensitizes mice with a mutation in Ccm1 (KRIT1) to development of cerebral vascular malformations. Am. J. Pathol. 2004;165:1509–1518. doi: 10.1016/S0002-9440(10)63409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reich P., Winkler J., Straube A., Steiger H.J., Peraud A. Molecular genetic investigations in the CCM1 gene in sporadic cerebral cavernomas. Neurology. 2003;60:1135–1138. doi: 10.1212/01.wnl.0000055470.62265.44. [DOI] [PubMed] [Google Scholar]
- 9.Bertalanffy H., Benes L., Miyazawa T., Alberti O., Siegel A.M., Sure U. Cerebral cavernomas in the adult. Review of the literature and analysis of 72 surgically treated patients. Neurosurg. Rev. 2002;25:1–53. doi: 10.1007/s101430100179. [DOI] [PubMed] [Google Scholar]
- 10.Petit N., Blecon A., Denier C., Tournier-Lasserve E. Patterns of expression of the three cerebral cavernous malformation (CCM) genes during embryonic and postnatal brain development. Gene Expr. Patterns. 2006;6:495–503. doi: 10.1016/j.modgep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Tanriover G., Boylan A.J., Diluna M.L., Pricola K.L., Louvi A., Gunel M. PDCD10, the gene mutated in cerebral cavernous malformation 3, is expressed in the neurovascular unit. Neurosurgery. 2008;62:930–938. doi: 10.1227/01.neu.0000318179.02912.ca. [DOI] [PubMed] [Google Scholar]
- 12.Eerola I., Plate K.H., Spiegel R., Boon L.M., Mulliken J.B., Vikkula M. KRIT1 is mutated in hyperkeratotic cutaneous capillary-venous malformation associated with cerebral capillary malformation. Hum. Mol. Genet. 2000;9:1351–1355. doi: 10.1093/hmg/9.9.1351. [DOI] [PubMed] [Google Scholar]
- 13.Sure U., Butz N., Schlegel J., Siegel A.M., Wakat J.P., Mennel H.D., Bien S., Bertalanffy H. Endothelial proliferation, neoangiogenesis, and potential de novo generation of cerebrovascular malformations. J. Neurosurg. 2001;94:972–977. doi: 10.3171/jns.2001.94.6.0972. [DOI] [PubMed] [Google Scholar]
- 14.Clatterbuck R.E., Eberhart C.G., Crain B.J., Rigamonti D. Ultrastructural and immunocytochemical evidence that an incompetent blood–brain barrier is related to the pathophysiology of cavernous malformations. J. Neurol. Neurosurg. Psychiatry. 2001;71:188–192. doi: 10.1136/jnnp.71.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong J.H., Awad I.A., Kim J.H. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46:1454–1459. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
- 16.Glading A., Han J., Stockton R.A., Ginsberg M.H. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell–cell junctions. J. Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogan B.M., Bussmann J., Wolburg H., Schulte-Merker S. ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum. Mol. Genet. 2008;17:2424–2432. doi: 10.1093/hmg/ddn142. [DOI] [PubMed] [Google Scholar]
- 18.Stahl S., Gaetzner S., Voss K., Brackertz B., Schleider E., Sürücü O., Kunze E., Netzer C., Korenke C., Finckh U., et al. Novel CCM1, CCM2, and CCM3 mutations in patients with cerebral cavernous malformations: in-frame deletion in CCM2 prevents formation of a CCM1/CCM2/CCM3 protein complex. Hum. Mutat. 2008;29:709–717. doi: 10.1002/humu.20712. [DOI] [PubMed] [Google Scholar]
- 19.Denier C., Labauge P., Bergametti F., Marchelli F., Riant F., Arnoult M., Maciazek J., Vicaut E., Brunereau L., Tournier-Lasserve E. Genotype–phenotype correlations in cerebral cavernous malformations patients. Ann. Neurol. 2006;60:550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 20.Wild P.J., Dietmaier W. Whole genome amplification by improved primer-extension pre-amplified PCR. In: Hughes S., Lasken R., editors. Whole Genome Amplification. The Methods Express Series. Bloxham, UK: Scion Publishing Ltd; 2005. pp. 23–32. [Google Scholar]
