Abstract
Glycans that are either N-linked to asparagine, or O-linked to serine or threonine, are the hallmark of glycoproteins, a class of protein that dominates the mammalian proteome. These glycans perform important functions in cells and in some cases are required for protein activity. Nuclear magnetic resonance (NMR) spectroscopy is a powerful tool for studying glycan structure and interactions, particularly in a form that exploits heteronuclei such as 13C. Here an approach is presented that that uses α-2,6-sialyltransferase (ST6Gal-I) to enzymatically add 13C-N-acetylneuraminic acid (NeuAc or sialic acid) to glycoproteins after their preparation using non-bacterial hosts. ST6Gal-I is itself a glycoprotein, and in this initial application labeling of its own glycans and observation of these glycans by NMR is illustrated. The catalytic domain from rat ST6Gal-I was expressed in mammalian HEK293 cells. The glycans from the two glycosylation sites were analyzed with mass spectrometry and found to contain sialylated biantennary structures. The isotopic labeling approach involved removal of the native NeuAc residues from ST6Gal-I with neuraminidase, separation of the neuramindase with a lectin affinity column, and addition of synthesized 13C-CMP-NeuAc to the desialylated ST6Gal-I. Chemical shift dispersion due to the various 13C-NeuAc adducts on ST6Gal-I was observed in a 3D experiment correlating 1H-13C3-13C2 atoms of the sugar ring.
Glycans that are either N-linked to asparagine, or O-linked to serine or threonine, are the hallmark of glycoproteins, a class of protein that dominates the mammalian proteome. These glycans insure quality of protein folding, dictate transport to various regions of the cell, mediate interactions of the cell with its environment, regulate clearance of spent proteins from the system, and in some cases are required for proper protein function.1 Nuclear magnetic resonance (NMR) spectroscopy is a powerful tool for studying glycan structure and interactions, particularly in a form that exploits heteronuclei such as 13C. It has been used effectively to determine the structure oligosaccharides, polysaccharides, and protein-bound oligosaccharides,2 but studies of glycans attached to their native proteins by NMR are rare.3 Here an approach to label glycans is presented that uses α-2,6-sialyltransferase (ST6Gal-I, EC 2.4.99.1) to add 13C-N-acetylneuraminic acid (NeuAc or sialic acid) to glycoproteins after their preparation using non-bacterial hosts. ST6Gal-I is itself a glycoprotein, and in this initial application labeling and observation of its own glycans by NMR is illustrated.
ST6Gal-I is a member of the glycosyltransferase family 29 of the CAZY database (www.cazy.org).4 Members of this family transfer NeuAc from the donor substrate, cytidine-5′-monophospho-N-acetylneuraminic acid (CMP-β-NeuAc), to terminal galactose (Gal) or N-acetylgalactosamine (GalNAc) residues of glycans to create α-2,6, α-2,3, and α-2,8 linkages. ST6Gal-I from most species has two N-glycosylation sites, which are necessary for normal function.5 For this reason, recombinant ST6Gal-I has been expressed primarily in cells that produce glycosylated forms.5,6 ,7
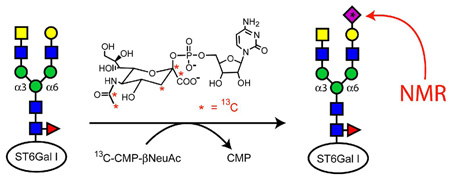
The isotopic labeling approach used here is illustrated in Figure 1. An NH2-terminal His-tagged form of the catalytic domain from rat ST6Gal-I (residues 97–403, GenBank P13721)8 was expressed in mammalian HEK293 cells as described previously, but without amino acid specific isotopic labeling.6 Native NeuAc residues were then removed from the glycans of ST6Gal-I with a general neuraminidase (New England Biolabs, Inc.) capable of catalyzing the hydrolysis of α-2,3, α-2,6, and α-2,8 linked NeuAc residues (38 µg ST6Gal-I, 50 units neuraminidase, 24 hrs, 0.21 mL, 37°C). Neuramindase was separated from the ST6Gal-I using a Concanavalin A sepharose affinity column (GE Healthcare, 1 mL), which has weak affinity for biantennary and truncated glycans like those of ST6Gal-I.9 The column was washed with 3 mL of buffer (20 mM bis-tris, pH 6.5, 0.5 M NaCl, 1 mM CaCl2, 1 mM MnCl2) to remove the neuraminidase.10 Desialylated ST6Gal-I was then eluted from the column with α-d-methylmannoside (Sigma-Aldrich, 15 mL, 0.2 M, buffer) and stored with 400 µM N-acetyl-2,3-dehydro-2-deoxyneuraminic acid (Sigma-Aldrich), a neuraminidase inhibitor. Before the addition of 13C-NeuAc, the desialylated ST6Gal-I was washed into NMR buffer (10 mM potassium phosphate, 200 mM NaCl, pH 6.5, 400 µM inhibitor, D2O). 13C-CMP-NeuAc (27 µg) was added to 176 µL of a 45 µM solution of the desialylated ST6Gal-I, and its addition to ST6Gal-I was monitored with 1H NMR.
