Abstract
Cytochrome c (cyt c) is a heme-containing protein that participates in electron transport in the respiratory chain and as a signaling molecule in the apoptotic cascade. Here we addressed the effect of removing mammalian cyt c on the integrity of the respiratory complexes in mammalian cells. Mitochondria from cyt c knockout mouse cells lacked fully assembled complexes I and IV and had reduced levels of complex III. A redox-deficient mutant of cyt c was unable to rescue the levels of complexes I and IV. We found that cyt c is associated with both complex IV and respiratory supercomplexes, providing a potential mechanism for the requirement for cyt c in the assembly/stability of complex IV.
The mitochondrial electron transport chain consists of four multisubunit complexes, namely, NADH-ubiquinone oxidoreductase (complex I),2 succinate-ubiquinone oxidoreductase (complex II), ubiquinone-cytochrome c oxidoreductase (complex III), and cytochrome c oxidase (complex IV, COX). Cytochrome c (cyt c) shuttles electrons from oxidative phosphorylation complex III to complex IV. Electrons are transferred from reduced cyt c sequentially to the CuA site, heme a, heme a3, and CuB binuclear center in the complex IV before being finally transferred to molecular oxygen to generate water (1). Respiratory complexes are assembled into supercomplexes (also called respirasomes). These contain complex I bound to dimeric complex III and a variable copy number of complex IV (2).
In Saccharomyces cerevisiae, cyt c is encoded by two genes: CYC1 and CYC7. Mutagenesis studies in yeast have shown that cyt c is required for the assembly of COX (3, 4). In yeast lacking both the cyt c genes (CYC1 and CYC7), COX assembly was absent. It was also shown that cyt c is only structurally required for COX assembly, because a catalytic mutant of cyt c (W65S) was sufficient to bring about near normal levels of COX. However, because yeast lacks complex I, they could not analyze the role of cyt c in the assembly/stability of complex I. Mammals possess two different isoforms of cyt c encoded on different chromosomes: the somatic (cyt cS)- and testis (cyt cT)-specific isoforms. In mouse, the cDNAs bear 74% homology, whereas the proteins possess 86% identity with most dissimilarity in the C terminus.
Cardiolipin (CL) is an anionic phospholipid present almost exclusively in the mitochondrial membranes and constitutes 25% of its total phospholipids (5). Work from several laboratories showed that CL is essential for the membrane anchorage of the respiratory supercomplexes. CL has two main roles in the mitochondrial structure and function, namely, stabilization of mitochondrial membranes and specific interactions with proteins. CL deficiency results in inefficient energy transformation by oxidative phosphorylation, swelling of mitochondria, decreased ATP/oxygen ratio, and reduced membrane potential (6, 7). In accordance, in S. cerevisiae lacking CL synthase, the supercomplex comprising complexes III and IV is unstable (8). Assembly mutants of COX had significantly reduced CL synthase activity, whereas assembly mutants of respiratory complex III and complex V showed less inhibition (9). Subsequently, the proton gradient across the inner mitochondrial membrane was found to be important for CL formation and that CL synthase was stimulated by alkaline pH at the matrix side (10). In this study, we investigated the role of cyt c depletion on CL levels by examining its content and composition in cyt c null cells.
Here we aimed to answer the following questions: What is the role of cyt c in the assembly and maintenance of the different respiratory complexes in mammals? Are there changes in the content/composition of lipids in the cyt c-ablated cells? Analysis of mouse fibroblasts revealed that cyt c is essential for the assembly/stability of COX, and a catalytically mutant form of cyt c cannot rescue the COX defect in the cyt c null cells. CL and triacylglycerols showed significant differences in the cyt c null cells, both in content and composition.
EXPERIMENTAL PROCEDURES
Genetically Modified Mice and Derived Cell Lines—The crosses performed to obtain mice with the genotype cyt cs-/- cyt ct-/- Transgeneflox/0 and the characterization of lung fibroblasts lacking cyt c were previously described (11).
Cell Lines—All cells were grown in Dulbecco's modified Eagle's medium with high glucose, supplemented with uridine and pyruvate. Clones L3, L4, and L7 lacked both the somatic and testis isoforms of cyt c and were designated as double knockout (dKO). LF represents the original lung fibroblasts before the deletion of the floxed cyt c. CL1 and CL15 were derived from L3, after the reintroduction of a wild-type cyt c cDNA. CL18 and CL25 were derived from L3 after the reintroduction of a mutant (W60S) cyt c cDNA.
Mouse LM(TK-) cells were obtained from ATCC (CCL 1.3), and the mitochondrial DNA (mitochondrial DNA)-less derivative was obtained by ethidium bromide treatment as described previously (12). Somatic cyt c cDNA was subcloned into pIRES-puro vector (Clontech, cyt c puro-8) and introduced into cyt cs dKO fibroblasts (L3 clone) by stable transfection to generate clones CL1 and CL15. A point mutation (W60S) was introduced into cyt c cDNA by PCR using the QuikChange site-directed mutagenesis kit (Stratagene) on the cyt c puro-8 template using mutant primers (forward: 5′-GCCAACAAGAACAAAGGCATCACCTcGGGAGAGGATACCCTGATGG-3′ and reverse: 5′-CCATCAGGGTATCCTCTCCCgAGGTGATGCCTTTGTTCTTGTTGGC-3′), with the modified nucleotides shown in lowercase. The presence of the mutation was confirmed by sequencing. After stable transfections in L3 (dKO) cells, two clones containing the desired mutation were obtained (CL18 and CL25).
