Abstract
Hyperphosphorylation of the Drosophila melanogaster origin recognition complex (DmORC) by cyclin dependent kinases (CDKs) allows nucleotide binding but inhibits the ATPase activity of Orc1, and ablates the ATP-dependent interaction of ORC with DNA. Here we present single particle electron microscopy (EM) studies of ORC bound to nucleotide in both the dephosphorylated and hyper-phosphorylated states. 3D image reconstructions show that nucleotide binding gives rise to an analogous conformation independent of phosphorylation state. At the intermediate resolution achieved in our studies, ATP promotes changes along the toroidal core of the complex with negligible differences contributed by phosphorylation. Thus, hyperphosphorylation of DmORC does not induce meso-scale rearrangement of the ORC structure. To better understand ORC’s role in origin remodeling, we performed atomic force microscopy (AFM) studies that show the contour length of a 688 bp linear DNA fragment shortens by the equivalent of ~130 bp upon ORC binding. This data, coupled with previous studies that showed a linking number change in circular DNA upon ORC binding, suggests that ORC may wrap the DNA in a manner akin to DnaA. Based on existing data and our structures, we propose a subunit arrangement for the AAA+ and winged helix domains, and in addition, speculate on a path of the 133 bp of DNA around the ORC complex.
Keywords: Single particle electron microscopy, Atomic force microscopy, DNA replication initiation, Origin recognition complex, Metazoan
1. Introduction
The ATP-bound initiator protein, the origin recognition complex (ORC), binds chromosomal DNA at sites potentially determined for DNA replication initiation. ORC (~390 kDa) is a heterohexamer that includes five proteins containing ATPases Associated with various cellular Activities (AAA+) domains (Clarey et al., 2006; Speck et al., 2005). Such AAA+ domains generally utilize nucleotide binding and hydrolysis to remodel protein:protein or protein:nucleic acid complexes (Davey et al., 2002; Neuwald et al., 1999). ORC recruits the replication factors Cdc6 and Cdt1 to the origin, which is then followed by loading of the pre-replicative helicase, the mini-chromosomal maintenance proteins 2–7 (MCM2–7) (Dutta and Bell, 1997; Kim and Kipreos, 2007). This complex of proteins is called the preRC and such nucleoprotein complexes can only form in G1 phase when cells have low levels of CDK activity (Diffley, 2004; Kelly and Brown, 2000; Stillman, 1996). After assembly of a preRC, transition to S phase in the budding yeast requires CDK phosphorylation of two proteins called Sld2 and Sld3, which together with the activity of another kinase, cdk7:Dbf4, activate duplex unwinding and DNA replication (Botchan, 2007). As CDK levels increase in the cell cycle, inhibition of re-replication is imposed at many levels, and, though replication initiation factors are generally conserved, the negative regulation of pre-RC proteins by CDKs is highly divergent throughout eukaryotes, with metazoans evolving novel regulatory mechanisms (Bell and Dutta, 2002; Diffley, 2004).
ORC is phosphorylated in a cell cycle-dependent manner in Saccharomyces cerevisiae, S. pombe, Xenopus laevis and humans, with the majority of the phosphorylation occurring after the beginning of S-phase and continuing until late M-phase (Carpenter et al., 1996; Lygerou and Nurse, 1999; Mendez et al., 2002; Nguyen et al., 2001; Tugal et al., 1998; Vas et al., 2001). It is known that phosphorylation of S. cerevisiae Orc2 and Orc6, and S. pombe Orc2 contributes to prevention of re-replication, but it is not clear how these modifications affect this regulation (Nguyen et al., 2001; Vas et al., 2001). Metazoans appear to control ORC activity through differential regulation of chromatin association and by proteolysis of the Orc1 subunit (Findeisen et al., 1999; Hua and Newport, 1998; Romanowski et al., 1996, 2000; Rowles et al., 1999; Tugal et al., 1998; Li et al., 2004). Interestingly, phosphorylation modification does not appear to directly affect ORC stability or chromatin association in yeast. Thus, phosphorylation modification may represent a significant difference between how yeast and metazoans regulate ORC function and provide a more complex level of re-replication control in higher eukaryotes (Diffley et al., 1994; Lygerou and Nurse, 1999; Remus et al., 2005).
How phosphorylation modification may alter ORC conformation to switch the complex from an active DNA-binding state to an inactive state is unknown. Biochemical analysis of the effect of CDK phosphorylation on the activities of Drosophila melanogaster ORC (DmORC) in vitro found that hyperphosphorylation of the DmOrc1 and DmOrc2 subunits inhibits ATPase activity of Orc1 but does not interfere with ATP binding to Orc1 within ORC (Remus et al., 2005). Additionally, it was discovered that hyperphosphorylation of DmORC eliminates its ability to form ATP-dependent ORC:DNA complexes. Phosphorylation did not affect the integrity of the DmORC complex and the inhibitory effects of CDK phosphorylation were reversed upon treatment with phosphatase, supporting a role for phosphorylation in regulating ORC’s affinity for DNA and there-fore chromosome association during the cell cycle. Further structural and biochemical studies are required to understand how ORC binds DNA for switching mechanisms that allow for either forward progression towards building a pre-RC or negative regulation of its activity to prevent re-initiation.
Understanding how phosphorylation modifications translate to conformational changes that govern ORC:DNA interaction and ATP-hydrolysis may further our understanding of the mechanism of origin activation and re-replication control in metazoa. Both electron microscopy (EM) and atomic force microscopy (AFM) single particle imaging methods were employed in an effort to learn more about the specific mechanistic details of origin processing during replication initiation. Our previous EM studies of DmORC were done with a sample containing a mixture of phosphorylation states. In an effort to increase sample homogeneity and learn more about potential conformational affects of phosphorylation, EM and image processing were used to create and compare 3D models of DmORC in various phosphorylation states. AFM was used to visualize ORC bound to DNA to investigate potential wrapping of DNA by DmORC. Based on our data and what is currently known about ORC subunit interactions, we propose a model for subunit arrangement within our DmORC structure arrangement. We also put forward a potential path of the DNA around the complex that supports a wrapping mechanism analogous to what has been proposed for the nucleotide-bound prokaryotic initiator, DnaA in complex with DNA (Fuller et al., 1984; Kornberg and Baker, 1992).