Figure 1.
Isotopic labeling of ST6Gal-I glycans.
13C-CMP-NeuAc was made in a four-step synthesis process following published procedures.11 Briefly, N-hydroxysuccinimide and 13C-acetyl chloride were combined to produce N-13C-[1,2]-acetoxy-succinimide, which was further reacted with d-mannosamine to make N-13C-[1,2]-acetyl-mannosamine (13C-ManNAc). The enzyme, NeuAc aldolase, was used to react 13C-ManNAc with 13C-pyruvate to produce 13C-[1,2,3,10,11]-NeuAc. 13C-[1,2,3,10,11]-CMP-β-NeuAc was then synthesized using the enzyme, CMP-NeuAc synthetase, and cytidine-5’-triphosphophate to produce the final product. Details are provided in supporting information.
The original glycans of ST6Gal-I as expressed in HEK293 cells were analyzed using PNGaseF digestion, permethylation, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), and exoglycosidase digestion. Two major glycoforms were identified as NeuAc-α-2-3/6GalNAc-β-1-4GlcNAc-β-1-2Man-α-1-3(NeuAc-α-2-3/6Gal-β-1-4GlcNAc-β-1-2Man-α-1-6)Man-β-1-4GlcNAc-β-1-4(Fuc-α-1-6)GlcNAc and a similar structure with a Gal instead of a GalNAc residue and without the fucose residue. The NeuAc-α-2-6GalNAc linkage of the major glycans is unusual, but not unprecedented.12 β-N-acetylhexosaminidase and α-1-2,3 mannosidase digestions, composition analysis, and MALDI-MS were used to confirm the GalNAc-β-1-4GlcNAc structure on the α3 arm of the glycan (details provided in supporting information). After neuraminidase treatment, the glycan structures of ST6Gal-I should have multiple sites for NeuAc addition.
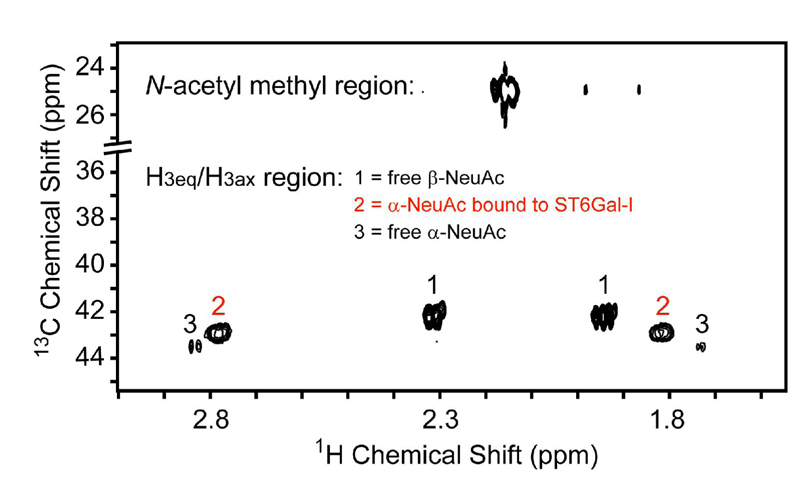
The addition of 13C-NeuAc to desialylated ST6Gal-I was monitored with a standard 2D 1H-13C heteronuclear single quantum coherence (HSQC) experiment (gChsqc, Varian Biopack, see Figure 2). The H3ax-C3 and H3eq-C3 cross peaks of the NeuAc bound to ST6Gal-I (labeled 2 in the figure) are distinct due to the chemical shift change on production of an α linkage as well as the increase in line width. Peaks from free NeuAc in α and β anomeric configurations, produced by the action of some residual neuraminidase, are labeled 3 and 1, respectively. The bound cross peak volumes measured from Figure 2 were used to determine a minimum level of addition of 13C-NeuAc to ST6Gal-I. The value determined (3 13C-NeuAc residues per molecule of ST6Gal-I) is reasonable, in view of the two glycosylation sites and the dominant glycan structures on ST6Gal-I, particularly if GalNAc terminated glycans can be substrates for ST6Gal-I.