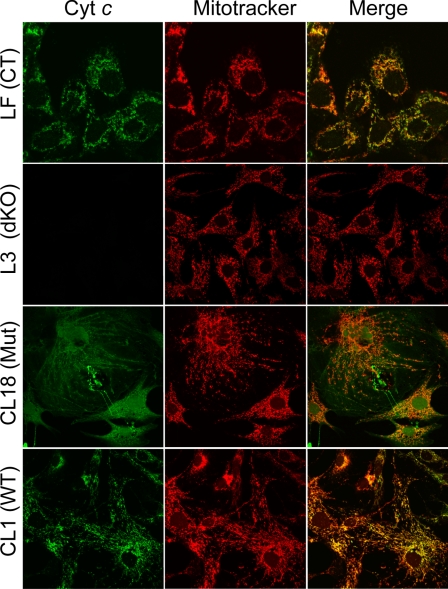
Immunostaining—Live cells were stained with MitoTracker red (200 nm, Invitrogen) and subsequently fixed and permeabilized in 4% paraformaldehyde and ice-cold methanol, respectively. Then, cells were incubated with a primary antibody against cyt c (BD Biosciences and Mitosciences) and subsequently with a secondary antibody tagged with fluorescent Alexa-fluor (Molecular Probes) and visualized by confocal microscopy.
Respiration Measurements—Cellular respiration was measured by polarography as described (13). The oxygen consumed by cells was measured both before and after addition of the complex III inhibitor, antimycin A, and the subsequent addition of ascorbate and N,N,N′,N′-tetramethyl-1,4-phenylenediamine dihydrochloride (TMPD).
Isolation of Mitochondria—Mitochondria were prepared by the nitrogen cavitation method as described previously (14) with some modifications. In brief, cells were washed in 10 ml of the ice-cold buffer A (100 mm sucrose, 1 mm EGTA, 20 mm MOPS, pH 7.4) and centrifuged at 1500 × g to pellet. Cells (∼1 ml pellet) were resuspended in 5 ml of hypotonic ice-cold buffer B (buffer A plus 10 mm triethanolamine, 5% Percoll, and protease inhibitor mixture (Complete, Roche Applied Science)), and subjected to nitrogen cavitation at 500 p.s.i. for 30 min. The mix of broken cells was centrifuged twice at 500 × g, and the final supernatant was centrifuged at 12,000 × g. The pellet was used as a crude mitochondrial preparation.
Complex Activities—Respiratory complex activities were measured on isolated mitochondria using spectrophotometric assays as described (13).
Western Blots—Cell lysates were prepared as described (11). In brief, cells were lysed in buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mm Tris, pH 8.0, supplemented with Complete protease inhibitor mixture), and protein concentrations were determined using the DC kit (Bio-Rad Laboratories). Cell lysates and mitochondrial preparation were resolved on SDS-PAGE gels, transferred onto a polyvinylidene difluoride (PVDF) membrane, and hybridized with the antibodies raised against cyt c, Ndufa9, COXI, SDH, Core2, ATPase-β, VDAC1 (BD Biosciences, Mitosciences, and Invitrogen), and α-tubulin (Sigma).
Southern Hybridization—Genomic DNA was isolated from cultured cells, digested with the SacI restriction endonuclease, and loaded onto a 0.8% agarose gel. The DNA was transferred onto Hybond membranes (Amersham Biosciences) and hybridized with [32P]dCTP-labeled probes specific to mouse mitochondrial and 18 S ribosomal DNA.
BN-PAGE Gels—Individual OXPHOS complexes were visualized, and their in-gel activities were determined after blue native (BN)-PAGE as described (15). To visualize supercomplexes, mitochondria were treated with digitonin (1:4 ratio) on ice and separated on a 4–10% BN-PAGE. Subsequently, individual lanes were excised from the gel and subjected to two-dimensional SDS-PAGE on a 12% gel. The gels from both the first and second dimension were blotted and probed with antibodies against different OXPHOS complexes.
35S Labeling of Mitochondrial Proteins—Cells were labeled with [35S]methionine and cysteine mixture as described (15). For pulse labeling, 1 million cells were first treated with 100 μg/ml emetine for 10 min, followed by labeling with [35S]methionine and cysteine mixture for 2 h. Fifty micrograms of the cell lysate was loaded on a 15% SDS-PAGE. For the pulse-chase experiment, 2 million cells were plated, and on the following morning, they were treated with 40 μg/ml chloramphenicol for 24 h. They were rinsed and treated with cycloheximide for 10 min, followed by labeling for 2 h with [35S]methionine and cysteine mixture and chased for 2, 4, and 24 h with normal medium. To visualize the respiratory complexes, 30 μg of the digitonin permeabilized cell lysate was loaded onto a 4–13% BN-PAGE, transferred onto a PVDF membrane, and subjected to autoradiography.
Proteinase K Sensitivity—Mitoplasts were prepared from isolated mitochondria (50 μg) by treatment with a hypotonic buffer (20 mm HEPES, pH 7.4). Both mitochondria and mitoplasts were incubated with either 3 or 15 μg/ml proteinase K for 60 min on ice. The reaction was stopped by the addition of phenylmethylsulfonyl fluoride to a final concentration of 2 mm, and the mitochondria/mitoplasts were recovered by centrifugation. The samples were suspended in Laemmli buffer and subjected to Western analysis with antibodies against cyt c, COX1, COXVb, and HSP60.
Heme Staining—250 μg of mitochondria were reduced with dithiothreitol (50 mm) on ice, resolved on a 12% lithium dodecyl sulfate-PAGE, and blotted onto a PVDF membrane. The heme-associated peroxidase activity was detected on the membrane using SuperSignal chemiluminescence reagent (Pierce) followed by autoradiography, as described (16, 17).
Electron Microscopy—Cultured cells were fixed in 2% phosphate-buffered glutaraldehyde (with 100 mm sucrose) overnight at 4 °C, followed by 2% buffered OsO4 for 1 h at room temperature. Cultures were further processed for EMbed plastic (Electron Microscopy Sciences) embedding. Areas were chosen for semi-thin sectioning; the 1-μm sections were stained with toluidine blue/methylene blue/sodium borate. Thin sections (90 nm) were stained with uranyl acetate and lead citrate and examined in a Philips CM-10 electron microscope (FEI Co.). Sections were cut perpendicularly to the plane of the coverslip (18).