2. Materials and methods
2.1. Hyperphosphorylation of ORC by cyclin E/CDK2
The starting material for all of the following protocols was DmORC purified as previously described (Chesnokov et al., 2001). Sf-9 insect cells were co-infected with baculoviruses expressing GST-tagged human cyclin E and untagged CDK2. Cyclin E/CDK2 complexes were purified from whole cell extracts using glutathione affinity chromatography followed by conventional chromatography. Hyper-phosphorylated DmORC was obtained by combining 50 µl of Mono S purified ORC with 25 µl Cyclin E/Cdk 2 in 225 µl of 20 mM HEPES, 4 mM MgCl2, 10% glycerol, 50 mM KCl, 0.4 mM PMSF, 2 mM β-ME, 0.1 mM EDTA, 1 mM ATP and incubated at RT for 1 h. The control experiment was run under the exact same conditions minus the Cyclin E/Cdk2. Parallel experiments were run with P32-radiolabeled ATP to ensure phosphorylation saturation of Orc1 and Orc2. The hyper-phosphorylated ORC was separated from the Cyclin E/Cdk2 by glycerol gradient purification. The total 300 µl solution was loaded on a 4 ml 15–35% glycerol gradient in 25 mM Hepes pH 7.6, 1 mM EDTA, 1 mM EGTA, 0.05% NP-40, 10% glycerol, 1 mM DTT, 0.2 mM PMSF, 150 mM KCl. ATPγS and MgCl2 were added to the glycerol gradient to a final concentration of 5 µM. Gradients were placed in a SW-60 rotor and centrifuged for 8 h at 42K rpm at 4 °C in a Beckman ultracentrifuge. Gradients were collect in 200 µl fractions from the top, run on SDS–PAGE and visualized by silver staining the gel. The fresh glycerol gradient purified ORC was immediately used to prepared EM sample grids to avoid any structural damage caused by freezing.
2.2. Dephosphorylation of ORC by λ-phosphatase
Dephosphorylated ORC was obtained by incubating 15 µg of Mono S purified ORC with 2000 units of λ-phosphatase from NEB in 60 µl phosphate buffer (50 mM Tris–HCl pH 7.5, 0.1 mM Na2ED-TA, 5 mM DTT, 0.01% Brij 35, 2 mM MnCl2) for 25 min at 30 °C. As a negative control, mock treated ORC was prepared in parallel under the same conditions substituting λ-phosphatase storage buffer for the protein. The dephosphorylated ORC was separated from the phosphatase by glycerol gradient purification. The total 60 µl solution was loaded on a 4 ml 15–35% glycerol gradient in 25 mM Hepes pH 7.6, 1 mM EDTA, 1 mM EGTA, 0.05% NP-40, 10% glycerol, 1 mM DTT, 0.2 mM PMSF, 150 mM KCl. For the ATP bound dephosphorylated volume, ATPγS and MgCl2 were added to the glycerol gradient to a final concentration of 5 µM. Gradients were placed in a SW-60 rotor and centrifuged for 8 h at 42 K rpm at 4 °C in a Beckman ultracentrifuge. Gradients were collect in 200 µl fractions from the top, run on SDS–PAGE and visualized by silver staining the gel. The peak fraction was immediately used to prepare EM sample grids.
2.3. Electron microscopy sample grid preparation
EM sample grids were prepared using a modified version of the sandwich stain method described by Ohi and coworkers (Ohi et al., 2004). Uranyl formate stain was prepared fresh immediately before making sample grids at a concentration of 0.75%. Holey carbon grids were glow discharged in air for 15 s. Immediately before grid preparation, three 200 µl drops of stain were placed on Parafilm. Additionally, a ceramic well was filled with ~500 µl stain and a small thin square of carbon was floated on the surface. Approximately 150 ng of glycerol gradient purified DmORC was pipetted onto glow discharged girds. Protein was allowed to absorb on the grid for 30 s before staining. The grid was then washed by sequentially touching it to the three 200 µl drops of stain. Immediately after washing, the grid was dipped into the stain well (protein side facing up) and used to lift up the carbon square floating on the surface. Excess stain was then blotted off.
2.4. Electron microscopy data collection and image processing
Particles were imaged at 49 K magnification using a FEI Tecnai 120 KeV microscope. Data was collected in low dose mode on Kodak SO-163 film at a defocus of approximately −1.5 µm. Micrographs were developed and first visually inspected for contrast, astigmatism and drift. High quality micrographs were then digitized at a resolution of 2.59 Å/pixel on a Nikon Super Coolscan 800. The power spectra of the digitized micrographs were checked for astigmatism and drift using the EMAN program CTFIT (Ludtke et al., 1999). Astigmatism and drift-free micrographs were used to pick a particle data set using the EMAN program BOXER, with a particle box size of 144 × 144 pixels. Reference-free two-dimensional alignment of the particles was done in IMAGIC using iterative cycles of Multivariant-Statistical Analysis (MSA) and Multi-Reference Alignment (MRA) (van Heel et al., 1996). Three-dimensional volumes were generated using SPIDER from the top third of the particle data sets by backprojection and iterative projection matching refinement (Frank et al., 1996) against a reference model, the original apo-ORC model (Clarey et al., 2006) filtered to 60 Å. Resolution of the final reconstruction was estimated by Fourier Shell correlation (FSC) using a cut-off of 0.5 (Harauz and van Heel, 1986; Saxton and Baumeister, 1982). The 3D volumes were thresholded to the calculated value for a molecular weight of ~390 kDa. Figures were generated using PyMol (http://pymol.sourceforge.net/).
2.5. Purification of 688 bp DNA fragments
A 688 bp region containing the ACE-3 Chorion gene sequence was PCR amplified from the pT2 plasmid. A reaction cocktail of 4 µl pT2 plasmid (5 mg/ml), 20 µl ACE-3 Forward primer (10pmol/µl), 20 µl ACE-3 Reverse primer (10pmol/µl), 50 µl dNTPs (2.5 mM), 5 µl Taq polymerase, 50 µl 10× Taq polymerase reaction buffer and 336ul PCR-grade water was split into ten 50 µl reactions. PCR product was loaded on 1% agarose gel (10 µg/lane). Bands were visualized by ethidium bromide staining. Bands corresponding to 688 bp fragment were excised and purified using the Qiagen Gel Extraction kit. The fragments were pooled and further purified by phenol/chloroform ethanol extraction and concentrated by ethanol precipitation.
