Abstract
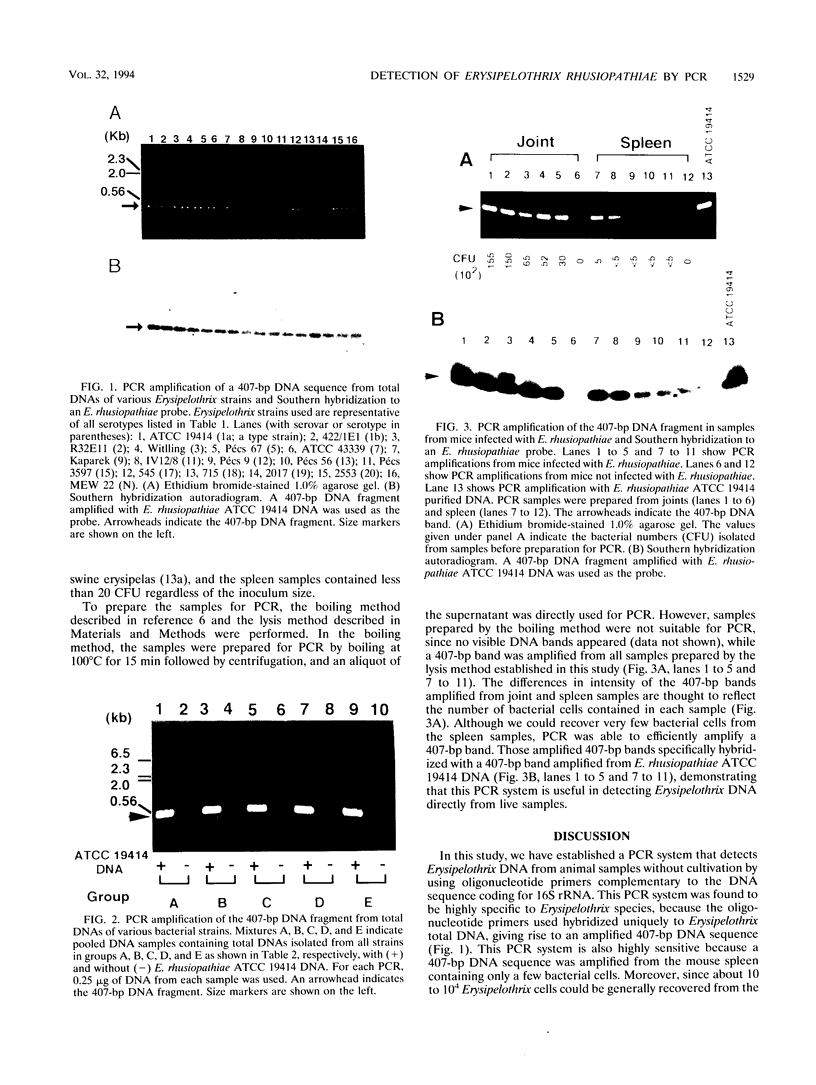
Erysipelothrix rhusiopathiae is a gram-positive rod capable of causing erysipelas in swine. To establish a method for specifically detecting E. rhusiopathiae for practical applications, such as for the inspection of slaughterhouses, the feasibility of using primers derived from the DNA sequence coding for 16S rRNA in a PCR-specific detection system was investigated. Oligonucleotide primers were designed to amplify a 407-bp DNA fragment by PCR. The amplification was specific to the Erysipelothrix DNA but not to that of other bacterial genera tested. This PCR-based method efficiently and specifically detected the Erysipelothrix DNA sequence in joint and spleen samples from mice within 6 h, and application of the 407-bp DNA segment from samples containing very low numbers of bacteria (< 20 bacteria per spleen from mice) was possible. Although this PCR amplification is specific for the Erysipelothrix genus, which contains at least two species, E. rhusiopathiae and E. tonsillarum, it can be concluded that all Erysipelothrix strains detected by this PCR system in diseased pigs are E. rhusiopathiae because only E. rhusiopathiae is virulent for pigs. These results show that this PCR amplification system using the DNA sequence coding for 16S rRNA is very rapid and reliable and avoids cumbersome and lengthy cultivation steps, demonstrating that this system could be used for practical applications.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brian M. J., Frosolono M., Murray B. E., Miranda A., Lopez E. L., Gomez H. F., Cleary T. G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992 Jul;30(7):1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S. I., Iinuma-Okada Y., Maruyama T., Ezaki T., Sasakawa C., Yoshikawa M. Direct detection of Bacillus anthracis DNA in animals by polymerase chain reaction. J Clin Microbiol. 1993 Mar;31(3):547–551. doi: 10.1128/jcm.31.3.547-551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Okada Y., Maruyama T., Kaneko S., Sasakawa C. PCR-based random amplified polymorphic DNA fingerprinting of Yersinia pseudotuberculosis and its practical applications. J Clin Microbiol. 1994 Jan;32(1):65–69. doi: 10.1128/jcm.32.1.65-69.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Sasakawa C., Kamata K., Kurata T., Yoshikawa M. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell. 1986 Aug 15;46(4):551–555. doi: 10.1016/0092-8674(86)90880-9. [DOI] [PubMed] [Google Scholar]