Determination of Reactive Oxygen Species—Reactive oxygen species were detected in live cells using the cell-permeable probe, dichlorofluorescein (Invitrogen) followed by flow cytometry. Cells were trypsinized and counted, and 0.2 million cells were stained with dichlorofluorescein, either untreated or upon treatment with hydrogen peroxide.
Mass Spectrometric Analysis of Lipids—Enhanced shotgun lipidomics analyses were performed as described before (19, 20). In brief, lipids were analyzed on a QqQ mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with an electrospray ion source. All electrospray ionization mass spectrometric analysis of lipids were conducted by direct infusion employing a Harvard syringe pump at a flow rate of 4 ml/min. Typically a 1-min period of signal averaging was employed for each mass spectrum, and a 2-min period of signal averaging was employed for each tandem mass spectrometric spectrum. A mass resolution of 0.4 Thomson was employed for acquisition of mass spectra with a QqQ instrument (21). The results are presented as the mean from two different cyt c dKO clones (L3 and L4) and two different cyt c reintroduced clones (CL1 and CL15). The error of the method, after standardization to internal controls, is 10% (22).
RESULTS
cyt c-/- Fibroblasts Are Deficient in Respiration—Lung fibroblasts (LFs) were derived from the conditional cyt c knockout mice as described (11). The cyt c transgene was deleted in cultured lung fibroblasts by the expression of Cre recombinase from a recombinant adenovirus. Because these fibroblasts lack both the cyt c isoforms, they were designated as double knockout (dKO). cyt c dKO cells were used for the reintroduction of wild-type or mutant cyt c. The expression of cyt c was assessed by both immunostaining and Western blots (Figs. 1 and 3A). Respiration was measured in the dKO and cells with reintroduced cyt c. Respiration in the dKO reintroduced with the wt cyt c was close to the values of control LF cells (Fig. 2A). The mutant cyt c cDNA had a single amino acid changed from tryptophan to serine (W60S), as described in S. cerevisiae (3, 23). The lines expressing the mutant form are clones CL18 and CL25. In yeast, the W65S mutant was catalytically inactive in electron transfer. As expected, the cyt c dKO cells with the mutant cyt c cDNA reintroduced did not respire. The cells lacked both endogenous and ascorbate plus TMPD-mediated respiration (Fig. 2A). We previously showed that the cyt c dKO cells maintained a mitochondrial membrane potential that could be dissipated with a protonophore (11), a phenomenon associated with a reverse function of complex V in OXPHOS-deficient cells (24). We could not detect differences in reactive oxygen species production between the different cell lines using dichlorofluorescein followed by flow cytometry analysis (data not shown). Other investigators made similar observations in the cyt c null cells (25).
FIGURE 1.
Expression and localization of cyt c in fibroblasts. Immunostaining of control (LF), dKO (L3), WT cyt c reintroduced (CL1) and W60S mutant cyt c reintroduced (CL18) cells with cyt c antibodies. Cells were co-stained with MitoTracker red to confirm the mitochondrial localization.
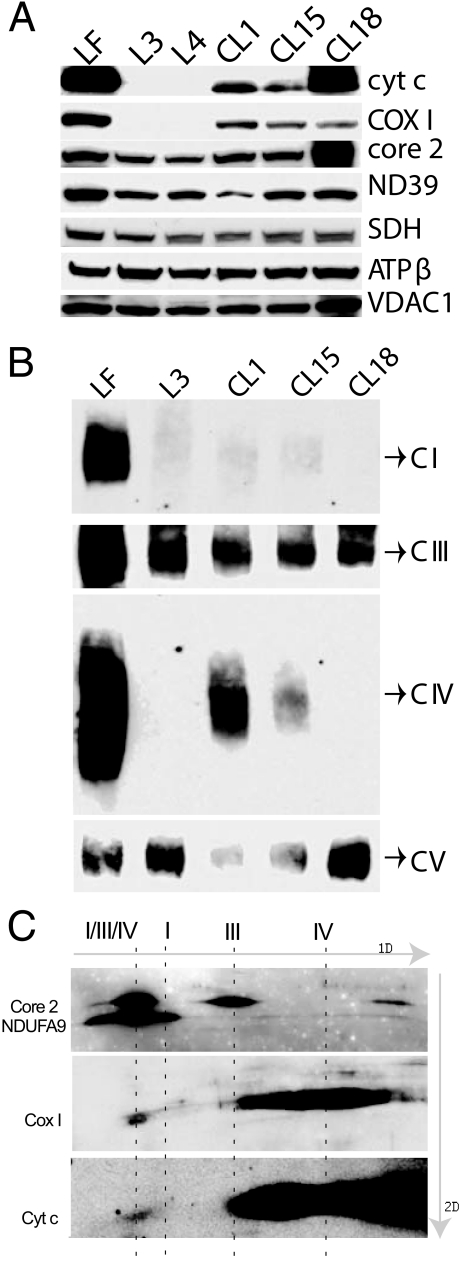
FIGURE 3.
cyt c-deficient cells lack OXPHOS complexes I and IV. A, Western analysis of mitochondria isolated from the different cell lines using antibodies raised against cyt c, Ndufa9, COX1, SDH, Core2, and ATPase-β. VDAC1 antibody was used as a loading control. cyt c dKO cells (L3 and L4) lacked COX1. B, BN-PAGE was performed on a 4–13% gel. The proteins were transferred onto a PVDF membrane and probed with antibodies raised against the different complex subunits, namely, Ndufa9 (complex I), Core2 (complex III), COX1 (complex IV), and ATPase-β (complex V). CI, CIII, CIV, and CV are complexes I, III, IV, and V, respectively. cyt c dKO cells (L3) and those reintroduced with the W60S mutant cyt c cDNA (CL18) lacked complexes I and IV. Complex I was also very reduced in CL1 and CL15. C, two-dimensional BN-PAGE of control (LF) mitochondria followed by Western analysis with antibodies against cyt c and complexes I (NDUFA9), III (core 2), and IV (COX 1).