Figure 2.
2D 1H-13C decoupled HSQC of ST6Gal-I reaction mixture after the addition of CMP-13C-β-NeuAc. A chemical shift table is included in the supporting information.
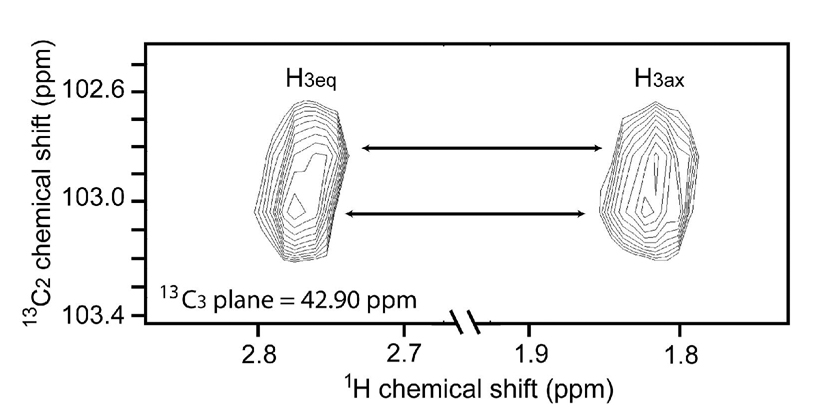
By exploiting chemical shift differences at the multiple 13C sites introduced in the NeuAc labels it is possible to demonstrate the presence of chemically or structurally distinct glycan sites in ST6Gal-I. 13C-NeuAc is 13C-labeled at the methyl and carbonyl carbons of the N-acetyl group and the C1, C2 and C3 carbons within the sugar ring. An 800 MHz Varian INOVA spectrometer was used to acquire several types of 2D and 3D 1H-13C correlation experiments. A 1H-13C HSQC experiment correlating the acetyl 1Hmethyl to the acetyl 13Cmethyl and 2D acetyl 1H(13Cmethyl)13CO experiments did not reveal detectable chemical shift dispersion. Chemical shift dispersion due to the various 13C-NeuAc adducts on ST6Gal-I was, however, observed in a 3D experiment correlating 1H-13C3-13C2 atoms of the sugar ring (shown in the 1H-13C2 plane of Figure 3). The C3 chemical shift has no detectable difference, but the C2 chemical shift is sensitive. At least two different 13C-NeuAc environments are resolved in the 2D 1H-13C2 plane as indicated by the arrows connecting the H3ax and H3eq correlation peaks. C2 is the anomeric linkage site for the α-2,6 glycosidic bond. The chemical shifts observed for C2 are expected to be sensitive to structural and chemical differences in the linked residue and the surrounding environment. In particular, they may be sensitive to linkage to Gal vs GalNAc residues as occurs in the two branches of the major glycoform depicted in Figure 1. Assignment of the peaks is not straightforward, but it may be possible using differential isotopic labeling combined with mass spectrometry, as illustrated in our recent work on 13C-methyl-lysine labeling.13
Figure 3.
1H-13C2 plane from a 3D 1H-13C3-13C2 correlation experiment. JC1C2 and JC2C3 were refocused during t1 evolution with selective π pulses.
Additional chemical shift dispersion would be desirable. Neither the chemical shift of the 13C1 of the carboxylate carbon, nor the potential of 4D experiments that include this carbon, have been exploited in the data presented. pKas of the carboxylate are expected to be sensitive to local environments and the 13C1 shifts would in turn be sensitive to differential protonation. Modern methods that allow efficient collection of multidimentional NMR including 13C data may provide sufficient resolution to undertake site specific assignments and monitor site specific interactions.14
Supplementary Material
Complete Ref. 7a., details of the synthesis of 13C-[1,2,3,10,11]-NeuAc, the analysis of glycans on ST6Gal-I, and chemical shifts of NeuAc compounds. This material is available free of charge via the Internet at (http://pubs.acs.org).
Acknowledgement
This work was supported by grants from the National Institutes of Health, RR024105, GM033225, and RR005351.