2.6. Aminopropyl silatrane (APS) synthesis
APS was synthesized as described in Shlyakhtenko et al. (2003). Identity of compound was confirmed by mass spectroscopy. Mica was derivatized immediately before use. Freshly exposed mica was immersed in 50 ml of a 1:300 APS- water solution (167 µl 50 mM APS stock solution plus 50 ml deionized water) and incubated for 30 min at RT. Mica was thoroughly rinsed with deionized water and dried with nitrogen gas.
2.7. Atomic force microscopy (AFM)
ORC+DNA samples for AFM were prepared by combining 20 µl glycerol gradient purified ORC (30 ng/µl) with 1 µl of DNA (0.6 µg/µl), 40 µl 5 mM ATP, and 340 µl AFM buffer (10 mM NaCl2, 2 mM MgCl2, 4 mM HEPES, 150 mM KCl). For DNA alone, 20 µl of ORC glycerol gradient buffer was added (25 mM Hepes pH 7.6, 1 mM EDTA, 1 mM EGTA, 0.05% NP-40, 10% glycerol, 1 mM DTT, 0.2 mM PMSF, 150 mM KCl). A 20 µl sample volume was applied to a freshly exposed APS-mica disc for 1 min. Disc was rinsed with a constant stream of deionized water for 30 s and then dried with nitrogen gas. AFM images were collected at nanometer and micrometer resolution using a Digital Instruments NanoScope III Atomic Force Microscope operating at 1 V in non-contact tapping mode in air.
The length of the DNA fragments were measured by hand tracing the fragments using the ScanPro 5 software. Each fragment was traced three times and the average of the measurements was recorded.
3. Results
3.1. The effect of phosphorylation state on DmORC structure
Electron microscopy and image analysis of negatively stained samples were used to investigate potential conformational differences in the regulation of DmORC structure by CDK phosphorylation. We had previously described the apo- and ATPγS-ORC EM structures (Clarey et al., 2006). Since DmOrc1 has seven phosphorylation sites and DmOrc2 has four, our samples are likely to have contained both biochemical and conformational heterogeneity (Remus et al., 2005). Our studies aimed to distinguish between phosphorylation states and thus potentially improve sample homogeneity. We generated both λ-phosphatase treated dephosphorylated DmORC and Cdk2/Cyclin E hyper-phosphorylated DmORC. A clear upward shift in the hyper-phosphorylated Orc1 and Orc2 subunits by SDS–PAGE and silver stain analysis is indicative of the phosphorylation state (Remus et al., 2005) (Fig. 1A). Saturating phosphorylation conditions for DmORC were determined by adding increasing amounts of Cdk2/Cyclin E with radio-labeled nucleotide and visualized by Audioradiogram (Fig. 1B). These samples were negatively stained for EM analysis using single particle methods, in order to generate 3D volumes of dephosphorylated DmORC+/−ATPγS and hyper-phosphorylated DmOR-C + ATPγS. Reference-free class averages (Fig. 1C) indicated that the overall architecture of the complexes was similar enough to that of our previous DmORC reconstructions to warrant use of the apo-ORC structure as a reference (after filtration see Section 2) in a projection matching approach. The resolution of the reconstructions, as judged from the 0.5 Fourier shell correlation (FSC) cut off, ranged from 24 to 27 Å (Supplemental data S1 and S2). For all the samples we observed preferential orientations around the long axis of the molecule, without any marked differences in the angular distribution of the different samples (Fig. 1C; Supplemental data S1 and S2).
Fig. 1. Hyper-phosphorylated versus dephosphorylated Drosophila melanogaster ORC.
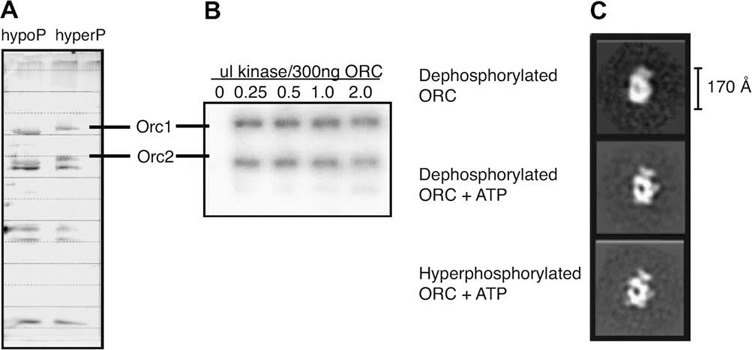
(A) Silver stain of SDS–PAGE exhibiting the molecular weight shift in the Orc1 and Orc2 subunits in dephosphorylated versus hyper-phosphorylated ORC. Orc1 has seven phosphorylation sites while Orc2 has four phosphorylation sites. (B) Audioradiogram of Cyclin E/CDK2 hyperphosphorylation of recombinant DmORC showing 32P signal versus increasing amounts of Cyclin E/CDK2. (C) Representative two-dimensional class averages from single particle electron microscopy reference-free image classification of dephosphorylated apo-ORC, ATPγS bound dephosphorylated ORC and ATPγS bound hyper-phosphorylated ORC. All three data sets show preferential orientations around the elongated axis of ORC.
A major conclusion of our analysis is that no large conformational changes can be observed due to phosphorylation of the nucleotide-bound form of the complex. The features of the dephosphorylated ORC volume without added nucleotide (Fig. 2A) closely resembled those of our previous apo-ORC structure (Clarey et al., 2006). A prominent toroidal core is defined by five main densities previous ascribed to the AAA+ domains (A to E in Fig. 2A, top row), encircling a central channel. Some of these densities extend back to define a collar of density on the reverse side of the view down the central channel (Fig. 2A, third row). The E density at the top of the core connects to a “shoulder”, which in turn connects to the “glove” defining the top of the structure. The only difference with our previous apo-ORC structure is that a thin density now partially closes off the channel in the dephosphorylated ORC, as density A in the toroidal core comes closer to the shoulder. As for the previous apo structure, the glove density at the top remains separate from the main body of the complex.