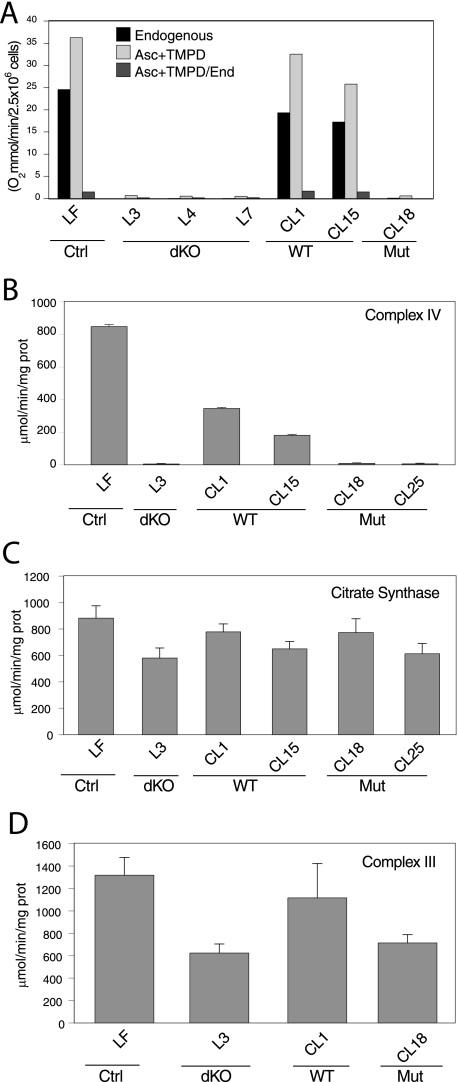
FIGURE 2.
Mouse fibroblasts with disrupted cyt c (somatic and testis) alleles are OXPHOS-deficient. A, control (LF), cyt c dKO (L3, L4, and L7), wild-type cyt c cDNA reintroduced (CL1 and CL15), and mutant cyt c cDNA reintroduced (CL18) intact cells were analyzed for KCN-sensitive oxygen consumption. All cell lines were also analyzed for the ratio of the ascorbate-TMPD respiration (where electrons are donated directly to cyt c) to endogenous respiration (starting at complexes I and II). cyt c dKO cells and those reintroduced with the mutant cyt c cDNA did not respire. B and D, respiratory complexes (III and IV) activities measured by spectrophotometric assays. Citrate synthase activity was measured in all the samples (C) as a measure of mitochondrial levels. cyt c dKO cells and those reintroduced with the W60S mutant cyt c cDNA (CL18 and CL25) lacked complex IV and had reduced complex III activities.
Activity of Respiratory Complexes in cyt c Null Cells—Respiratory complex activities were measured by spectrometric assays on mitochondria isolated from the cells. We observed that the cyt c dKO fibroblasts were completely deficient in complex IV activity (Fig. 2B), whereas the complex III activity was reduced to 50% of the control levels (Fig. 2D). Citrate synthase activity was measured to normalize the other activities and found to be similar in all samples (Fig. 2C). To verify that the mitochondrial DNA content was reduced in the cyt c-ablated cells, we performed Southern hybridization. We found that mitochondrial DNA levels were the same in the control and cyt c null cells (data not shown).
Steady-state Levels of Respiratory Complexes in cyt c Null Cells—Western blot analyses were performed to assess the steady-state levels of different respiratory complex subunits. Antibodies against Ndufa9, SDH, Core2, COXI, and ATPase-β were used (Fig. 3A). Cyt c dKO fibroblasts had undetectable levels of COXI protein. There were no major changes in subunits of other OXPHOS complexes. When we reintroduced either wild-type or mutant cyt c cDNA (W60S) into the cyt c null fibroblasts, COX I protein was observed (Fig. 3A).
The assembled respiratory complexes were analyzed by BN-PAGE (15) followed by Western analysis. Interestingly, the cyt c dKO cells had undetectable levels of not only complex IV, but also complex I (Fig. 3B and supplemental Fig. S1). Complex III steady-state levels were only mildly reduced in most cells, whereas the relative levels of the complexes II and V were not changed (Fig. 3B and supplemental Fig. S1). Upon reintroduction of cyt c cDNA into the cyt c dKO fibroblasts, we found that fully assembled COX could be restored, and to a lesser extent, also complex I (Fig. 3B).
The W60S mutant clone (CL18), though showing the presence of COX1, lacked detectable levels of both complexes IV and I (Fig. 3B). In-gel activity results were in agreement with the Western blots (supplemental Fig. S2).
Two-dimensional BN-PAGE analysis was performed to address the role of cyt c in the stability of OXHOS complexes. Digitonin-treated mitochondria from control fibroblasts (LFs) were resolved on a 4–10% BN-PAGE followed by a second dimension SDS-PAGE (12%) and Western analysis. This revealed that a fraction of cyt c was associated with a supercomplex containing complexes I, III, and IV (Fig. 3C). This fraction correlated with the levels of complex IV detected in the respirasome. Accordingly, the bulk of the cyt c was associated with complex IV (Fig. 3C).
Newly Synthesized Complex IV Cannot Be Detected in the Absence of cyt c—We wanted to determine whether the lack of complexes I and IV in the cyt c dKO fibroblasts was due to defective synthesis or degradation. Control, dKO, and dKO cells reintroduced with cyt c cDNA were labeled with [S35]methionine and cysteine in the presence of emetine, a cytosolic protein synthesis inhibitor. We observed that all the cell types synthesized COX1, COX2, and COX3 at comparable levels, indicating that they were capable of synthesizing the COX subunits (supplemental Fig. S3).