- Makino S., Sasakawa C., Uchida I., Terakado N., Yoshikawa M. Cloning and CO2-dependent expression of the genetic region for encapsulation from Bacillus anthracis. Mol Microbiol. 1988 May;2(3):371–376. doi: 10.1111/j.1365-2958.1988.tb00041.x. [DOI] [PubMed] [Google Scholar]
- Makino S., Uchida I., Terakado N., Sasakawa C., Yoshikawa M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J Bacteriol. 1989 Feb;171(2):722–730. doi: 10.1128/jb.171.2.722-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Fujisawa T., Tamura Y., Suzuki S., Muramatsu M., Sawada T., Benno Y., Mitsuoka T. DNA relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int J Syst Bacteriol. 1992 Jul;42(3):469–473. doi: 10.1099/00207713-42-3-469. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Sawada T., Muramatsu M., Tamura Y., Fujisawa T., Benno Y., Mitsuoka T. Serotype, antimicrobial susceptibility, and pathogenicity of Erysipelothrix rhusiopathiae isolates from tonsils of apparently healthy slaughter pigs. J Clin Microbiol. 1987 Mar;25(3):536–539. doi: 10.1128/jcm.25.3.536-539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Sawada T., Takagi M., Seto K., Kanzaki M., Maruyama T. Serotypes of Erysipelothrix rhusiopathiae strains isolated from slaughter pigs affected with chronic erysipelas. Nihon Juigaku Zasshi. 1984 Apr;46(2):149–153. doi: 10.1292/jvms1939.46.149. [DOI] [PubMed] [Google Scholar]
- Uchida I., Makino S., Sasakawa C., Yoshikawa M., Sugimoto C., Terakado N. Identification of a novel gene, dep, associated with depolymerization of the capsular polymer in Bacillus anthracis. Mol Microbiol. 1993 Aug;9(3):487–496. doi: 10.1111/j.1365-2958.1993.tb01710.x. [DOI] [PubMed] [Google Scholar]
- Varela P., Rivas M., Binsztein N., Cremona M. L., Herrmann P., Burrone O., Ugalde R. A., Frasch A. C. Identification of toxigenic Vibrio cholerae from the Argentine outbreak by PCR for ctx A1 and ctx A2-B. FEBS Lett. 1993 Jan 2;315(1):74–76. doi: 10.1016/0014-5793(93)81136-n. [DOI] [PubMed] [Google Scholar]
- Wegmüller B., Lüthy J., Candrian U. Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl Environ Microbiol. 1993 Jul;59(7):2161–2165. doi: 10.1128/aem.59.7.2161-2165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernars K., Heuvelman C. J., Chakraborty T., Notermans S. H. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J Appl Bacteriol. 1991 Feb;70(2):121–126. doi: 10.1111/j.1365-2672.1991.tb04437.x. [DOI] [PubMed] [Google Scholar]