Fig. 2. Three-dimensional comparison of ORC indicates ATP gives rise to analogous conformational changes in the core of ORC independent of phosphorylation state. All volumes were filtered to 30 Å and are displayed in four identical views rotated 90 ° counter-clockwise around their elongated axis.
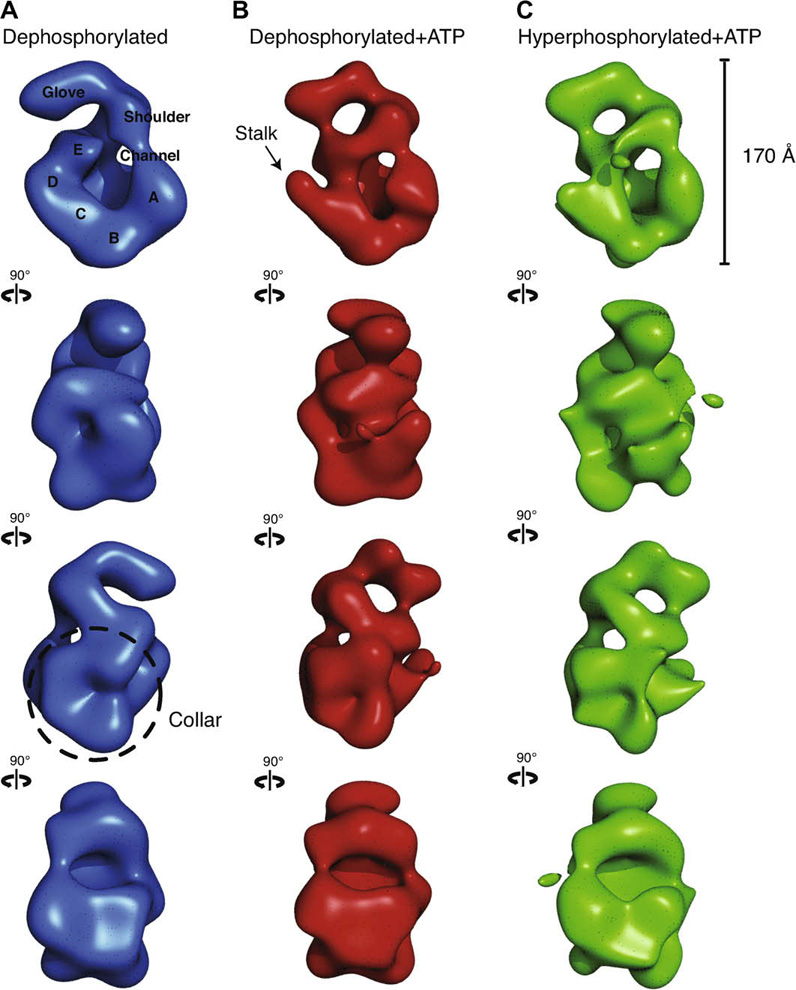
(A) Dephosphorylated ORC is rendered as a blue isosurface. The five lobes of density composing the core are labeled A–E. A thin density closes the central channel and fuses the shoulder to lobe A. The glove density at the top remains separate from the main body of the complex. (B) Dephosphorylated ORC bound to ATPγS is rendered as a red isosurface. An arrow points to the region where upon nucleotide-binding a discernable stalk density separates lobe D from E. The shoulder is again fused with lobe A and the glove is now connected to lobe E. The density labeled lobe E was previously identified as Orc5 by immuno-labeling. (C) Hyper-phosphorylated ORC bound to ATPγS is rendered as a green isosurface. The separation of lobes D and E as well as the fusing of the glove with lobe E is also observed, indicating that nucleotide binding promotes a very similar conformational state in the core of ORC regardless of the phosphorylation state of the complex.
Previous studies with the S. cerevisiae ORC have shown that ATPase activity of the complex requires canonical AAA+ interactions between the arginine finger domain of Orc4 and the Walker ATP binding motifs of Orc1 (Bowers et al., 2004). Based on the previous ATP-bound ORC volume, we speculated that the coordination of ATP by the AAA+ domains of Orc1 and Orc4 propagates a tightening of the toroidal core, likely giving rise to the proper DNA binding conformation. It is interesting to note that in previous studies by Liu and coworkers it was not possible to obtain a stable 3D EM reconstruction of S. cerevisiae ORC without the stabilizing addition of nucleotide (Speck et al., 2005), an effect that has also been observed biochemically for human ORC (Ranjan and Gossen, 2006), suggesting that in some organisms ATP-induced changes include overall stabilization/ordering of the complex.
The volume of the dephosphorylated ORC bound to ATPγS (Fig. 2B) is compacted around the central channel with respect to the complex without added nucleotide, as also observed in our previous nucleotide-bound reconstruction. The boxing glove density has closed down on the E density that was previously identified as corresponding to Orc5 (Clarey et al., 2006). The channel is closed off by the contact of the shoulder with both the E and A densities at opposite ends of the toroidal core, resulting in a tightening of the channel. Noticeably, an apparent stalk is seen protruding off from the central core at the level of the D toroidal density, which was first observed in the original ATPγS bound ORC model, and that results from the separation of the D and E densities.
The hyper-phosphorylated ORC volume with added nucleotide is strikingly similar to the ATPγS bound dephosphorylated ORC volume (Fig. 2C). The channel is again closed off by the interaction of the shoulder with the E and A densities around the toroidal core. Also shared with the ATP-bound dephosphorylated structure is the clear stalk protruding off from the mid-core area due to the separation of the D and E densities. Thus, the observed changes in the core and specifically the presence of the stalk density, make the nucleotide added structures more similar to one another than to the dephosphorylated no-added nucleotide condition.