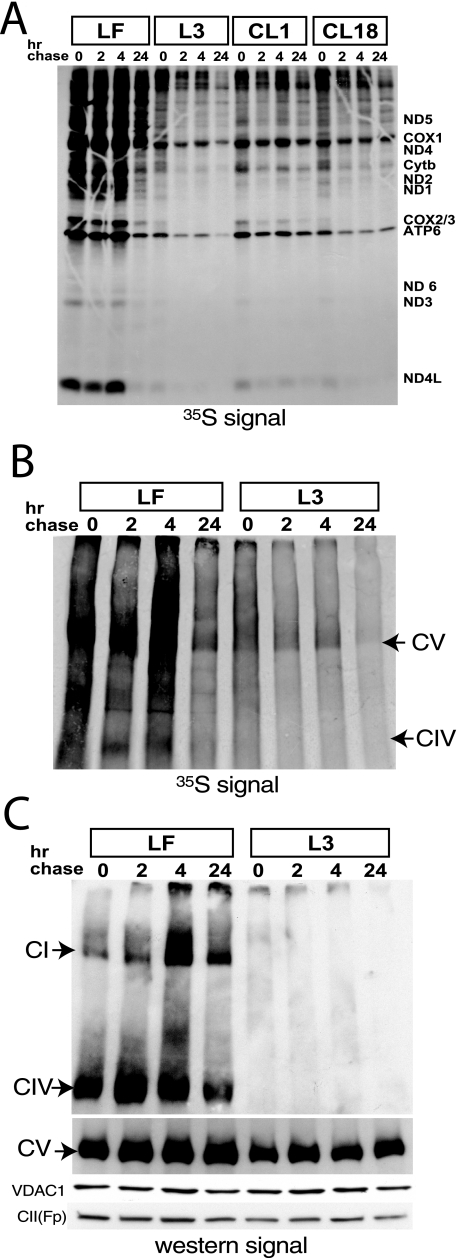
We next examined the stability of the different OXPHOS complexes by pulse-chase experiment. Cells were labeled with [35S]methionine in the presence of cycloheximide, a reversible cytosolic protein synthesis inhibitor (15) for 2 h followed by a chase period of 2, 4, or 24 h. The samples were resolved on both SDS-PAGE and BN-PAGE to visualize the individual mitochondrial subunits and assembled complexes, respectively (Fig. 4, A–C). In the SDS-PAGE, we observed that the COX subunits were synthesized in all cell lines tested (in the zero time point), but they were degraded faster during the chase in the dKO and W60S mutant cyt c reintroduced cells (more clearly seen in COX2/3). ATP6 protein was more stable during the chase in all cells tested indicating that the lack of cyt c does not affect complex V assembly/stability (Fig. 4A).
FIGURE 4.
Complexes I and IV are not synthesized or are extremely unstable in cyt c dKO cells. Cells were labeled with [35S]methionine and cysteine, followed by a chase for 2, 4, and 24 h, respectively. Lysates from the different cell lines were resolved on an SDS-PAGE (A). Lysates of LF and L3 (dKO) were also analyzed by BN-PAGE (B) and subjected to autoradiography. In C, the same samples shown in B were transferred onto a membrane and probed with antibodies raised against the different complex subunits, namely, Ndufa9 (complex I), COXI (complex IV), and ATPase-β (complex V). VDAC1 and CII(Fp) antibodies were used as loading controls.
BN-PAGE analysis of the pulse-chase experiment revealed that the cyt c-ablated cells did not assemble COX (or had it extremely unstable). We were only able to detect a 35S signal in complexes IV and V, and the latter was not affected in the dKO cells. Analyses of these same samples by BN-PAGE followed by probing with different antibodies showed once more that complexes IV and I were essentially absent in the cyt c null cells, whereas complex V was not affected (Fig. 4C). A similar lack of complexes I and IV was observed in cells with the W60S mutant cyt c reintroduced (not shown). Surprisingly, the levels of complex I in the dKO cells with reintroduced cyt c were still very low. This could be a reflection of the relatively low levels of complex IV in these cells, but the reason for these low levels of complexes IV and I are not clear.
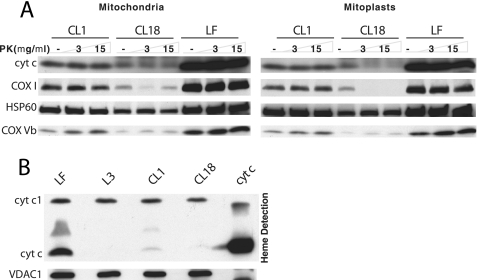
Localization of Mutant cyt c in Mitochondria—Isolated mitochondria and mitoplasts were treated with proteinase K and subjected to Western analysis. Wild-type and mutant cyt c were detected in both mitochondria and mitoplasts. The wild-type cyt c (CL1 and LF) was resistant to proteinase K, probably because it was mostly protected within cristae folds (Fig. 5A). However, the mutant cyt c (CL18) was more sensitive to the protease (Fig. 5A, right panel). This increased sensitivity can be due to incorrect folding and/or altered localization in the intermembrane space. Surprisingly, we found that COX I was present at higher levels when the mutant cyt c was reintroduced (it was undetectable in the dKO, Fig. 3A). However, it was also very sensitive to proteinase K in mitoplasts (Fig. 5A, right panel). This sensitivity has been consistently observed in different experiments. We conclude that a functional cyt c is required for the normal localization of COX I within the inner membrane.
FIGURE 5.
Mutant cyt c does not localize normally in the mitochondria. A, mitochondria and mitoplasts from LF, CL1, and CL18 were treated with proteinase K and subjected to Western analysis using antibodies against cyt c, COX1 (inner membrane protein), COXVb (inner membrane protein), and HSP60 (matrix protein). B, 250 μg of mitochondria from LF, L3 (dKO), CL1, and CL18 were resolved on a lithium dodecyl sulfate-PAGE, blotted, and subjected to heme staining to detect mitochondrial c-type cytochromes. Horse cyt c (200 pmol, Sigma) was loaded as a positive control.