- 21.Werther M., Saure C., Pahl R., Schorr F., Rüschoff J., Alles J.U., Heinmöller E. Molecular genetic analysis of surveillance biopsy samples from Barrett’s mucosa–significance of sampling. Pathol. Res. Pract. 2008;204:285–294. doi: 10.1016/j.prp.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Sürücü O., Sure U., Gaetzner S., Stahl S., Benes L., Bertalanffy H., Felbor U. Clinical impact of CCM mutation detection in familial cavernous angioma. Childs Nerv. Syst. 2006;22:1461–1464. doi: 10.1007/s00381-006-0202-8. [DOI] [PubMed] [Google Scholar]
- 23.Sürücü O., Sure U., Stahl S., Gaetzner S., Miller D., Bertalanffy H., Felbor U. A novel CCM1 mutation in a 2-year old child. Cerebral and cutaneous manifetations of familial cavernoma. Monatsschr. Kinderheilkd. 2007;155:1161–1165. [Google Scholar]
- 24.Brouillard P., Vikkula M. Genetic causes of vascular malformations. Hum. Mol. Genet. 2007;16(Spec no. 2):R140–R149. doi: 10.1093/hmg/ddm211. [DOI] [PubMed] [Google Scholar]
- 25.Brouillard P., Boon L.M., Mulliken J.B., Enjolras O., Ghassibe M., Warman M.L., Tan O.T., Olsen B.R., Vikkula M. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (‘glomangiomas’) Am. J. Hum. Genet. 2002;70:866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limaye N., Wouters V., Uebelhoer M., Tuominen M., Wirkkala R., Mulliken J.B., Eklund L., Boon L.M., Vikkula M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2008 doi: 10.1038/ng.272. doi:10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaetzner S., Stahl S., Surucu O., Schaafhausen A., Halliger-Keller B., Bertalanffy H., Sure U., Felbor U. CCM1 gene deletion identified by MLPA in cerebral cavernous malformation. Neurosurg. Rev. 2007;30:155–159. doi: 10.1007/s10143-006-0057-1. [DOI] [PubMed] [Google Scholar]
- 28.Felbor U., Gaetzner S., Verlaan D.J., Vijzelaar R., Rouleau G.A., Siegel A.M. Large germline deletions and duplication in isolated cerebral cavernous malformation patients. Neurogenetics. 2007;8:149–153. doi: 10.1007/s10048-006-0076-7. [DOI] [PubMed] [Google Scholar]
- 29.Liquori C.L., Berg M.J., Squitieri F., Leedom T.P., Ptacek L., Johnson E.W., Marchuk D.A. Deletions in CCM2 are a common cause of cerebral cavernous malformations. Am. J. Hum. Genet. 2007;80:69–75. doi: 10.1086/510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liquori C.L., Penco S., Gault J., Leedom T.P., Tassi L., Esposito T., Awad I.A., Frati L., Johnson E.W., Squitieri F., et al. Different spectra of genomic deletions within the CCM genes between Italian and American CCM patient cohorts. Neurogenetics. 2008;9:25–31. doi: 10.1007/s10048-007-0109-x. [DOI] [PubMed] [Google Scholar]
- 31.Notelet L., Houtteville J.P., Khoury S., Lechevalier B., Chapon F. Proliferating cell nuclear antigen (PCNA) in cerebral cavernomas: an immunocytochemical study of 42 cases. Surg. Neurol. 1997;47:364–370. doi: 10.1016/s0090-3019(96)00248-0. [DOI] [PubMed] [Google Scholar]
- 32.Voss K., Stahl S., Schleider E., Ullrich S., Nickel J., Mueller T.D., Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 33.Wildeboer D., Naus S., Amy Sang Q.X., Bartsch J.W., Pagenstecher A. Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J. Neuropathol. Exp. Neurol. 2006;65:516–527. doi: 10.1097/01.jnen.0000229240.51490.d3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.