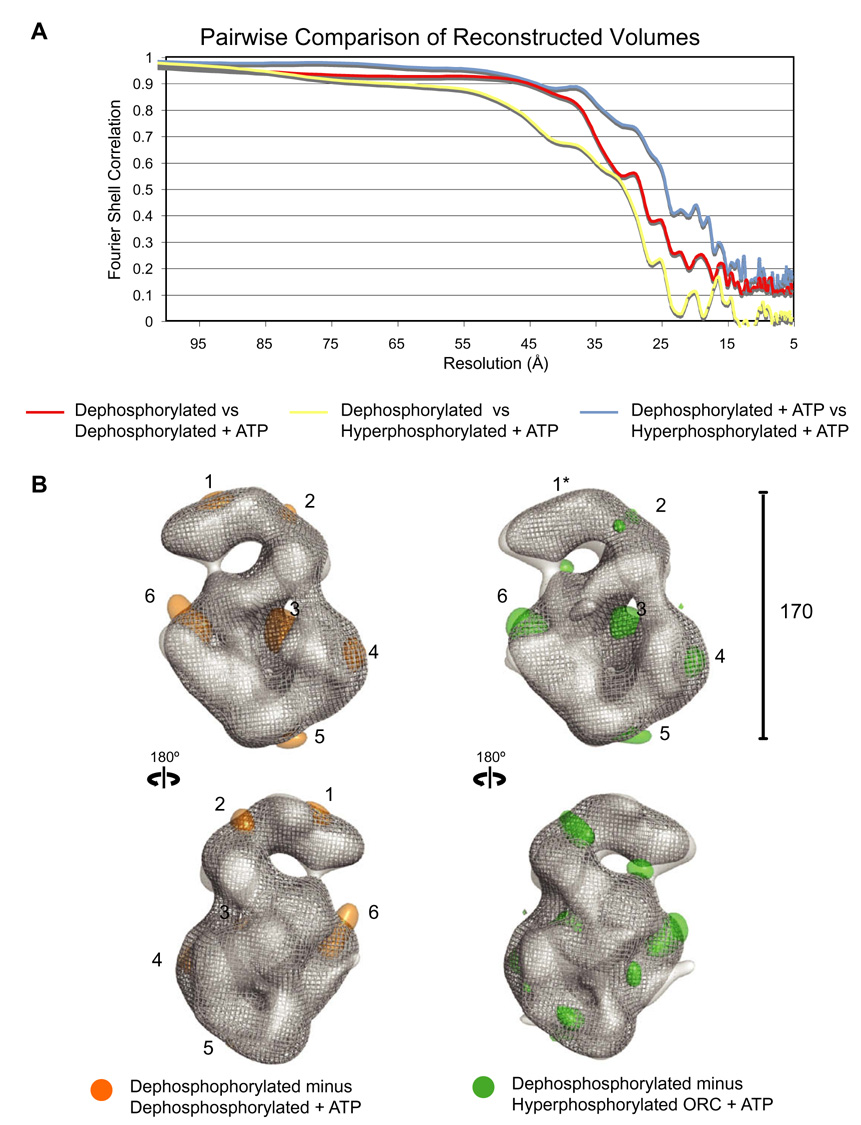
Cross correlation analysis was used in an attempt to evaluate more quantitatively and objectively the relative differences between the reconstructed volumes. The analysis strongly supported the visual conclusion that the structure of the dephosphorylated ORC without added nucleotide has a distinctive conformation with respect to the other two nucleotide-bound samples, which closely resemble one another (Fig. 3A). Correlation between the apodephosphorylated volume and the hyperphosphorylated volume with added nucleotide begins to fall off almost immediately (Fig. 3A, yellow line). A similar trend occurs when comparing the apo-dephosphorylated volume with its nucleotide-added counter-part, further supporting the notion that ATP-binding promotes rearrangements in the core (Fig. 3, red line). The nucleotide-added versions of hyperphosphorylated and dephosphorylated ORC had a stronger correlation that only falls off close the resolution limit of the individual reconstructions (Fig. 3, blue line). Based on the correlation data, ATP-binding leads to increased similarity between the structures independent of phosphorylation state. It should be reiterated here that the analysis again emphasizes that differences between the volumes are relatively small yet clearly detectable between the apo dephosphorylated state and the two ATP-bound phosphorylation states, which themselves look very similar.
Fig. 3. ATP-binding leads to similar ORC conformations, independent of phosphorylation state.
(A) Pairwise fourier shell correlation comparison of dephosphorylated ORC versus dephosphorylated ORC bound to ATPγS (red line), dephosphorylated ORC versus hyper-phosphorylated ORC bound to ATPγS (yellow line), and dephosphorylated ORC bound to ATPγS versus hyper-phosphorylated ORC bound to ATPγS (blue line). Apo-dephosphorylated ORC has a distinctly different conformation as compared to the ATP-bound volumes, as is indicated by the highest correlation between the two ATPγS volumes. The correlation between dephosphorylated ORC and nucleotide-bound hyperphopshorylated ORC is the weakest, suggesting additional small differences due to the phosphorylation state, in addition to those larger differences caused by nucleotide binding. (B) Difference maps showing the main regions of difference between the complexes. Only positive differences are shown for clarity. Left, the volume of the ATP-bound dephosphorylated ORC (grey mesh isosurface) was subtracted from that of the dephosphorylated ORC without added nucleotide (transparent grey isosurface) with the mass differences highlighted in orange. The main areas of observed differences are, from the top and moving clockwise: (1) on the top of the glove density, (2) in the glove-shoulder connection, (3) inside the central channel, (4) the movement of A towards the shoulder, (5) minor changes at the bottom of the complex in the connection of the toroidal AAA+ core and the collar, and (6) the separation of D and E that gives rise to the stalk. Right, the ATP-bound hyper-phosphorylated ORC volume (grey mesh isosurface) was subtracted from the dephosphorylated ORC volume (transparent grey isosurface) with the mass differences highlighted in green. The similarity in the position of the mass differences corresponding to positions 2, 3, 5 and 6 exemplifies the analogous effect of nucleotide addition independent of hyperphosphorylation in ORC. While positions 1* and 4* do not have large mass differences, they have conformations distinct from the apo-dephosphorylated volume that are more closely related to the nucleotide-bound dephosphorylated volume.