W60S cyt c Has Impaired Heme Addition—We determined the presence of heme groups in cyt c by detecting the peroxidase activity associated with heme. Proteins were resolved on a lithium dodecyl sulfate-PAGE gel and transferred onto a PVDF membrane. The c-type mitochondrial cytochromes were detected on the blot after the addition of a chemiluminescence substrate (Fig. 5B). The signal corresponding to cyt c-heme in W60S mutant was ∼30% of the signal in the wild-type cyt c, which is similar to what was reported in the yeast (3).
cyt c KO Cells Have Abnormal Mitochondria—As expected from an OXPHOS-deficient cell, cyt dKO fibroblasts showed the presence of enlarged mitochondria, which displayed irregular cristae organization. Abundant lipid droplets (gray) and lipofuscin (darkly stained) were detected in the dKO fibroblasts, but not in the control cells (supplemental Fig. S4). The abnormal lipids, disorganized cristae, and absence of complexes I and IV suggested that the lipid composition of the cyt c null mitochondria was altered.
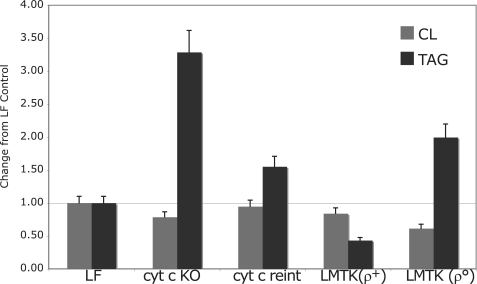
cyt c-ablated Cells Have Altered Mitochondrial Lipid Composition—Shotgun lipidomics analyses on cellular extracts of cyt c null fibroblasts was performed as previously described (19, 20). We demonstrated two marked changes in lipid content in cyt c dKO fibroblasts. First, we found lower CL content in the dKO fibroblasts (Fig. 6). The total CL content in the dKO fibroblasts was 22% lower than in the control. The reduction was mainly present in CL species containing oleic acid (18:1) (supplemental Fig. S5A). Secondly, we found that the total content of triacylglycerols (TAG) in the cyt c null fibroblasts was substantially elevated (over 3-fold), probably reflecting the impaired OXPHOS system (Fig. 6 and supplemental Fig. S5B). The accumulation of TAG content in the dKO fibroblasts was also revealed by the presence of abundant lipid droplets using electron microscopy (supplemental Fig. S4).
FIGURE 6.
Lipid composition of mitochondria lacking cyt c. Quantitation of the total cardiolipin (CL) and triacylglycerol (TAG) content in the different cell lines by electrospray ionization-mass spectrometry was performed as shown in supplemental Figs. S5A and S5B. The results are expressed as a ratio to the respective lipid in control cells (LF).
After the reintroduction of wt cyt c cDNA, the total amount of CL and TAG, as well as the composition of CL and TAG molecular species were restored to near the levels of control cells (Fig. 6). We also measured the content of lipids in a cell line depleted of mitochondrial DNA (rho zero (ρ0) cells) and its control cell line (LM(TK-)). Mitochondrial DNA deficiency in ρ0 cells resulted in a marked reduction of CL content, and a substantial accumulation of TAG content (Fig. 6). These results suggest that the altered lipid content and/or composition are likely related to the OXPHOS assembly and/or function and not with the specific loss of cyt c.
DISCUSSION
The function of cyt c in the assembly of mammalian OXPHOS complexes has not been explored. We found that both the function and the physical presence of cyt c were required for the assembly/stability of respiratory complexes. Pulse-chase experiments revealed that the mitochondrially encoded COX subunits 1, 2, and 3 were reduced in the KO and the mutant cyt c introduced cells as early as 2 h post-synthesis, possibly due to increased turnover of the unassembled subunits. Therefore, our results demonstrate that, in mammals, a functional cyt c is required for the assembly/stability of COX.
In S. cerevisiae, cyt c is encoded by two genes: CYC1, which encodes Iso-1-cytochrome c that accounts for 95% of the total cyt c in mitochondria, and the homologous and less abundant iso-2-cytochrome c encoded by CYC7. Even though iso-2-cytochrome c represents only 5% of the total cyt c, it is sufficient to support respiration and growth, although at a reduced rate on non-fermentable carbon sources. Mutations in both isoforms lead to a respiratory defect (26). Previously, Barrientos et al. (3) reported that yeast lacking both the cyt c alleles, namely, cyc1 and cyc7 failed to assemble COX. The cyc1 null mutant had ∼40% of the COX activity of the wild-type cells, even though they had only 12% of the levels of cyt c compared with the control cells. The possibility that cyt c could be required for heme a biosynthesis was excluded (27). It was also shown that cyt c is only structurally required for COX assembly, because a catalytic mutant of cyt c (W65S) was sufficient to bring about near normal levels of COX (3). In contrast, our catalytically inactive mutant was not able to restore complex IV assembly. Work in yeast revealed that COX contains three redox centers: Cox1p contains two redox centers, one formed by heme A, and another by heme a3 and CuB. The third redox center of the enzyme is formed by the two copper ions present in the CuA site of Cox2p. Electrons are transferred from reduced cyt c sequentially to CuA site, heme a, heme a3, and CuB binuclear center in the complex IV (1). We speculate that the reduction of COX1 by cyt c may be important for its correct insertion in the inner membrane, because COX1 was susceptible to proteinase K digestion when mitoplasts derived from the W60S mutant cyt c-expressing cells were treated with the protease. It is not clear why COX1 was more stable in the mutant than in the null cell line, but the physical presence of cyt c could have a stabilizing effect on unassembled COX1. In addition, based on our observations that cyt c is a part of respirasomes and is associated with complex IV, and the lack of cyt c results in the loss of this complex, we hypothesize that cyt c is required for the stability of the complex IV. Recently it was also reported by others that cyt c is a component of the respirasome (38).