References
- 1.(a) Moremen KW, Molinari M. Curr. Opin. Struct. Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]; (c) Paulson JC, Blixt O, Collins BE. Nat. Chem. Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 2.(a) Sayers EW, Prestegard JH. Biophys. J. 2000;79:3313–3329. doi: 10.1016/S0006-3495(00)76563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kjellberg A, Weintraub A, Widmalm G. Biochem. 1999;38:12205–12211. doi: 10.1021/bi9910629. [DOI] [PubMed] [Google Scholar]; (c) Blundell CD, DeAngelis PL, Day AJ, Almond A. Glycobiol. 2004;14:999–1009. doi: 10.1093/glycob/cwh117. [DOI] [PubMed] [Google Scholar]; (d) Azurmendi HF, Vionnet J, Wrightson L, Trinh LB, Shiloach J, Freedberg DI. PNAS. 2007;104:11557–11561. doi: 10.1073/pnas.0704404104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Macnaughtan MA, Kamar M, Alvarez-Manilla G, Venot A, Glushka J, Pierce JM, Prestegard JH. J. Mol. Biol. 2007;366:1266–1281. doi: 10.1016/j.jmb.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Gilhespymuskett AM, Partridge J, Jefferis R, Homans SW. Glycobiol. 1994;4:485–489. doi: 10.1093/glycob/4.4.485. [DOI] [PubMed] [Google Scholar]; (b) Wyss DF, Choi JS, Wagner G. Biochem. 1995;34:1622–1634. doi: 10.1021/bi00005a019. [DOI] [PubMed] [Google Scholar]; (c) Miyazaki T, Sato H, Sakakibara T, Kajihara Y. J. Am. Chem. Soc. 2000;122:5678–5694. [Google Scholar]
- 4.(a) Coutinho PM, Henrissat B. In: Recent Advances in Carbohydrate Bioengineering. Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]; (b) Campbell JA, Davies GJ, Bulone V, Henrissat B. Biochem. J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Coutinho PM, Deleury E, Davies GJ, Henrissat B. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Colley KJ. Glycobiol. 2000;10:531–583. doi: 10.1093/glycob/10.5.531. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Venot A, Meng L, Tian F, Moremen KW, Boons G-J, Prestegard JH. Chem. Biol. 2007;14:409–418. doi: 10.1016/j.chembiol.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Hamilton SR, et al. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]; (b) Hill DR, Aumiller JJ, Shi X, Jarvis DL. Biotechnol Bioeng. 2006;95:37–47. doi: 10.1002/bit.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein J, Lee EU, Mcentee K, Lai PH, Paulson JC. J. Biol. Chem. 1987;262:17735–17743. [PubMed] [Google Scholar]
- 9.Creus S, Chaia Z, Pellizzari EH, Cigorraga SB, Ulloa-Aguirre A, Campo S. Mol. Cell. Endocrinol. 2001;174:41–49. doi: 10.1016/s0303-7207(00)00453-6. [DOI] [PubMed] [Google Scholar]
- 10.Chromatographic resolution could very likely be improved by using a lectin affinity column specific to terminal Gal and GalNAc residues.
- 11.(a) Heidlas JE, Lees WJ, Pale P, Whitesides GM. J. Org. Chem. 1992;57:146–151. [Google Scholar]; (b) Aubin Y, Prestegard JH. Biochem. 1993;32:3422–3428. doi: 10.1021/bi00064a028. [DOI] [PubMed] [Google Scholar]; (c) Knorst M, Fessner W-D. Adv. Synth. Catal. 2001;343:698–710. [Google Scholar]
- 12.Ohkura T, Seko A, Hara-Kuge S, Yamashita K. J. Biochem. 2002;132:891–901. doi: 10.1093/oxfordjournals.jbchem.a003302. [DOI] [PubMed] [Google Scholar]
- 13.Macnaughtan MA, Kane A, Prestegard JH. J. Am. Chem. Soc. 2005;127:17626–17627. doi: 10.1021/ja056977r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Mandelshtam VA, Taylor HS, Shaka AJ. J. Magn. Reson. 1998;133:304–312. doi: 10.1006/jmre.1998.1476. [DOI] [PubMed] [Google Scholar]; (b) Kim S, Szyperski T. J. Am. Chem. Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete Ref. 7a., details of the synthesis of 13C-[1,2,3,10,11]-NeuAc, the analysis of glycans on ST6Gal-I, and chemical shifts of NeuAc compounds. This material is available free of charge via the Internet at (http://pubs.acs.org).