In a further effort to describe the reorganization of the complex upon either nucleotide binding or hyperphosphorylation, we calculated difference maps to more explicitly position the main regions of difference between the dephosphorylated ORC and the two modified complexes (Fig. 3B, only positive differences shown for clarity). The volume of the ATP-bound dephosphorylated ORC was subtracted from that of the dephosphorylated ORC without added nucleotide (Fig. 3B, left). Main areas of observed differences are, from the top and moving clockwise: (1) on the top of the glove density (due to a reshaping from a straighter to a more curved shape), (2) in the glove-shoulder connection, (3) inside the central channel, (4) the movement of A towards the shoulder, (5) minor changes at the bottom of the complex in the connection of the toroidal AAA+ core and the collar, and (6) the separation of D and E that gives rise to the stalk in the ATP-added samples. Areas corresponding to 2,3,5 and 6 were found when the ATPγS-bound hyper-phosphorylated ORC volume was subtracted from the dephosphorylated ORC volume (Fig. 3B, right). Although the shape of densities are distinct and of slightly different significance in the two calculated difference maps, their similarity in number and position underlies the analogous effect of nucleotide addition irrelevant of hyperphosphorylation in ORC.
In conclusion, our results show that structural changes due to the phosphorylation state of drosophila ORC must be small, even smaller that the relatively subtle changes we see with nucleotide state, and thus indicate that higher resolution data will be required to define the molecular basis of how phosphorylation inhibits ATP-hydrolysis by Orc1 and prevents the formation of ORC:DNA complexes.
3.2. ORC wraps ~130 bp of linear DNA
DmORC binds DNA without sequence specificity and this makes difficult studies focused on how many subunits interact directly with the template, or the size of the ORC footprint on a DNA molecule. Nevertheless, such detailed information, coupled with knowledge of where DNA binds with regard to the phosphorylated residues in Orc1 and Orc2 will be important in understanding how hyperphosphorylation cripples DNA:ORC interaction. To continue investigation of ORC’s interaction with DNA, a linear 688 bp DNA template containing the known DmORC-origin ACE-3 sequence from the Chorion gene locus (Remus et al., 2004, 2005) was used to assess the extent of DNA wrapping by DmORC using AFM. Studies have shown that functionalizing the mica surface with 1-(3-aminopropyl) silatrane (APS) tolerates high ionic strengths and allows reliable imaging of DNA in 10–200mM salt (Shlyakhtenko et al., 2003). For our studies, the mica surface was treated with APS to allow for imaging of ORC:DNA at the optimal salt concentration (150 mM KCl) for binding which typically disrupts DNA attachment to the unmodified mica surface. Control images of the linear 688 bp DNA template were taken in the absence of ORC. For the ORC binding experiment, DmORC:DNA complexes were formed in the presence of ATP before being deposited on the APS-mica (Fig. 4A). The DmORC used for the DNA binding studies was not treated with phosphatase and is thus heterogeneous as in previous studies, although the hyper-phosphorylated form was not detected by SDS–PAGE analysis.
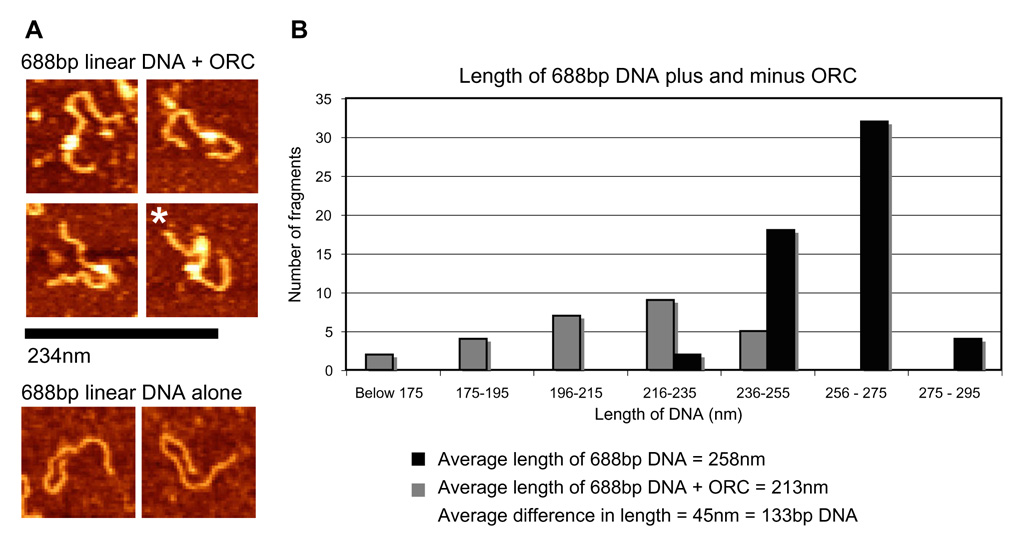
Fig. 4. ORC wraps ~130 bp of linear DNA. Atomic force microscopy images of linear 688 bp DNA containing the ACE-3 Chorion gene sequence plus and minus DmORC on APS-mica.
(A) representative 688 bp linear DNA fragments bound to a DmORC (top four panels) and alone (bottom two panels). Asterisk (*) indicates a DNA fragment with two ORCs bound. Scale bar correlating to the theoretical length of 688 bp of DNA or 234 nm. (B) Graph of the average length of a linear 688 bp DNA fragment with (black columns) and without (grey columns) ATPγS DmORC bound. The measured lengths of linear 688 bp DNA were binned into 10 nm groups. The average measured length of an unbound 688 bp DNA fragment was ~ 258 nm while the average length of the 688 bp DNA fragment with DmORC bound was 213 nm. The average difference in length of DNA with DmORC bound is 45 nm or ~133 bp.
The contour length of both the DNA fragments alone, and the ORC-bound DNA fragments were measured (Fig. 4B). DNA fragments with multiple ORCs bound were observed but only fragments with one ORC molecule were measured (Fig. 4A, multiple ORCs bound marked by *). The average measured length of the 688 bp DNA template alone was 258 nm as compared to 213 nm with a single DmORC bound. This average difference of 45 nm between ORC bound DNA fragments and unbound fragments corresponds to a shortening equivalent to ~133 bp. As a point of reference, the DmORC model measures approximately 17 nm by 8.5 nm, or in the longest dimension a length corresponding to ~50 bp of DNA. These points together suggest that ORC is not merely “sitting” on the DNA but is gripping it in such a manner that involves wrapping an extensive amount of the DNA around the complex. A model that would accommodate such a wrap and physical shortening of the DNA and be consistent with the dimensions of ORC is discussed in the following section.