Previous results from our group (15) and others (28) have shown that complex I is not stable in the absence of complex IV. Similar results were observed when complex III was absent (29, 30). Complexes I, III, and IV exist together as supramolecular assemblies called respirasomes. The assembly of a respirasome might have advantages in substrate channeling of quinones and/or cyt c, sequestration of reactive intermediates, and stabilization of individual complexes (2). It is likely that the lack of complex I is related to the lack of complex IV, as previously described (15). However, our results showed that the requirement for complex I assembly/stability is not of a complex IV subunit, but rather the presence of an intact complex IV. In addition, our findings indicate that respirasome formation depends not only on OXPHOS complexes, but also on single molecular entities, such as cyt c.
Results from our experiments designed to probe protein localization in mitochondria and heme-association to cyt c revealed indirect evidence that the W60S mutant cyt c is misfolded. Englander's group did extensive work on cyt c folding using the two-dimensional NMR technique (31, 32). They reported that cyt c is composed of five cooperatively folding units, called foldons, that continually unfold and refold even under native conditions. Foldons are generally coincident with secondary structural elements. Based on the cyt c structure (33), the tryptophan residue at position 60, which we mutated (Trp-60), lies in the C-terminal α-helix. During cyt c folding, the C-terminal α-helix together with the N-terminal α-helix forms the first foldon in a sequential unfolding-refolding pathway (31). Hence, introducing the W60S mutation might impair the subsequent steps in the cyt c folding pathway.
Import of apocytochrome c into the mitochondrial intermembrane space is distinct from that of proteins containing a presequence. Using proteoliposomes, apocytochrome c was shown to require the protease-resistant part of the TOM complex (translocase in outer membrane), involving TOM40 and cytochrome c heme lyase (CCHL) (34). cyt c import does not require ATP hydrolysis and is driven by the interaction of cyt c with its “trans-side receptor” CCHL (35, 36). The W60S apocytochrome c-CCHL interaction might be weak, instead of the normal strong and stable binding. Hence, the covalent attachment of heme by CCHL was found to occur only in a low percentage of the mutant cyt c apoprotein. The heme addition renders cyt c translocation process irreversible, which is followed by folding to the native conformation and dissociation from CCHL. In addition, the cyt c structure reveals that tryptophan at position 60 is one of the residues that interact with the heme, mutation of which could result in improper folding.
CL is an anionic phospholipid present exclusively in the mitochondrial membrane and constitutes 25% of its total phospholipids (5). Work from several laboratories showed that CL is not only essential for membrane anchorage of the respiratory supercomplexes but also possesses multiple other roles in the mitochondrial structure and function, including stabilization of mitochondrial membranes, contribution to the very dynamic mitochondrial fusion and fission, and facilitating specific interactions with mitochondrial membrane proteins. In S. cerevisiae lacking cardiolipin synthase, the supercomplex comprising complexes III and IV was unstable (8). Additionally, assembly mutants of complex IV have significantly reduced CL synthase activity, whereas assembly mutants of respiratory complex III and complex V are less inhibitory (9). Subsequently, the proton gradient across the inner mitochondrial membrane was shown to be important for CL formation, and the authors conclude that CL synthase is stimulated by alkaline pH at the matrix side (10).
In the current study, we found both cyt c ablation and mitochondrial DNA depletion lead to a deficiency in CL content. Because the proton gradient is important for CL synthesis, the disruption of the OXPHOS respiration chain, or complex IV, could be sufficient to explain our findings of CL deficiency in the cyt c dKO fibroblasts and ρ0 cells. Although not a specific effect, the changes in cardiolipin may also affect complex and supercomplex assembly/stability.
In conclusion, cyt c is essential for the assembly/stability of complex IV in mammals. In the absence of cyt c, complex IV cannot assemble or is extremely unstable. We also showed that the W60S cyt c mutant cannot restore COX assembly, even though it can somehow stabilize COX1, enough to protect it from degradation from endogenous proteases. The presence of cyt c in association with complex IV and the respirasome suggests a structural function for this small protein. In summary, our findings show that, in mammalian cells, not only OXPHOS multisubunit complexes are interdependent, but also small single polypeptide carriers, such as cyt c.
Supplementary Material
Acknowledgments
We are grateful to Dayami Hernandez for technical assistance. We are also grateful to the input from Dr. Antoni Barrientos during the course of this work.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA85700 and R01-NS41777 (to C. T. M.) and P01 HL57278 (to X. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: complex I, NADH-ubiquinone oxidoreductase; complex II, succinate-ubiquinone oxidoreductase; complex III, ubiquinone-cytochrome c oxidoreductase; complex IV, cytochrome c oxidase (COX); cyt c, cytochrome c; TMPD, N,N,N′,N′-tetramethyl-1,4-phenylenediamine dihydrochloride; MOPS, 4-morpholinepropanesulfonic acid; CL, cardiolipin; dKO, double knockout; PVDF, polyvinylidene difluoride; BN, blue native; LF, lung fibroblast; wt, wild type; TAG, triacylglycerol; CCHL, cytochrome c heme lyase.
References
- 1.Moraes, C. T., Diaz, F., and Barrientos, A. (2004) Biochim. Biophys. Acta 1659 153-159 [DOI] [PubMed] [Google Scholar]
- 2.Schagger, H., and Pfeiffer, K. (2000) EMBO J. 19 1777-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrientos, A., Pierre, D., Lee, J., and Tzagoloff, A. (2003) J. Biol. Chem. 278 8881-8887 [DOI] [PubMed] [Google Scholar]
- 4.Pearce, D. A., and Sherman, F. (1995) J. Biol. Chem. 270 20879-20882 [DOI] [PubMed] [Google Scholar]
- 5.Robinson, B. H. (2000) Pediatr. Res. 48 581-585 [DOI] [PubMed] [Google Scholar]
- 6.Koshkin, V., and Greenberg, M. L. (2000) Biochem. J. 347 687-691 [PMC free article] [PubMed] [Google Scholar]
- 7.Koshkin, V., and Greenberg, M. L. (2002) Biochem. J. 364 317-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer, K., Gohil, V., Stuart, R. A., Hunte, C., Brandt, U., Greenberg, M. L., and Schagger, H. (2003) J. Biol. Chem. 278 52873-52880 [DOI] [PubMed] [Google Scholar]
- 9.Zhao, M., Schlame, M., Rua, D., and Greenberg, M. L. (1998) J. Biol. Chem. 273 2402-2408 [DOI] [PubMed] [Google Scholar]
- 10.Gohil, V. M., Hayes, P., Matsuyama, S., Schagger, H., Schlame, M., and Greenberg, M. L. (2004) J. Biol. Chem. 279 42612-42618 [DOI] [PubMed] [Google Scholar]
- 11.Vempati, U. D., Diaz, F., Barrientos, A., Narisawa, S., Mian, A. M., Millan, J. L., Boise, L. H., and Moraes, C. T. (2007) Mol. Cell Biol. 27 1771-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey, R., and Moraes, C. T. (2000) J. Biol. Chem. 275 7087-7094 [DOI] [PubMed] [Google Scholar]
- 13.Barrientos, A., Kenyon, L., and Moraes, C. T. (1998) J. Biol. Chem. 273 14210-14217 [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb, R. A., and Adachi, S. (2000) Methods Enzymol. 322 213-221 [DOI] [PubMed] [Google Scholar]
- 15.Diaz, F., Fukui, H., Garcia, S., and Moraes, C. T. (2006) Mol. Cell Biol. 26 4872-4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernard, D. G., Gabilly, S. T., Dujardin, G., Merchant, S., and Hamel, P. P. (2003) J. Biol. Chem. 278 49732-49742 [DOI] [PubMed] [Google Scholar]
- 17.Dutta, C., and Henry, H. L. (1990) Anal. Biochem. 184 96-99 [DOI] [PubMed] [Google Scholar]
- 18.Plant, G. W., Currier, P. F., Cuervo, E. P., Bates, M. L., Pressman, Y., Bunge, M. B., and Wood, P. M. (2002) J. Neurosci. 22 6083-6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, X., and Gross, R. W. (2005) Mass Spectrom. Rev. 24 367-412 [DOI] [PubMed] [Google Scholar]
- 20.Han, X., Yang, K., Yang, J., Cheng, H., and Gross, R. W. (2006) J. Lipid Res. 47 864-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancuso, D. J., Sims, H. F., Han, X., Jenkins, C. M., Guan, S. P., Yang, K., Moon, S. H., Pietka, T., Abumrad, N. A., Schlesinger, P. H., and Gross, R. W. (2007) J. Biol. Chem. 282 34611-34622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng, H., Mancuso, D. J., Jiang, X., Guan, S., Yang, J., Yang, K., Sun, G., Gross, R. W., and Han, X. (2008) Biochemistry 47 5869-5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweingruber, M. E., Stewart, J. W., and Sherman, F. (1979) J. Biol. Chem. 254 4132-4143 [PubMed] [Google Scholar]
- 24.Buchet, K., and Godinot, C. (1998) J. Biol. Chem. 273 22983-22989 [DOI] [PubMed] [Google Scholar]
- 25.Mansfield, K. D., Guzy, R. D., Pan, Y., Young, R. M., Cash, T. P., Schumacker, P. T., and Simon, M. C. (2005) Cell Metab. 1 393-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downie, J. A., Stewart, J. W., Brockman, N., Schweingruber, A. M., and Sherman, F. (1977) J. Mol. Biol. 113 369-384 [DOI] [PubMed] [Google Scholar]
- 27.Barros, M. H., and Tzagoloff, A. (2002) FEBS Lett. 516 119-123 [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., D'Aurelio, M., Deng, J. H., Park, J. S., Manfredi, G., Hu, P., Lu, J., and Bai, Y. (2007) J. Biol. Chem. 282 17557-17562 [DOI] [PubMed] [Google Scholar]
- 29.Acin-Perez, R., Bayona-Bafaluy, M. P., Fernandez-Silva, P., Moreno-Loshuertos, R., Perez-Martos, A., Bruno, C., Moraes, C. T., and Enriquez, J. A. (2004) Mol. Cell 13 805-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schagger, H., de Coo, R., Bauer, M. F., Hofmann, S., Godinot, C., and Brandt, U. (2004) J. Biol. Chem. 279 36349-36353 [DOI] [PubMed] [Google Scholar]
- 31.Krishna, M. M., Maity, H., Rumbley, J. N., Lin, Y., and Englander, S. W. (2006) J. Mol. Biol. 359 1410-1419 [DOI] [PubMed] [Google Scholar]
- 32.Maity, H., Maity, M., and Englander, S. W. (2004) J. Mol. Biol. 343 223-233 [DOI] [PubMed] [Google Scholar]
- 33.Rumbley, J., Hoang, L., Mayne, L., and Englander, S. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 105-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diekert, K., de Kroon, A. I., Ahting, U., Niggemeyer, B., Neupert, W., de Kruijff, B., and Lill, R. (2001) EMBO J. 20 5626-5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumont, M. E., Cardillo, T. S., Hayes, M. K., and Sherman, F. (1991) Mol. Cell Biol. 11 5487-5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer, A., Driessen, A., Neupert, W., and Lill, R. (1995) Methods Enzymol. 260 252-263 [DOI] [PubMed] [Google Scholar]
- 37.Deleted in proof
- 38.Acin-Perez, R., Fernandez-Silva, P., Peleato, M. L., Perez-Martos, A., and Enriquez, J. A. (2008) Mol. Cell 32 529-539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.