Further supporting the notion that ORC wraps DNA is the finding that a linking number change was measured upon binding to circular molecules and that the superhelicity of the DNA plays an important role in ORC’s affinity for DNA (Clarey et al., 2006; Remus et al., 2004). It is interesting to point out that our value closely agrees with the shortening of ~140 bp of linear DNA by SpORC as observed by AFM (Gaczynska et al., 2004). SpORC is known to contain an AT-hook on the Orc4 subunit that binds to DNA, and though this is not conserved in the DmORC, many of the structural interactions with DNA seem similar, including a strong preference for negative supercoiled DNA. The distribution of the DNA length measurements we observe here is spread out over a similar range of values, in the case of SpORC ~75 and ~80 nm for DmORC.
It should be noted that the extend of DNA interaction inferred from our results for DmORC is substantially longer than what has been observed for the ScORC footprint, which has been shown to protect a stretch of DNA equivalent to ORC’s length of ~48–49 bp of DNA in vivo and in vitro (Donovan et al., 1997; Santocanale and Diffley, 1996; Speck et al., 2005). Thus many physico–chemical binding properties we describe here and elsewhere (Remus et al., 2004, 2005) are similar to those found for fission yeast ORC, and perhaps the differences with the DNA binding properties of the budding yeast point to real mechanistic divergences. Alternatively, future studies may show that supercoiling and additional replication proteins (Cdc6 and Hu like factors) may help the S. cerevisiae ORC wrap and tightly interact with the template.
4. Discussion
At the current resolution achieved by our EM studies, hyper-phosphorylation does not lead to a large conformational change in ORC. However, small differences do exist, and these alter the chemical properties of interacting surfaces to result in the large change in affinity observed by biochemical assays. The data do support previous observations that nucleotide binding promotes conformational flexion in the core of ORC. Furthermore, the present data demonstrates that this effect occurs independently of phosphorylation state. This result suggests that rearrangements in this core region are important for origin DNA engagement and/or processing. Orc1 has seven Cdk2/Cyclin E phosphorylation sites located in the linker region between the N-terminus bromo adjacent homology (BAH) domain and one in the C-terminus AAA+ domain (Remus et al., 2005). The four Cdk2/Cyclin E phosphorylation sites on Orc2 are N-terminal while the AAA+ domain is located at the C-terminus. Thus, in contrast to nucleotide binding, phosphorylation modification occurs in residues outside the AAA+ domains, raising the question of how the addition of phosphates can communicate inhibitory effects to Orc1 and/or Orc4 in the central toroid, if in fact the AAA+ domains form the backbone of the ORC complex. Structurally, the inhibition of hydrolysis may be realized by these flexible domains subtly holding the AAA+ domain of Orc1 in a position such that it can bind ATP but cannot coordinate ATP-hydrolysis with the arginine finger domain of the neighboring Orc4 AAA+ domain. On the other hand, the additional negative charges contributed by the 11 phosphate groups on Orc1 and Orc2 may reduce ORC’s affinity for DNA and prevent the formation of ORC:DNA complexes.
Based on these core rearrangements observed in ORC, it is reasonable to assume that DNA processing events are occurring in this central region of the complex. If the AAA+ domains are indeed situated in this area and are making direct contact with the DNA as has recently been observed for archaeal initiators (Dueber et al., 2007; Gaudier et al., 2007), the tightening and increase in pitch of the ATP-bound core may be part of a series of events that act to distort the helix during replication initiation. In a mechanism parallel to that of DnaA, a certain degree of flexibility in the dephosphorylated apo- state may allow for initial ORC contact with DNA, upon which ATP-binding would lock ORC in the observed nucleotide-bound conformation, which in turn would promote rearrangements along the core AAA+ domains, remodeling the bound DNA. Hyperphosphorylation may then hold ORC, by means of its inhibition of hydrolysis, in this later conformation, which could be inactive for the initial DNA binding step, and thus aid in preventing rereplication.
ATP has been found to be also important for the integrity of the ORC complex, and has been proposed to serve as a structural cofactor (Ranjan and Gossen, 2006; Siddiqui and Stillman, 2007). It is likely that these “structural” nucleotides are incorporated into ORC during its assembly, and that little or no turn over and/or exchange occurs after assembly. Ranjan and Gossen have shown a direct interaction between human Orc 2 and Orc3, and the formation of a trimer with Orc5, this subunit being needed for further stable interaction with Orc4. The interaction of Orc1 further requires both Orc4 and Orc5. Interestingly, incorporation of Orc4 and Orc1 into ORC is dependent on the presence of ATP, and in particular on the binding of ATP by Orc4 (Ranjan and Gossen, 2006). Studies in budding yeast by Siddiqui and Stillman also found that ATP binding to Orc4 and 5, but not Orc1, were required for formation of a full ORC complex by budding yeast proteins (Siddiqui and Stillman, 2007).
Previous localization of the Orc5 subunit in DmORC using specific antibodies directed to this subunit (Clarey et al., 2006) place it in the region labeled E in Fig. 2. Based on the available biochemical data and our own structural work, we propose a subunit arrangement for DmORC that shows the AAA+ domain of Orc1 at the toroidal base, directly contacting Orc4 (Fig. 5A), corresponding, respectively, to densities A and B. Previous studies have shown that the AAA+ domains of Orc 1 and Orc4 must interact for ORC hydrolysis (Bowers et al., 2004) motivating a proximal placement of these domains in a putative structure. The proposed architecture would also provide an open Orc1 AAA+ interaction surface for the AAA+ domain of Cdc6 to dock with, concurring with the closure of the channel observed upon Cdc6 binding to ScORC (Speck et al., 2005). Orc4 would contact Orc2 or Orc3, as motivated by the biochemistry indicating an Orc2/3/4 subassembly. Hence we ascribe the Orc2 and Or3 AAA+ domains to densities C and D, with the AAA+ domain of Orc5 being at the end of the AAA+ toroid (density E). The collar, as previously proposed (Clarey et al., 2006), would be composed of the wing helix domains (WHD) of Orc1, 4 and 5 with Orc1 and Orc5 interacting through the WHD and not their AAA+ domains. These non-AAA+ contacts would explain the required presence of Orc5 for interaction of Orc4 with the Orc2, 3, 5 subcomplex, as well as the presence of both Orc4 and Orc5 for incorporation of Orc1. Although there is not direct evidence, the shoulder and glove densities could belong to additional non-AAA+ domains in ORC1.
Fig. 5. Speculative model of ORC subunit architecture and the path of 133 bp of DNA.
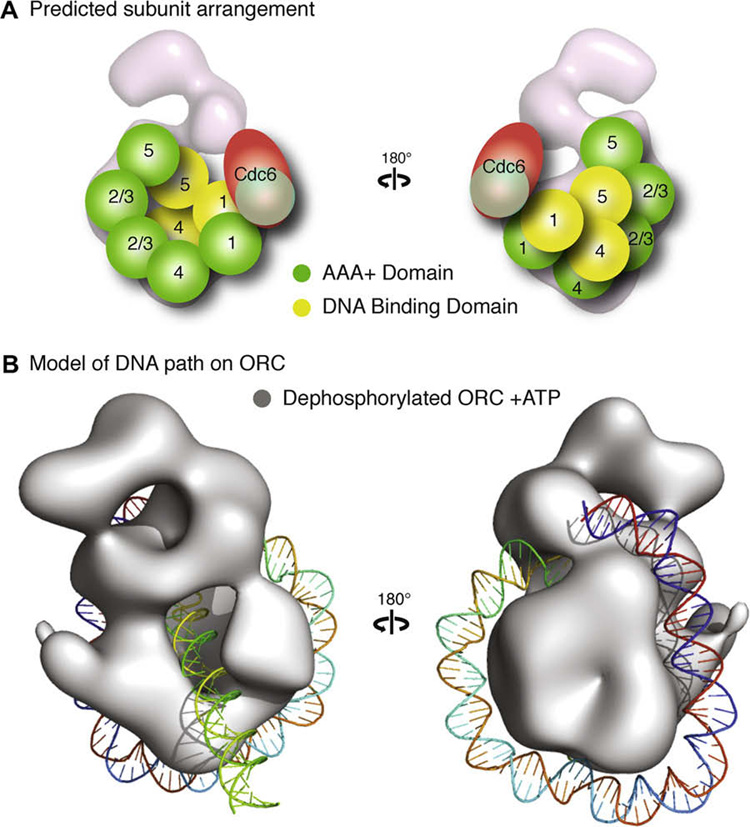
(A) A potential arrangement of the subunits would have the five AAA+ domains (illustrated as green spheres) of Orc1–5 composing the core of the complex, with Orc1 situated at the base directly contacting Orc4, Orc2 and Orc3 in the central region where the stalk density shifts upon nucleotide binding, followed by Orc5. The position of Orc2 and Orc3 are interchangeable and are labeled “2/3” to indicated this in the figure. The winged helix domains (illustrated as yellow spheres) of Orc1, 4 and 5 would form the collar. This position would allow for the AAA+ domain of Cdc6 to dock at the AAA+ interface of Orc1. The mass of the other domains of Orc1–5 and Orc6 would compose the remainder of the density. (B) A model of dephosphorylated ORC bound to ATPγS (rendered as a grey isosurface) with 133 bp of a linear DNA wrapped around the complex starting at the wing-helix domains in the collar and following a right-handed path along the AAA+ backbone to loop back through the channel.
We have not place the Orc6 subunit in our model, as its size is very small and could be incorporated in a number of locations within the structure, and no biochemical information as to its partners is available. It is of note that a stable ORC1–5 complex has been observed both in vivo (Gossen et al., 1995) and with recombinant complexes (Chesnokov et al., 2001). Furthermore, DmOrc6 is known to be a DNA binding protein and is apparently important for DNA interaction in the full DmORC (Chesnokov et al., 2001).
A speculative path of the DNA on the ORC complex that would accommodate ~130 bp of DNA would be one in which the nucleic acid is wrapped along the outer surface of the toroidal core. In this model the DNA enters a crevice between the WHDs predicted to be located in the collar adjacent to the AAA+ backbone, allowing for contacts to be made with both the DNA-binding domains and the AAA+ domains (Fig. 5B). The DNA would then continue along the base of the complex before making a sharp, right-handed turn where upon it is looped back through the channel, enclosed and potentially bent by Cdc6 closure of the central channel.
Supplementary Material
Acknowledgments
The authors acknowledge Carlos Bustamante, Dirk Remus, Patricia Grob and Hong Wei Wang for technical assistance and helpful discussion. This work was supported by US National Institutes of Health grant R39 CA 30490 to M.B., and by a Biomedicine chair from the BBVA Foundation to E.N. E.N. is a Howard Hughes Medical Institute Investigator.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jsb.2008.08.006.
References
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–274. doi: 10.1038/445272a. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr. Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Findeisen M, El-Denary M, Kapitza T, Graf R, Strausfeld U. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem. 1999;264:415–426. doi: 10.1046/j.1432-1327.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Gaczynska M, Osmulski PA, Jiang Y, Lee JK, Bermudez V, Hurwitz J. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proc. Natl. Acad. Sci. USA. 2004;101:17952–17957. doi: 10.1073/pnas.0408369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell. Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Kelly TJ, Brown GW. Regulation of chromosome replication. Annu. Rev. Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kipreos ET. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2007;2:18. doi: 10.1186/1747-1028-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. New York, NY: W.H. Freeman and Co.; 1992. [Google Scholar]
- Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol. Cell. Biol. 2004;24:5875–5886. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Nurse P. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 1999;112(Pt 21):3703–3712. doi: 10.1242/jcs.112.21.3703. [DOI] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc. Natl. Acad. Sci. USA. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Blanchette M, Rio DC, Botchan MR. CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex. J. Biol. Chem. 2005;280:39740–39751. doi: 10.1074/jbc.M508515200. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Marr J, Madine MA, Rowles A, Blow JJ, Gautier J, Laskey RA. Interaction of Xenopus Cdc2 x cyclin A1 with the origin recognition complex. J. Biol. Chem. 2000;275:4239–4243. doi: 10.1074/jbc.275.6.4239. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell. Sci. 1999;112(Pt 12):2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 1982;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J. Biol. Chem. 2007;282:32380–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.